Abstract
The balance between parental genome dosage is critical to offspring development in both animals and plants. In some angiosperm species, despite the imbalance between maternally and paternally inherited chromosome sets, crosses between parental lines of different ploidy may result in viable offspring. However, many plant species, like Arabidopsis thaliana, present a post-zygotic reproductive barrier, known as triploid block which results in the inability of crosses between individuals of different ploidy to generate viable seeds but also, in defective development of the seed. Several paternal regulators have been proposed as active players in establishing the triploid block. Maternal regulators known to be involved in this process are some flavonoid biosynthetic (FB) genes, expressed in the innermost layer of the seed coat. Here we explore the role of selected flavonoid pathway genes in triploid block, including TRANSPARENT TESTA 4 (TT4), TRANSPARENT TESTA 7 (TT7), SEEDSTICK (STK), TRANSPARENT TESTA 16 (TT16), TT8 and TRANSPARENT TESTA 13 (TT13). This approach allowed us to detect that TT8, a bHLH transcription factor, member of this FB pathway is required for the paternal genome dosage, as loss of function tt8, leads to complete rescue of the triploid block to seed development.
Similar content being viewed by others
Introduction
In angiosperms, seeds are genetically chimeric structures formed upon double fertilization. The maternally-derived diploid seed coat (differentiated from ovule integuments), encloses the embryo resulting from the fusion of an haploid sperm cell with the haploid egg cell (diploid tissue), while the endosperm derives from the fusion of the diploid central cell with another haploid sperm cell1,2,3. To ensure proper seed development, complex crosstalk signalling between different seed compartments is required4. In fact, the innermost layer of the seed coat, the endothelium, which is in direct contact with the endosperm, has been reported to play a key role in this process5,6, being also, the only cell layer of the seed where the synthesis of proanthocyanidins occurs6,7. The proper development of the seed, after fertilization, is a process tightly controlled by several regulators such as genes specific to each compartment of the seed, hormones, and genes with maternal or paternal imprint2,4,8,9,10. This implies that changes in the ploidy of the parental lines can lead to defects in seed development and in several cases to seed unviability3. In fact, the triploid endosperm has a genomic ratio of 2 m:1p between the maternal and paternal genomes and several studies have demonstrated that alterating the 2 m:1p genomic ratio of endosperm leads to dramatic consequences11,12, that can cause failure of the development of F1 seed11,13,14. This phenomenon is known as the triploid block, which is a post-zygotic reproductive barrier tightly linked to parental genome dosage effects in the F1 seeds arising from interploidy crosses3.
Although polyploidization plays a prominent role in plant evolution15,16,17,18 and despite the agronomic importance of interploidy crosses, the molecular mechanisms responsible for the successful formation of triploid seeds are not yet fully understood19. Evidence suggests that molecular mechanisms regulating the triploid block are likely related to parent-of-origin small RNA dosage and to histone modifications in the developing endosperm20,21. In Arabidopsis thaliana the response to parental genome imbalance, resulting from interploidy crosses, depends on the genetic background of the accessions used in the crosses. Whereas interploidy crosses within the Columbia ecotype (Col-0 or Col) genetic background result in a strong triploid block, interploidy crosses within the Landsberg erecta (Ler) or C24 genetic backgrounds show only a weak or partial triploid block22.
In addition, maternal versus paternal genome dosage excess affects seed size23,24,25. Precocious endosperm cellularization and reduced F1 seed size are correlated with F1 seeds having maternal-excess genome dosage, which in some cases results in seed abortion23,24. Excess in the paternal genomic dosage, on the other hand, is associated with endosperm over-proliferation and cellularization failure, leading to larger F1 seeds, including collapsed and/or aborted seeds25,26,27.
Two main molecular mechanisms have been suggested to explain such effects in seed development as result of the unbalanced ratio of maternal/paternal genomic dosage in F1 offspring6,28. First, that the altered expression of imprinted genes in the endosperm would lead to cellularization defects, and possibly to seed abortion11,19,29,30,31,32, and the second mechanism, involves a maternal-zygotic crosstalk possibly involving flavonoid biosynthetic genes.
Endosperm development is also under the sporophytic maternal control of the seed integuments, alteration of such sporophytic maternal–zygotic crosstalk in interploidy crosses can interfere with developmental programs of the embryo, endosperm and seed coat, resulting in defective seed formation6,33,34,35,36,37.
Here we focused on the maternal regulators and their possible role in seed development, flavonoids and in particular proanthocyanidins (PAs) which are synthesized in the endothelium, the innermost layer of the seed coat, of maternal origin, where they accumulate and give the characteristic brown colour of Arabidopsis thaliana seeds upon oxidation38,39. PAs accumulation provides many benefits to seeds such as protection against UV-light, microbes and herbivores, in addition to antioxidant and allelopathic activities40. It has been suggested also, that PAs are involved in seed development resulting from interploidy crosses6. Mutation of maternally-expressed genes, such as TRANSPARENT TESTA GLABRA 2 (TTG2) and TRANSPARENT TESTA 4 (TT4), involved in PA biosynthesis, a branch of the flavonoid biosynthesis pathway (FBP), can partially suppress F1 seed lethality caused by paternal-excess interploidy crosses in Arabidopsis thaliana6,22,41. Although much progress has been made in identifying genetic modifiers of the triploid block, most of the advances to date have focused on the paternal elements responsible for (i) the high sensitivity of Arabidopsis thaliana to increased paternal genome dosage and, (ii) the triploid block31,42,43. Thus, to have a better understanding of the possible role of flavonoids in triploid block, here we explore the role of several genes in the flavonoid biosynthetic pathway (i.e., TT4, TT7, STK, TT16, TT8 and TT13; Fig. 1) in balanced and unbalanced crosses, by analyzing maternal parents carrying loss-of-function mutations and crossing them with paternal excess genomic ratio dosage plants (4x). Our results indicate that mutants of most of these genes, partially rescue triploid block unlike TT8 which completely rescues the triploid block, which occurs in response to increased paternal genome dosage in Arabidopsis thaliana Col-0 ecotype. Also suggesting, that the maternal contribution of TT8 transcription factor plays a major role for post-zygotic barriers and that the flavonoid biosynthetic pathway is not entirely responsible for triploid block, besides that the function of TT8 in triploid block could be independent of the flavonoid biosynthetic pathway. TT8 which belongs to the bHLH gene family and, together with members of the MYB, bHLH and WD40 gene families forms the MBW complex44,45,46, also appears to interact with Aux/IAA proteins (http://bar.utoronto.ca). Our results from the study of triploid block, this complex mechanism that is still poorly understood and for which additional studies are required, allow us to propose novel hypotheses about the role of endothelium specific transcription factors in the interplay between maternal, paternal and zygotic control in F1 polyploid seeds.
Schematic representation of the flavonoid biosynthetic pathway in Arabidopsis thaliana seeds. Genes encoding enzymes for each step are indicated as follows: (1) Early biosynthesis genes: TRANSPARENT TESTA 4 (TT4), TRANSPARENT TESTA 5 (TT5), TRANSPARENT TESTA 6 (TT6) and TRANSPARENT TESTA 7 (TT7), which are involved in the biosynthesis of PAs precursors and other classes of Arabidopsis flavonoids. (2) Later biosynthesis genes (LBG): TRANSPARENT TESTA 3 (TT3), TRANSPARENT TESTA 18 (TT18) and BANYULS/ANTHOCYANIDIN REDUCTASE (BAN/ANR). 3) TRANSPARENT TESTA 12 (TT12, MATE transporter), TT10 (laccase 15), TT19 (Glutathione-S-transferase) and TRANSPARENT TESTA 13 (TT13). The regulatory control of LBGs requires the action of a MBW transcriptional regulation complex (MYB-bHLH-WDR), formed by a specific R2R3-MYB (TT2), bHLH transcription factors (EGL3, TT8) and a WD repeat protein TRANSPARENT TESTA GLABRA 1 (TTG1). Other transcription factors belonging to different families such as C2H2 and C2HC zinc finger (TT1/WIP1), MADS-box (ABS/TT16/AGL32, STK) and WRKY (TTG2/DSL1/WRKY44) also participate in their regulation.
Results
Maternal-specific mutations in FBP genes rescue the post-zygotic triploid block to seed development
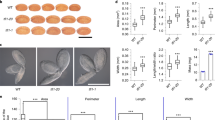
To explore the possible role of flavonoids in the control of the post-zygotic triploid block in F1 seed development, we used loss-of-function mutants for genes involved in different steps of the flavonoid biosynthetic pathway, which also allows us to determine if there is any difference along this pathway in the control of triploid block. Thus, we tested mutants of several genes such as TT4, FLAVANONE 3 HYDROXYLASE/ TRANSPARENT TESTA 7 (F3’H/TT7), ARABIDOPSIS BSISTER/TRANSPARENT TESTA 16 (ABS/TT16), SEEDSTICK (STK), TT8 and AUTOINHIBITED H( +)-ATPASE ISOFORM 10/TRANSPARENT TESTA 13 (AHA10/TT13)47,48,49 (Fig. 1). TT4, acts in the first step of flavonoid biosynthesis, and it has already been reported that it partially rescues the post-zygote triploid block resulting from unbalanced crosses22. Homozygous loss-of-function mutants in each of the genes were used as maternal parents in interploidy (and euploidy) crosses with pollen from 2x (diploid) and 4x (tetraploid) plants50. The percentage of plump F1 seeds obtained in interploidy crosses of the 2 × mutants X 4 × Col were compared to the percentage of plump F1 seeds produced by 2 × Col X 2 × Col, 4 × Col X 4 × Col euploidy controls, and also in 2 × Col X 4 × Col (paternal-excess control) and 4 × Col X 2 × Col (maternal-excess control) (Fig. 2). The loss of function for both TT4 and TT7 mutants displayed a statistically significant increase in the number of F1 plump seeds (64% and 56%, respectively) when compared to the 35% of F1 plump seeds produced by the paternal-excess control crosses 2 × Col X 4 × Col (Fig. 2b). The plump F1 tt4 seed percentage was equivalent to what was previously reported for ttg2 (64%), which works upstream of TT422 (Fig. 2b). In addition, the disruption of TT13, partially rescued the percentage of plump F1 seed phenotype (56% of plump seeds) (Fig. 2b).
F1 seed shape of different FBP related mutants in interploidy crosses. (a) Mature F1 seeds obtained crossing FBP mutants in balanced and paternal-excess crosses. Wildtypes are shown on the first row. (b) Percentage of plump F1 seeds. Data are presented as means ± standard error. 200–300 F1 seeds were analyzed for each cross. Pooled data of three independent assays performed with 3 replicates for each measurement. One-way ANOVA followed by Tukey HSD test was used for analyzing significance. tt4 is tt4-11; tt7 is tt7-2 ; stk is stk-2; tt16 is tt16-7; tt8 is tt8-6; tt13 is tt13-6. Crosses marked with the same lowercase letter display no statistical difference. p: plump seeds; s: shriveled seeds, Col = Col-0. Scale: 1 mm (a).
Compared to the 2 × Col X 4 × Col cross, neither 2 × stk (Col) X 4 × Col nor 2 × tt16 (Col) X 4 × Col, have shown any statistical difference from the paternal-excess control. In contrast, the 2 × tt8 (Col) X 4 × Col cross rescued almost completely the percentage of plump F1 seeds (97%) (Fig. 2b).
To verify the role of TT8 in maternal control of the triploid block, we analyzed four different tt8 alleles namely tt8-1, tt8-5, tt8-4, tt8-8 (Supplementary Fig. S1a), which allowed us to corroborate that all the tt8 alleles fully recover the F1 plump seed phenotype from unbalanced crosses (Supplementary Fig. S1).
The bypass of the triploid block was further corroborated with germination tests on all alleles, showing that tt8 mutations fully rescue seed viability after interploidy crosses (Supplementary Fig. S2). Indicating that the bypass of the triploid block may arise due to the lack of functional TT8 protein (Fig. 2).
Overall, these results indicate that loss-of-function mutations of sporophyte-expressed genes involved in the flavonoid biosynthetic pathway may partially, as occurs with TT4, TT7, TT13 or completely, as occurs with TT8, bypass the triploid block triggered by an unbalanced zygotic genome dosage (Fig. 2).
While the Col accession exhibited a strong triploid block to seed development in 2 × X 4 × crosses (paternal excess), the prevalence of shrivelled seeds generated was significant around 90%. Paternal-excess interploidy crosses with the accessions Ler and C24, generated almost 100% plump seeds51,52,53 (Fig. 3a,b).
Interploidy crosses in wildtype and tt8 using three accessions of Arabidopsis thaliana (Col, Ler and C24). (a) Mature F1 seeds obtained crossing diploid Col and tt8 mutant with diploid or tetraploid pollen from three Arabidopsis thaliana. (b) Percentage of plump F1 seeds from interploidy crosses using different Arabidopsis accessions as pollen donors to pollinate diploid Col and tt8 mutants. Data presented as means ± standard error. Around 200 F1 seeds were analyzed for each cross. Pooled data of three independent assays performed with 3 replicates for each measurement. (c) Seed size of F1 seeds resulting from interploidy crosses using three accessions of Arabidopsis pollen donors to pollinate diploid Col and 2 × tt8 (Col) mutants. Data is presented as means ± standard error. Pooled data of three independent assays performed with 3 replicates for each treatment (200–300 seeds/each). tt8 corresponds to tt8-6. One-way ANOVA followed by Tukey HSD test was used for analyzing significance. Crosses marked with the same lowercase letter display no statistical difference. Scales: 1 mm (a).
Paternal-excess interploidy crosses (2 × Col X 4 × Col) generated larger F1 seeds than seeds from 2 × Col X 2 × Col crosses25, mainly due to over-proliferation and defective cellularization of the endosperm, and abnormal development of the seed coat6,24,54. Consistent with previous studies25, we found an increase of approx. 34% of F1 seed size derived from 2 × Col X 4 × Col crosses, compared to F1 seeds derived from 2 × Col X 2 × Col crosses (here defined as the control) (Fig. 3b). All tt8 alleles tested displayed a similar increase in F1 seed size and this was slightly lower than the control (Fig. 3c; Supplementary Fig. S1d).
The resulting F1 seeds from crosses with 4 × Ler and 4 × C24, displayed a significant increase in seed size which was slightly lower when we pollinated 2 × tt8 (Col) plants compared to the 2 × Col plants (Fig. 3c).
Eventhough maternal excess in wildtype Arabidopsis thaliana does not exhibit a post-zygotic barrier (i.e., 4 × Col X 2 × Col; produces viable seeds)37, it is unclear whether loss-of-function tt8 results in any seed defects. To find out if TT8 impacts seed development in maternal-excess interploidy crosses, we generated colchicine-induced nulliplex autotetraploid (4x) mutant lines of tt8 (Col) that were then crossed with pollen from 2 × Col and 4 × Col wildtype (WT) lines (Supplementary Fig. S3), tt4 analyses were also included (Supplementary Fig. S3). It was observed that 4 × tt8 (Col) X 2 × or 4 × tt4 (Col) X 2 × maternal-excess interploidy crosses had no influence on F1 seed development (Supplementary Fig. S3a,b).
Regarding seed size, the maternal-excess triploid seeds from 4 × Col X 2 × Col crosses exhibited smaller F1 seed size compared37 to 4 × Col X 4 × Col seeds (Supplementary Fig. S3c). The 4 × tt8, and 4 × tt4 mutations showed a similar trend in seed size as the control situation (4 × Col X 2 × Col), showing a 40% reduction in seed size compared to balanced situation (4 × Col X 4 × Col) (Supplementary Fig. S3c).
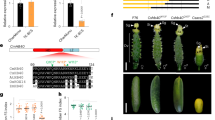
TT8 expression was strongly upregulated in paternal-excess crosses 2 × Col X 4 × Col, respect 2 × Col X 2 × Col (Fig. 4). TT8 is reported to be specifically expressed in the endothelium after fertilization54. Indeed, in seeds obtained by crossing tt8 with 2 × or 4 × pollen, TT8 expression is not detectable, indicating that paternal TT8 (wildtype) allele is not expressed in embryo or in the endosperm at the analyzed stages (Fig. 4). These results suggest that pollen ploidy might influence, at least in Col, the expression of seed coat specific genes such as TT8.
TT8 mRNA expression levels in seeds at 72 h after pollination from balanced and paternal excess crosses. tt8 corresponds to tt8-6. Data was normalized to ACTIN, with error bars indicating standard deviations based on four independent technical replicates. Statistical analysis was performed using Student’s t test (**p < 0.01). Three qPCR (biological replicates) were performed and generated similar results. Three independent biological replicates were performed.
TT8 controls the rate of endosperm proliferation to balance interploidy incompatibility
In Arabidopsis thaliana, endosperm proliferation begins after double fertilization, in coordination with the first mitotic divisions of the embryo55. Once endosperm development reaches its final volume, approximately 5 DAP the cellularization process begins56, simultaneously several genes repressing cell wall formation are silenced and active cytokinesis occurs in the syncytial endosperm24,57.
To provide a more in-depth analysis of the role of TT8 in endosperm development, detailed measurement of endosperm proliferation, endosperm area, and characterization of embryo development was performed using a clearing protocol (Fig. 5a). Having found that embryo development appears relatively normal in paternal excess (2 × Col X 4 × Col) compared to control (2 × Col X 2 × Col). However, at 3 DAP, a slight delay in the development of the F1 seeds with paternal excess was observed, compared to the balanced cross (Fig. 5b). This delay was later compensated on the fourth day after pollination, showing embryos in the medium and late globular stage (Fig. 5b). The progeny of the F1 seed of the tt8 mutant crossed with 4 × Col, also showed a minor delay compared to the progeny of the relative isogenic cross at 3 DAP, delay that appeared recovered at fourth day after pollination, then appearing similar to the control) (Fig. 5b).
Timing of endosperm and embryo development following balanced and interploidy crosses. (a) Optical microscope image of cleared seeds from balanced and unbalanced crosses. (b) Percentage of embryo stages following balanced and interploidy crosses (percentages obtained from 20–25 seeds). (c) Endosperm proliferation following balanced and interploidy crosses at 3 and 4 DAP. Endosperm nuclei were counted in whole-mount seeds after clearing treatment. The mean number of endosperm nuclei ± standard error (n = 10 to 15) is shown for each cross. (d) Endosperm cavity area measurement following balanced and interploidy crosses at 4 DAP. Endosperm cavity areas were measured through whole-mount seeds after clearing treatment. The mean number of endosperm cavity area (µm2) ± standard error (n = 10 to 15) is shown for each cross. One-way ANOVA followed by Tukey HSD test was used for analyzing significance. tt8 corresponds to tt8-6. Crosses marked with the same lowercase letter display no statistical difference. Scale: 20 μm (a).
Interestingly, at 4 DAP, our analyses indicated that paternal-excess crosses 2 × Col X 4 × Col showed an increment of approx. 25% in endosperm nuclei in Col wildtype, compared to 2 × Col X 2 × Col (Fig. 5c). The tt8 mutant presented a lower number of endosperm nuclei when compared to F1 seed progeny from isogenic crosses, showing an approx. 20% reduction compared to 2 × Col X 2 × Col and from interploidy crosses 25% reduction compared to 2 × Col X 4 × Col (Fig. 5c). With this phenotype, the tt8 mutant was found to display a reduced endosperm area in F1 seeds, both from balanced crosses 25% approx. reduction compared to 2 × Col X 2 × Col and unbalanced crosses 25% reduction compared to 2 × Col X 2 × Col (Fig. 5d).
Overall, we consider that the small endosperm area (Fig. 5d) may balance the developmental program leading to the maternal rescue observed in the tt8 mutant interploidy crosses paternal-excess.
Discussion
The triploid block resulting from interploidy crosses has been reported for over than five decades58,59. Recent studies, using the model species Arabidopsis thaliana, have mainly focused on the role of the fertilization product: endosperm and embryo in the establishment of the triploid block29,30,31, providing valuable information on the genetic regulatory factors that control it29,30,31,59. However, the potential role of maternally derived sporophytic tissues (seed coat) needs to be further explored.
It has been proposed that the flavonoid biosynthetic pathway, acting on the endothelium of the seed coat, may control triploid block. Genes such as TT4 and TTG2 that have been reported to impact the viability of seeds after unbalanced crosses. The results of this study show that the “lethal effect” observed in interploidy paternal excess crosses, in the Col ecotype can be bypassed by TT8 loss-of-function mutations in all the tt8 alleles assessed (Fig. 2, Supplementary Figs S1, S2). While the tt8 mutant fully rescues the triploid block, our findings also show that loss-of-function mutations of other flavonoid biosynthetic pathway genes such as TT4, TT7, TT13, can only partially bypass the triploid block (Fig. 1, 2). Thus, triploid block rescue mediated by tt8 knockout differs from the previously reported partial seed recovery in F1 seeds from paternal-excess crosses resulting from mutating FBP related genes6,23.
The assessment based on the percentages of plump seed phenotype, in both balanced and unbalanced F1 seeds shows that 98% of tt8 knocked-out paternal excess seeds are plump, resembling wildtype balanced seed (2 × Col X 2 × Col; Fig. 3b), whereas only 65% of tt4 mutant paternal excess seeds (2 × tt4 Col x 4 × Col) are plump seeds. Seed size in tt8 mutants is slightly reduced compared to wildtype seeds in both balanced and unbalanced F1 seeds (Fig. 3c) as has been reported for other seed coat specific genes such as TTG222. These phenotypes are similar when using different Arabidopsis thaliana ecotypes (Ler and C24) as pollen donors (Fig. 3).
Moreover, the high expression of TT8 observed in unbalanced F1 seeds with paternal excess (2 × Col X 4 × Col), raises two probabilities: (1) that 2 × Col X 4 × Col seeds trigger the activation of TT8 transcription and, (2) that in these unbalanced crosses there has been a repression removed of TT8.
Embryo development is arrested in unbalanced seeds resulting from paternal excess crosses37 (Fig. 5) leading to unviability of seeds. In fact, tt8 mutants show normal embryo development at the stages analysed here (Fig. 5). The effects of the triploid block are manifested in the endosperm since the seeds resulting from paternal excess do not show endosperm cellularization13,28,60. The cellularization process beginning 5 DAP while our observations, after having analysed seeds up to 4 DAP, allow us to determine that there is not premature cellularization detected. Our developmental characterization of triploid seeds corroborates a lack of cellularization in the balanced-ploidy crosses (2 × Col X 2 × Col and 4 × Col X 4 × Col) at 3 DAP13,28,60 (Figs. 5). In the tt8 mutants resulting from balanced and imbalanced crosses it appears that endosperm development is not significantly affected (Fig. 5a,b). However, the reduced growth of the endosperm cavity in tt8 mutants (Fig. 5c), translated into a reduction in the number of nuclei in the endosperm (Fig. 5d) that could possibly lead to an anticipated cellularization in paternal excess crosses. Also, the F1 seeds of the tt8 mutant crossed with 4 × Col presented a reduced delay in embryo development compared to the progeny of the relative control exposed to unbalanced crosses. Therefore, these traits could be considered as part of the mechanisms responsible to bypass the triploid block in seeds from crosses with paternal excess13,60. This leads us to support the hypothesis that seed development is tightly coordinated between the seed coat, the embryo and endosperm developmental programs during normal seed development35. Given that the final seed size of the 2 × tt8 (Col) X 4xCol is bigger in relation to the wildtype 2 × Col X 2 × Col (Fig. 3c) whereas the endosperm cavity 3 DAP is smaller (Fig. 5d), suggests that 1) the bigger seed size may be due to the width of the seed coat, or 2) the endosperm enlarges at later stages of seed development; a more detailed morphological characterization of the tt8 mutant phenotype, will uncover its precise role in each tissue of the seed.
It is important to note that genes, involved in FBP, play a role in seed size regulation6. In balanced ploidy crosses, it is common for loss-of-function mutations of some FBP-related genes to produce smaller seeds35,38,61,63. The effect on seed size observed in these mutants, is considered to be associated with the precocious onset of endosperm cellularization64. The timing of endosperm proliferation is critical for the size of the embryo sac, which in turn is critical for defining the final size of the seed58,65. From the seed size assessment for tt4 and tt8 in interploidy crosses with maternal-excess (4 × Col X 2 × Col), we found no major differences (Supplementary Fig. S3).
Focusing on the triploid block that occurs in the endosperm, this study highlights seed coat-specific genes that play an important role in triploid block as they affect the endosperm. Underlying our findings, a cross-talk between the different tissues of the seed is evidenced2,35.
-
Our findings, together with previous studies, show that although there are genes, members of the flavonoid biosynthetic pathway, that partially rescue triploid block, the function of TT8 is crucial by fully rescuing it, and suggest in further that TT8 function on triploid block may be independent of that in flavonoid biosynthesis.
-
According to the Arabidopsis thaliana interactions viewer 2.0 (http://bar.utoronto.ca; Supplementary Fig. S4), TT8 interacts not only with members of the MBW complex of the FBP45,46,66, but also with other genes that are strongly expressed in the seed coat, including the repressor IAA27/PAP2 (Supplementary Fig. S4) a canonical Aux/IAA67. This suggests that IAA27 might repress TT8. Protein–protein interaction essays, are still required to corroborate this hypothesis however, in an unbalanced F1 seed (2 × Col X 4 × Col), endosperm cellularization does not occur as result of high auxin levels in the endosperm68 whereas in the seed coat, TT8 is highly expressed (Fig. 4), and it has been previously shown that after fertilization auxin is transported from the endosperm to the seed coat2,4,60. Therefore, in F1 2 × Col X 4 × Col F1 seeds, higher levels of auxin in the endosperm activate TT8 transcription via ubiquitination and degradation of IAA27. Corroborating previous results that auxin is involved in the coordinated development of the different seed compartments4,60,67.
-
The early members of the flavonoid biosynthetic pathway such as TT4, which encodes the structural protein chalcone synthase, are involved in auxin transport via PIN and PGPs69,70,71, further affecting TT8 expression, via auxin signaling, which could explain the partial rescue of triploid block observed in tt4 mutants (Fig. 2). And suggesting that the role of these genes in triploid block is not due to their function in the FBP per se, but rather to their role in auxin flux and signaling.
Altogether, our findings provide a better understanding of the role of seed coat-specific genes in seed development and triploid block, shedding light on novel mechanisms, and on TT8, an important regulator for proper seed development.
Materials and methods
Plant material and growth conditions
For this study the following Arabidopsis thaliana accessions and lines were used: wildtype (2 × ecotype Columbia-0, Col-0, 2 × ecotype Ler (Landsberg erecta), 2 × ecotype C24), as well as (4 × ecotype Columbia-0, Col-0, 4 × ecotype Ler (Landsberg erecta), 4 × ecotype C24) (obtained from Spillane Lab, National University of Ireland Galway, Ireland), tt16-7 (T-DNA SALK_077737), tt13-6 (T-DNA GK-170A0747), tt7-2 (T-DNA SALK_05339472; later called tt7-573), stk-274, tt4-11 (T-DNA SALK_02058375 later called tt4-1373), tt8 (reported in TAIR as tt8-6) (T-DNA GK-241D05; 2nd intron). Other tt8 alleles (Supplementary Fig. S1) that were used were the following: tt8-4 (T-DNA SALK_030966; 1st exon76), tt8-5 (T-DNA SALK_048673; 3rd exon), tt8-14 (T-DNA SALK_063334; 5th intron), tt8-15 (T-DNA SALK_121609; 3’UTR; new report according to conventional nomenclature45). Alleles tt8-7 to tt8-13 were not used in this study77. Plants were germinated at 22 °C under short-day conditions (8 h light/16 h darkness) and transferred to long day conditions (16 h light/ 8 h darkness). The mutant lines were already corroborated homozygous mutant plants. Plants were cultivated and collected following the relevant institutional, national, and international guidelines and legislation, with all the permissions and licenses for collec tion of plants/seeds.
Colchicine treatments
The, tt4 and tt8 tetraploid mutant lines in Col background were generated by applying 0.1% (w/v) colchicine solution as described in to two-week-old diploid seedlings. The newly generated tetraploids were self-pollinated for three generations by single seed descent and the ploidy level was validated by flow cytometry using a BD Accuri™ C6 Plus flow device following the manufacturer’s instructions.
Expression analysis
Flower buds that were just about to open were emasculated. After 24 h, emasculated pistils were manually pollinated by depositing the pollen from the donor plant on the stigmatic papillae of the receptor pistils. RNA from 20 to 25 manually dissected siliques at 3 DAP (Day After Pollination) after being pollinated were collected in NucleoProtect RNA stabilization reagent (Macherey–Nagel). 3 biological replicates were collected and RNA was obtained using the NucleoSpin RNA, Mini kit for RNA purification (Macherey–Nagel) following the supplier's instructions. Total RNA was retro-transcribed using the iScriptTM gDNA Clear cDNA Synthesis kit (Bio-Rad) following the supplier’s instruction. cDNAs were used as templates in the qRT-PCR reactions containing the iQ SYBR Green Supermix (Bio-Rad). The qRT-PCR assay was conducted in triplicate on different biological replicates, with three technical replicates for each sample, and was performed in a Bio-Rad iCycler iQ Optical System (software version 3.0a). Relative transcript enrichment of genes of interest was calculated normalizing the amount of mRNA against control fragments (ACT). Gene expression analysis was performed with the 2ˆ − ∆∆Ct method using the specific tool at Bio-Rad CFX Maestro software v.4.0 (Bio-Rad). The primers used for this analysis are listed in Suplementary Table S1.
Seed Size analysis
Seeds were photographed using a Leica MZ6 stereomicroscope, and seed images were measured using smartgrain software78. Measurements were statistically analyzed by one-way ANOVA with a post-hoc Tukey honestly significant difference (HSD) comparison test.
Morphological analysis of interploidy crosses
Microscopic observations were performed using a Zeiss Axiophot D1 microscope (http://zeiss.com/) equipped with differential interface contrast (DIC) optics. Images were recorded with an Axiocam MRc5 camera (Zeiss) using the Axiovision program (version 4.1). In A. thaliana floral stems, the self-pollinated siliques in formation and the already opened flowers were removed. The flower buds that were just about to open were emasculated. With the help of tweezers and under a stereomicroscope, all the floral organs were removed, except for the pistil, which contained the ovary. Within a period of 24 h after emasculation a first pollination was performed by depositing the pollen from the donor plant in the stigma of the receptor pistils. Within 24 h after first pollination, a second one was performed in the same way with the purpose of increasing the possibilities of obtaining a successful fecundation. It was considered that the crosses were successful when a few days after pollinating, the pollinated pistils grew until maturity and the resulting siliques contained seeds. Endosperm analyses were performed as previously reported22, 20 seeds per genotype at 3 DAP and 4 DAP were cleared75 and immediately photographed using a microscope. Endosperm size was measured in ImageJ (https://imagej.nih.gov/ij/) using the ‘analyze particles’ function.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data bases under the following accession numbers: TT13 = At1g17260; CHS/TT4 = At5g13930; TT7 = At5g07990; TT8/bHLH042 = At4g09820; ABS/TT16/AGL32 = At5g23260; STK = At4g09960.
Data availability
The authors declare that all data supporting the findings in this study are available within the paper, Supplementary information and Source data. All data are available upon request to the corresponding author.
References
Moïse, J. A., Han, S., Gudynaitę-Savitch, L., Johnson, D. A. & Miki, B. L. A. Seed coats: Structure, development, composition, and biotechnology. Vitro Cell. Dev. Biol. - Plant 41, 620–644 (2005).
Figueiredo, D. D. & Köhler, C. Signalling events regulating seed coat development. Biochem. Soc. Trans. 42, 358–363 (2014).
Haig, D. & Westoby, M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 134, 147–155 (1989).
Figueiredo, D. D., Batista, R. A., Roszak, P. J., Hennig, L. & Köhler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. Life 5, e20542 (2016).
Mizzotti, C. et al. The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development. Plant J. 70, 409–420 (2012).
Doughty, J., Aljabri, M. & Scott, R. J. Flavonoids and the regulation of seed size in Arabidopsis. Biochem. Soc. Trans. 42, 364–369 (2014).
Debeaujon, I. et al. Proanthocyanidin-accumulating cells in arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15, 2514–2531 (2003).
Köhler, C. et al. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553 (2003).
Köhler, C. & Makarevich, G. Epigenetic mechanisms governing seed development in plants. EMBO Rep. 7, 1223–1227 (2006).
Rodrigues, J., Luo, M., Berger, F. & Koltunow, A. M. Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex. Plant Reprod. 23, 123–133 (2010).
Gehring, M. & Satyaki, P. R. Endosperm and imprinting, inextricably linked. Plant Physiol. 173, 143–154 (2017).
Pennington, P. D., Costa, L. M., Gutierrez-Marcos, J. F., Greenland, A. J. & Dickinson, H. G. When genomes collide: Aberrant seed development following maize interploidy crosses. Ann. Bot. 101, 833–843 (2008).
Köhler, C., Mittelsten Scheid, O. & Erilova, A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 26, 142–148 (2010).
Song, Q. & Chen, J. Z. Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24, 101–109 (2015).
Ramsey, J. & Schemske, D. W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1, 467–501 (1998).
Bretagnolle, F. & Thompson, J. D. Gametes with the somatic chromosome number: Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 129, 1–22 (1995).
Otto, S. P. & Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437 (2000).
Adams, K. L. & Wendel, J. F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141 (2005).
Lafon-Placette, C., Vallejo-Marín, M., Parisod, C., Abbott, R. J. & Köhler, C. Current plant speciation research: Unravelling the processes and mechanisms behind the evolution of reproductive isolation barriers. New Phytol. 209, 29–33 (2016).
Kirkbride, R. C. et al. Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 116, 2761–2766 (2019).
Wang, L. et al. Rice interploidy crosses disrupt epigenetic regulation, gene expression, and seed development. Mol. Plant 11, 300–314 (2018).
Dilkes, B. P. et al. The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol. 6, e308 (2008).
Erilova, A. et al. Imprinting of the Polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 5, 1 (2009).
Kradolfer, D., Hennig, L. & Köhler, C. Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in arabidopsis seed development. PLoS Genet. 9, e1003163 (2013).
Scott, R. J., Tratt, J. L. & Bolbol, A. Seed Development in Interploidy Hybrids. in Polyploid and Hybrid Genomics 271–290 (John Wiley & Sons, Inc., 2013). https://doi.org/10.1002/9781118552872.ch17.
Jiang, H. & Köhler, C. Evolution, function, and regulation of genomic imprinting in plant seed development. J. Exp. Bot. 63, 4713–4722 (2012).
Rodrigues, J. A. & Zilberman, D. Evolution and function of genomic imprinting in plants. Genes Dev. 29, 2517–2531 (2015).
Lafon-Placette, C. & Köhler, C. Endosperm-based postzygotic hybridization barriers: Developmental mechanisms and evolutionary drivers. Mol. Ecol. 25, 2620–2629. https://doi.org/10.1111/mec.13552 (2016).
Borges, F. et al. Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50, 186–192 (2018).
Jiang, H. et al. Ectopic application of the repressive histone modification H3K9me2 establishes post-zygotic reproductive isolation in Arabidopsis Thaliana. Genes Dev. 31, 1272–1287 (2017).
Martinez, G. et al. Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet. 50, 193–198 (2018).
Moreno-Romero, J., Jiang, H., Santos-González, J. & Köhler, C. Parental epigenetic asymmetry of PRC 2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 35, 1298–1311 (2016).
Adamski, N. M., Anastasiou, E., Eriksson, S., O’Neill, C. M. & Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 106, 20115–20120 (2009).
Ohto, M., Fischer, R. L., Goldberg, R. B., Nakamura, K. & Harada, J. J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. 102, 3123–3128 (2005).
Orozco-Arroyo, G., Paolo, D., Ezquer, I. & Colombo, L. Networks controlling seed size in Arabidopsis. Plant Reprod. 28, 17–32 (2015).
Robert, H. S. et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 4, 548–553 (2018).
Scott, R. J., Spielman, M., Bailey, J. & Dickinson, H. G. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341 (1998).
Debeaujon, I., Lepiniec, L., Pourcel, L., & Routaboul, J. M. Seed Coat Development and Dormancy. in Seed Development, Dormancy and Germination 25–49 (wiley, 2007). https://doi.org/10.1002/9780470988848.ch2.
Shirley, B. W. et al. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671 (1995).
Koes, R., Verweij, W. & Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242 (2005).
Li, C. et al. TOP1?, UPF1, and TTG2 regulate seed size in a parental dosage dependent manner. PLoS Biol. 18, e3000930 (2020).
Schatlowski, N. et al. Hypomethylated pollen bypasses the interploidy hybridization barrier in arabidopsis. Plant Cell 26, 3556–3568 (2014).
Lafon-Placette, C. et al. Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrata and Arabidopsis arenosa in Central Europe. Proc. Natl. Acad. Sci. USA 114, E1027–E1035 (2017).
Appelhagen, I. et al. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene 484, 61–68 (2011).
Gonzalez, A., Zhao, M., Leavitt, J. M. & Lloyd, A. M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 (2008).
Gonzalez, A., Mendenhall, J., Huo, Y. & Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325, 412–421 (2009).
Appelhagen, I. et al. Update on transparent testa mutants from Arabidopsis thaliana: Characterisation of new alleles from an isogenic collection. Planta 240, 955–970 (2014).
Nesi, N. et al. The transparent testa 16 locus encodes the arabidopsis bsister mads domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14, 2463–2479 (2002).
Ezquer, I. et al. The developmental regulator SEEDSTICK controls structural and mechanical properties of the arabidopsis seed coat. Plant Cell 28, 2478–2492 (2016).
Miller, M., Zhang, C. & Chen, Z. J. Ploidy and hybridity effects on growth vigor and gene expression in arabidopsis thaliana hybrids and their parents. G3 Genes Genom. Genet. 2, 505–513 (2012).
Redei, G. Crossing experiments with polyploids. Arabidops. Electron. Inf. Serv. 1, 380–382 (1962).
Stoute, A. I., Varenko, V., King, G. J., Scott, R. J. & Kurup, S. Parental genome imbalance in Brassica oleracea causes asymmetric triploid block. Plant J. 71, 503–516 (2012).
Wolff, P., Jiang, H., Wang, G., Santos-González, J. & Köhler, C. Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. eLife 4, (2015).
Xu, W. et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression. Arabidopsis. New Phytol. 198, 59–70 (2013).
Hands, P., Rabiger, D. S. & Koltunow, A. Mechanisms of endosperm initiation. Plant Reprod. 29, 215 (2016).
Cheng, Z. J. et al. Abscisic acid regulates early seed development in arabidopsis by ABI5-mediated transcription of short hypocotyl under blue1. Plant Cell 26, 1053–1068 (2014).
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J. & Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 102, 17531–17536 (2005).
Marks, G. E. The origin and significance of intraspecific polyploidy: experimental evidence from Solanum chacoense. Evolution 552–557 (1966).
Nishiyama, I. & Inomata, N. Embryological studies on cross-incompatibility between 2x and 4x in Brassica. Jpn. J. Genet. 41, 27–42 (1966).
Figueiredo, D. D. & Köhler, C. Bridging the generation gap: Communication between maternal sporophyte, female gametophyte and fertilization products. Curr. Opin. Plant Biol. 29, 16–20 (2016).
Garcia, D., Fitz Gerald, J. N. & Berger, F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17, 52–60 (2005).
Huc, J. et al. Bypassing reproductive barriers in hybrid seeds using chemically induced epimutagenesis. Plant Cell 34, 989–1001 (2022).
Paolo, D. et al. The arabidopsis mads-domain transcription factor seedstick controls seed size via direct activation of e2fa. Plants 10, 192 (2021).
Ehlers, K. et al. The MADS box genes ABS, SHP1, and SHP2 are essential for the coordination of cell divisions in ovule and seed coat development and for endosperm formation in Arabidopsis thaliana. PLoS ONE 11, e0165075 (2016).
Wang, A. et al. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679 (2010).
Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8, 272–279 (2005).
Dreher, K. A., Brown, J., Saw, R. E. & Callis, J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18(3), 699–714 (2006).
Batista, R. A., Figueiredo, D. D., Santos-González, J. & Köhler, C. Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev. 33, 466–476 (2019).
Brown, D. E. et al. Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol. 126, 524–535 (2001).
Peer, W. A. & Murphy, A. S. Flavonoids and auxin transport: modulators or regulators?. Trends Plant Sci. 12, 556–563 (2007).
Murphy, A., Peer, W. A. & Taiz, L. Regulation of auxin transport by aminopeptidases and endogenous flavonoids. 10.
Lewis, D. R. et al. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156(1), 144–164. https://doi.org/10.1104/pp.111.172502 (2011).
Bowerman, P. A., Ramirez, M. V., Price, M. B., Helm, R. F. & Winkel, B. S. Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia. BMC Res. Notes 5(1), 1–9 (2012).
Pinyopich, A. et al. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88 (2003).
Buer, C. S., Sukumar, P. & Muday, G. K. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 140(4), 1384–1396 (2006).
Jia, L. et al. Class III peroxidases are activated in proanthocyanidin-deficient Arabidopsis thaliana seeds. Ann. Bot. 111, 839–847 (2013).
Jiang, N. et al. Diversity of genetic lesions characterizes new Arabidopsis flavonoid pigment mutant alleles from T-DNA collections. Plant Sci. 291, 110335 (2020).
Tanabata, T., Shibaya, T., Hori, K., Ebana, K. & Yano, M. SmartGrain: High-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160, 1871–1880 (2012).
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie (Poliploid-Grant Agreement No 101007438 and SexSeed-Grant Agreement No 690946). CZ-C is founded by EMBO Fellowship ALTF 975-2021. The research was supported by Science Foundation Ireland (SFI; grants 02/IN.1/B49 and 08/IN.1/B1931), and by Università degli Studi di Milano (Piano di sviluppo di ateneo) and PRIN 2017 (20175R447S). We thank A. Cavalleri for helpful discussion and B. Grieco, T. Tawhida, E. Cavalera and D. Boffelli for technical support. Part of this work was carried out at the Botanical Garden of the Università degli Studi di Milano and at NOLIMITS, the advanced imaging facility of the Università degli Studi di Milano. The flow cytometry experiments were performed in the NUI Galway Flow Cytometry Core Facility which is supported by funds from NUI Galway, Science Foundation Ireland, the Irish Government’s Programme for Research in Third Level Institutions, Cycle 5 and the European Regional Development Fund. The authors thank the two anonymous reviewers for the careful review and suggestions, that helped improved the final version of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: I.E., M.A., R.C., L.C., C.S. Performed the experiments: C.Z.-C., I.E., M.A.M., M.A., M.D.M., C.B., C.M., R.C. Analyzed the data: C.Z.-C., I.E., M.A.M., M.A., C.M., R.C. Contributed reagents/materials/analysis tools: I.E., C.S., L.C. Wrote and revised the paper: C.Z.-C., I.E., M.A., C.S., L.C., R.C. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zumajo-Cardona, C., Aguirre, M., Castillo-Bravo, R. et al. Maternal control of triploid seed development by the TRANSPARENT TESTA 8 (TT8) transcription factor in Arabidopsis thaliana. Sci Rep 13, 1316 (2023). https://doi.org/10.1038/s41598-023-28252-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28252-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.