Abstract
Mercaptopurine intolerance is an adverse effect of mercaptopurine administration in pediatric patients with acute lymphoblastic leukemia (ALL). NUDT15 variants have emerged as major determinants of mercaptopurine intolerance, especially in the Asian population. Two variants, c.55_56insGAGTCG in exon 1 and c.415C > T in exon 3, were commonly detected in the same allele, named NUDT15*1/*2. Although rare, compound heterozygous mutations also occur, with the two variants on different alleles (NUDT15*3/*6), which may confer tolerance to considerably lesser mercaptopurine dosage. Sanger sequencing or pyrosequencing can determine the NUDT15 variants but not the phase. Here, we designed an allele-specific PCR (AS-PCR) with locked nucleic acid-modified primers. A cohort of 63 patients harboring heterozygous c.55_56insGAGTCG and c.415C > T NUDT15 variations was selected for haplotyping using AS-PCR. Of the 63 patients, 60 harbored the NUDT15*1/*2 variant and three harbored compound heterozygous mutations, including two NUDT15*3/*6 and one NUDT15*2/*7 variants. These findings suggest that AS-PCR can determine NUDT15 diplotype and identify patients with compound heterozygous NUDT15 variants, which may enable precise genetic diagnosis of NUDT15. Nevertheless, a larger clinical trial is required to understand the clinical significance of NUDT15*3/*6 in pediatric patients with ALL because of its low incidence rate and challenges in detecting this variant.
Similar content being viewed by others

Introduction
Thiopurines, including azathioprine, 6-thioguanine (6-TG), and 6-mercaptopurine (6-MP), are effective anti-inflammatory, anticancer, and immunosuppressive drugs that are widely administered for different clinical indications1. In pediatric acute lymphoblastic leukemia (ALL), 6-MP is widely used in the induction, consolidation (with high-dose methotrexate), and maintenance phases2,3,4,5. It is a prodrug that is converted to the active form upon metabolization6. Therefore, the proteins involved in the metabolic pathways might prolong the half-life of 6-MP, thereby augmenting its side effects, especially neutropenia7. Polymorphisms in the thiopurine methyltransferase (TPMT) gene have been linked to susceptibility to thiopurine-induced marrow suppression in patients. Preemptive TPMT genotype-guided dosing is a successful example of genetics-based precision medicine for treating childhood ALL, which has been used in Western countries for more than 30 years8,9,10,11.
Asian pediatric patients with ALL experience more thiopurine-induced toxicity than Europeans, despite a lower frequency of TPMT mutations. A genome-wide association study reported a missense variant in NUDT15 (rs116855232, referred to as c.415C > T or p.Arg139Cys) strongly associated with thiopurine-related myelosuppression in patients with inflammatory bowel disease in a Korean population12. In addition, NUDT15 c.415C > T was demonstrated to be associated with mercaptopurine intolerance in childhood ALL by Yang et al.13. NUDT15 encodes a nucleoside diphosphatase that dephosphorylates thioguanine nucleotides, thereby preventing their incorporation into DNA14. Moriyama et al. sequenced all three coding regions of NUDT15 and identified more comprehensive genetic variants of this gene. Several NUDT15 variants associated with decreased diphosphatase activity have been reported, and haplotypes with different combinations of the variants have been assigned star allele numbers proposed by Moriyama et al.15. The loss-of-function NUDT15*2 allele is characterized by a missense variant in exon 3 (NM_018283.4:c.415C > T, p.Arg139Cys, rs116855232; also present in NUDT15*3) and an in-frame insertion in exon 1 (NM_018283.4:c.55_56GAGTCG, p.V18_V19insGV, rs746071566; also present independently as NUDT15*6)11,15.
Patients with heterozygous c.415C > T and c.55_56GAGTCG are either heterozygous (NUDT15*1/*2) or compound heterozygous (NUDT15*3/*6) for this mutation16. Patients with NUDT15*3/*6 are possibly intermediate metabolizers11. In theory, patients with two defective alleles might be poor metabolizers; for example, one patient with NUDT15*3/*6 was reported to tolerate a very low dose of mercaptopurine16. However, the currently available common methods cannot define diplotypes in complex genetic alterations. Hence, a simple genetic diagnostic method is required to identify the patients with NUDT15*3/*6 in daily clinical practice. In our previous study, we elucidated the NUDT15 diplotypes in patients with multiple heterozygous variants by performing next-generation sequencing (NGS) using NUDT15 cDNA. However, targeted sequencing of NUDT15 cDNA cannot be used in routine clinical practice because of the cost and long turn-around time (TAT)16. In contrast, allele-specific PCR (AS-PCR) is a conceptually simple, single nucleotide polymorphism (SNP)-based genotyping strategy based on the position of the 3′ base of a PCR primer to match one SNP allele and accurately extend only the correctly matched primer under stringent conditions. Locked nucleic acid (LNA) is an RNA nucleotide analog with a methylene bridge connecting the 2' oxygen and 4' carbon. This bridge restricts the ribose in the C3′-endo conformation, thereby enhancing the stability of LNA and increasing the hybridization melting temperature (Tm)17. LNA hybridizes with its complementary sequence, with high affinity over DNA or RNA oligonucleotides, which makes LNA oligos an excellent tool for distinguishing single nucleotides18.
In this study, AS-PCR was performed using LNA primers to determine the diplotype of NUDT15 in pediatric ALL patients who harbor heterozygous c.415C > T and c.55_56GAGTCG mutations. Although most patients were NUDT15*1/*2, there were two patients with NUDT15*3/*6 identified by AS-PCR.
Methods
Patients
In total, 449 patients with pediatric ALL (aged < 18 years) were enrolled in this study between April 1997 and December 2021. We performed Sanger sequencing to screen for NUDT15 variants and selected 63 patients with heterozygous c.55_56insGAGTCG and c.415C > T mutations16,19. The study was performed in accordance with the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the National Taiwan University Hospital (201510016RIND). Written informed consent was obtained from parents, guardians, or patients.
Genomic DNA extraction
White blood cells were purified from whole blood samples using RBC lysis buffer according to the manufacturer’s instructions (Thermo Fisher Scientific, Piscataway, NJ, USA). Genomic DNA was extracted from white blood cells using a phenol/chloroform-based method as described previously20, and the concentration of the extracted DNA was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific).
NUDT15 genotyping
The method of NUDT15 genotyping is published elsewhere19 and is shown in Fig. 1A. The patients harboring heterozygous c.55_56GAGTCG and c.415C > T were further analyzed using AS-PCR.
Primer designs for NUDT15 genotyping and allele-specific PCR. (A) Two primer sets, E1-F/E1-R and E3-F/E3-R, were used to amplify exons 1 and 3 of NUDT15, respectively. (B) Two sets of locked nucleic acid (LNA) primers were used to amplify samples with heterozygous NUDT15 c.55_56insGAGTCG and c.415C > T to differentiate between NUDT15*1/*2 and NUDT15*3/*6.
AS-PCR analysis of NUDT15
The NUDT15 c.415C and c.415T alleles were specifically amplified using AS-PCR. Each PCR reaction included 10 ng genomic DNA, 800 nM dNTP, 1 × PrimeSTAR GXL buffer, 200 nM forward (common primer, CP-F), 200 nM reverse primer (415C-R for c.415C allele or 415T-R for c.415T allele), and 0.75 U PrimeSTAR GXL DNA polymerase (Takara Bio, San Jose, CA, USA) (Fig. 1B). Thermocycling was performed under the following conditions: 98℃ for 5 min, 35 cycles of 98 ℃ for 10 s and 68 ℃ for 1.5 min, and final extension at 68℃ for 5 min. The PCR products were analyzed by performing gel electrophoresis on 1% agarose gel, and an 8.2 kb band was observed in case of successful PCR amplification. Gels were imaged by SmartView Pro Imager (Major Science, UVCI-1100). Images were acquired with 312 nm UV light at 3 to 5 s exposure times with the SmartView Pro 1100 Imager system (Major Science) and stored in a .JPEG format. The .JPEG files were directly used for presentation. PCR clean-up reaction was performed as follows prior to sequencing: 5 μL PCR product was mixed with 1 U FastAP (Thermo Fisher Scientific) and 10 U exonuclease I (Thermo Fisher Scientific) and incubated at 37℃ for 15 min, followed by 15 min incubation at 85℃. The purified PCR products were sequenced using the E1-S primer. The sequencing results were aligned to the NCBI GenBank entry NG_047021.1 using the SnapGene 4.1.3 software (GSL Biotech LLC, San Diego, CA, USA). The NUDT15 diplotype was determined to be NUDT15*1/*2 when c.55_56insGAGTCG and c.415C > T were located on the same allele or NUDT15*3/*6 when c.55_56insGAGTCG and c.415C > T were located on the different alleles. Detailed information on the primers is listed in Table 1.
Results
Sanger sequencing for screening NUDT15 variants
Sanger sequencing to analyze heterozygous c.55_56insGAGTCG and c.415C > T variants (Fig. 2) identified a total of 63 patients harboring the heterozygous c.55_56insGAGTCG and c.415C > T variants (14%, 63/449). These patients included the patients (n = 37) evaluated in our previous publications16,19. The NUDT15 diplotypes of these patients in the previous study were NUDT15*1/*2 (n = 35), NUDT15*3/*6 (n = 1, ID898) and NUDT15*2/*7 (n = 1, ID341)16,19,21. The genotype based on NUDT15 expression of all 63 patients is shown in Table 2.
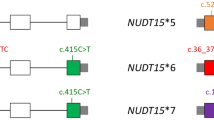
Results of Sanger sequencing of samples with both heterozygous c.55_56insGAGTCG and c.415C > T. The NUDT15 exon 1 (red line) and exon 3 (blue line) regions are amplified separately, and the corresponding sequencing results are shown below. Both NUDT15*1/*2 and NUDT15*3/*6 showed identical Sanger sequencing signals.
Distinguishing NUDT15*1/*2 from *3/*6 using AS-PCR
To define the NUDT15 diplotype of patients harboring heterozygous c.55_56GAGTCG and c.415C > T, an AS-PCR assay was used for the c.415C or c.415T alleles. The common forward primer (CP-F) was specific for the 5′ UTR and the reverse primer was specific for c.415C (415C-R) or c.415T (415T-R) (Fig. 1B). To improve the mismatch discrimination, the 3′ nucleotide of the reverse primer was substituted with LNA. Several nucleotides of the forward and reverse primers were also replaced with LNA to increase the melting temperature (Tm) to meet the parameters of PCR. Unlike PCR with LNA primers, AS-PCR using DNA primers cannot distinguish between the c.415C and c.415T alleles (Fig. 3). To test the efficacy of AS-PCR assay, samples with homozygous c.415C (*1/*1), homozygous c.415T (*2/*3), and heterozygous c.415C > T (*1/*2 and *3/*6) were used. The *1/*1 and *2/*3 samples could only be amplified using 415C AS-PCR or 415T AS-PCR, respectively, while *1/*2 and *3/*6 could be amplified using both AS-PCRs (Fig. 4). The sequencing of the AS-PCR products using the E1-S primer to determine the NUDT15 diplotype, located the c.55_56insGAGTCG variant on the c.415T allele of the *1/*2 sample or the c.415C allele of the *3/*6 sample (Fig. 4).
Comparison of allele-specific PCR (AS-PCR) using LNA or DNA primers. NUDT15 was amplified using AS-PCR with LNA or DNA primers. Agarose gel electrophoresis of equal volumes of PCR products amplified from NUDT15*1/*1 (homozygous c.415C) and NUDT15*2/*3 (homozygous c.415T) samples; M, 1 kb DNA marker; kb, kilobase.
NUDT15 diplotype analysis using allele-specific PCR (AS-PCR). (A) Samples with NUDT15*1/*2, *3/*6, *1/*1, and *2/*3 were amplified using 415C AS-PCR and 415T AS-PCR, and the PCR products were analyzed using 1% agarose gel electrophoresis. (B) Results of Sanger sequencing of AS-PCR product amplified from NUDT15*1/*2 and *3/*6 samples (ID1233). The c.55_56insGAGTCG insertion is shown by a red dotted line; M, 1 kb DNA marker; kb, kilobase; NTC, no template control.
Three patients with compound heterozygous variants were identified
Of the 63 patients, 60 harbored NUDT15*1/*2, and two samples were identified to harbor NUDT15*3/*6 (Fig. 4, Fig. S1) and one NUDT15*2/*7 (Fig. S2). The two patients harboring NUDT15*3/*6 identified in this study (ID898 and ID1233) included the patient (ID898) reported in our previous study16. Unfortunately, the patient (ID1233) identified in this study died of fungal infection during the induction chemotherapy and took 6-MP only for 3 days. Therefore, we are not able to determine her tolerable dose of 6-MP.”
Discussion
The emerging role of NUDT15 in precision medicine has been indicated in several guidelines, which has rendered the evaluation of this gene important11,22. Most laboratories perform Sanger sequencing to screen the genetic variants of NUDT1522. If c.55_56insGAGTCG and c.415C > T are identified simultaneously, the mutation is labeled as NUDT15*1/*2. However, NUDT15*3/*6 may also exist in some cases. Our study showed that AS-PCR targeting the C or T nucleotide amplifies the individual allele and can be used to differentiate NUDT15*1/*2 from NUDT15*3/*6. This approach might help to precisely identify the genetic variants of NUDT15 and determine the allele type. Therefore, it could be more appropriate to determine the dosing of mercaptopurine before prescribing it for treating childhood ALL.
The NUDT15*6 allele is associated with uncertain functions and is characterized by the in-frame insertion of a 6 base pair repeat (GAGTCG) in exon 1 of NUDT15, which results in four copies of the GAGTCG repeat. The NUDT15*6 allele is present in approximately 1.3% of the East Asian population23. Detection of the duplication is required to accurately detect and distinguish NUDT15*2 from NUDT15*322. Earlier, three studies investigated the diplotype of NUDT15. Yu et al. used targeted sequencing of NUDT15 cDNA to determine the diplotype, and two patients harboring compound heterozygosity were identified, one with NUDT15*3/*6 and the other with NUDT15*2/*716. Tsujimoto et al. defined the diplotypes of 14 patients harboring the two variants (i.e., c.415C > T and c.55_56insGAGTCG) using droplet digital PCR (ddPCR); however, they could not identify any patients with NUDT15*3/*624. Recently, although the robustness of a long-read HiFi amplicon sequencing assay has been reported for phased full gene characterization and the discovery of pharmacogenomic allele25, the method may not conveniently elucidate the phasing of NUDT15 in daily clinical practice due to its higher cost and unavailability in most laboratories in Taiwan.
AS-PCR has several advantages in identifying the NUDT15 diplotype in daily clinical practice. Usually, physicians need to send one sample to the laboratory to assess the mutations in NUDT15 before prescribing mercaptopurine or related drugs. Sanger sequencing is a rapid method for screening variants of the NUDT15 coding region. If variants are not identified in the coding region, the genotype should be reported as wild type and exempted from further analysis. However, if c.415C > T and the c.55_56insGAGTCG are identified simultaneously, AS-PCR with LNA primers can be used to determine the diplotype, which will be cost-effective (approximately 20 USD per sample) and fast (TAT within 5 working days). Importantly, the process is lesser labor-intensive than ddPCR or NGS-based methods16,24,25.
The patients with NUDT15*3/*6 are believed to have intermediate-poor metabolism and might require a dose reduction of 6-MP11. A recent publication of NUDT15 by several joint committees suggested NUDT15*3 as a Tier 1 variation22. Tier 1 alleles include the most common variant alleles that can be used to maximize disease detection rate in most populations. Variant alleles associated with the loss-of-function present in 0.01% or greater percentage of any subpopulation are included in Tier 2 for more comprehensive genotyping. NUDT15*2, *4, *6, *9 and *14 are listed as Tier 2 variations22. Many laboratories that perform NUDT15 genotyping only target the c.415C > T variant present in both NUDT15*2 and *3 and may not differentiate between these two alleles, as it is technically challenging to assess c.55_56insGAGTCG. However, c.55_56insGAGTCG can be identified using Sanger sequencing. AS-PCR using LNA primers can differentiate between NUDT15*1/*2 or *3/*6. As patients harboring NUDT15*3/*6 can be identified in high variant allele frequency populations, differentiating between these two genotypes might be important if more cases were identified and their lower tolerable 6-MP doses were verified. In our previous study, we identified 3 patients with NUDT15 compound heterozygous variant16. The tolerable 6-MP doses were lower for NUDT15*3/*6 (2.5 mg/m2, one patient died before 6-MP dose evaluation) and NUDT15*2/*7 (6.7 mg/m2) than that for NUDT15*1/*2 (median, 12.5 mg/m2)16,19. The patients with NUDT15*3/*6 or NUDT15*2/*7 still experienced frequent neutropenia or neutropenic fever even though they were given a low dose of mercaptopurine (12 mg/m2). Because white blood cell count-based dose adjustments are regularly performed for known NUDT15- deficient patients and result in a reduced risk of neutropenia and febrile neutropenia26, the accurate determination of patients with NUDT15*3/*6 may avoid them from receiving a high dose of mercaptopurine; even if the mercaptopurine dose was already reduced based on the results of NUDT15*1/*2 genetic analysis. The clinical significance of NUDT15*3/*6 needs further evaluation in future international clinical trials27 due to the lower incidence rate and challenges in detecting this variant in Asian populations.
In conclusion, Sanger sequencing can be used to screen NUDT15 variants. In contrast, AS-PCR with LNA primers can be applied to determine the true diplotype of NUDT15 and simultaneously detect heterozygous c.55_56insGAGTCG and c.415C > T. This approach would enable the diagnosis of compound heterozygous mutations (NUDT15*3/*6). However, because of the limited number of NUDT15*3/*6 cases, and as most methods are unable to differentiate them from NUDT15*1/*2, the tolerable dosing of 6-MP remains to be determined for this small subset of patients. In the future, large international clinical trials are required to evaluate the adjusted dosing of mercaptopurine in pediatric ALL patients with NUDT15*3/*6.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Karran, P. & Attard, N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat. Rev. Cancer 8, 24–36 (2008).
Pui, C. H. et al. Childhood acute lymphoblastic Leukemia: Progress through collaboration. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. https://doi.org/10.1200/JCO.2014.59.1636 (2015).
Pui, C. H., Nichols, K. E. & Yang, J. J. Somatic and germline genomics in Paediatric acute lymphoblastic Leukaemia. Nat. Rev. Clin. Oncol. 16, 227–240. https://doi.org/10.1038/s41571-018-0136-6 (2019).
Teachey, D. T., Hunger, S. P. & Loh, M. L. Optimizing therapy in the modern age: differences in length of maintenance therapy in acute lymphoblastic leukemia. Blood 137, 168–177. https://doi.org/10.1182/blood.2020007702 (2021).
Toksvang, L. N., Lee, S. H. R., Yang, J. J. & Schmiegelow, K. Maintenance therapy for acute lymphoblastic leukemia: Basic science and clinical translations. Leukemia 36, 1749–1758. https://doi.org/10.1038/s41375-022-01591-4 (2022).
Aarbakke, J., Janka-Schaub, G. & Elion, G. B. Thiopurine biology and pharmacology. Trends Pharmacol Sci 18, 3–7. https://doi.org/10.1016/s0165-6147(96)01007-3 (1997).
Moriyama, T., Relling, M. V. & Yang, J. J. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood 125, 3988–3995. https://doi.org/10.1182/blood-2014-12-580001 (2015).
Krynetski, E. Y. et al. Genetic polymorphism of thiopurine S-methyltransferase: Clinical importance and molecular mechanisms. Pharmacogenetics 6, 279–290. https://doi.org/10.1097/00008571-199608000-00001 (1996).
Relling, M. V. et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther 93, 324–325. https://doi.org/10.1038/clpt.2013.4 (2013).
Relling, M. V. et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. JNCI J. Nat. Cancer Inst. 91, 2001–2008. https://doi.org/10.1093/jnci/91.23.2001 (1999).
Relling, M. V. et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin. Pharmacol. Ther. 105, 1095–1105. https://doi.org/10.1002/cpt.1304 (2019).
Yang, S.-K. et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat. Genet. 46, 1017–1020. https://doi.org/10.1038/ng.3060 (2014).
Yang, J. J. et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 33, 1235–1242. https://doi.org/10.1200/jco.2014.59.4671 (2015).
Valerie, N. C. et al. NUDT15 hydrolyzes 6-thio-deoxyGTP to mediate the anticancer efficacy of 6-thioguanine. Cancer Res. 76, 5501–5511. https://doi.org/10.1158/0008-5472.Can-16-0584 (2016).
Moriyama, T. et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 48, 367–373. https://doi.org/10.1038/ng.3508 (2016).
Yu, C. H. et al. Determination of NUDT15 variants by targeted sequencing can identify compound heterozygosity in pediatric acute lymphoblastic leukemia patients. Sci. Rep. 10, 14400. https://doi.org/10.1038/s41598-020-71468-y (2020).
Koshkin, A. A. et al. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54, 3607–3630. https://doi.org/10.1016/S0040-4020(98)00094-5 (1998).
Latorra, D., Campbell, K., Wolter, A. & Hurley, J. M. Enhanced allele-specific PCR discrimination in SNP genotyping using 3’ locked nucleic acid (LNA) primers. Hum. Mutat. 22, 79–85. https://doi.org/10.1002/humu.10228 (2003).
Wang, D.-S. et al. Childhood acute lymphoblastic leukemia mercaptopurine intolerance is associated with NUDT15 variants. Pediatr. Res. 89, 217–222. https://doi.org/10.1038/s41390-020-0868-8 (2021).
Yang, Y. L. et al. IKZF1 deletions predict a poor prognosis in children with B-cell progenitor acute lymphoblastic leukemia: A multicenter analysis in Taiwan. Cancer Sci. 102, 1874–1881. https://doi.org/10.1111/j.1349-7006.2011.02031.x (2011).
Moriyama, T. et al. Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry. Blood 130, 1209–1212. https://doi.org/10.1182/blood-2017-05-782383 (2017).
Pratt, V. M. et al. TPMT and NUDT15 genotyping recommendations: A joint consensus recommendation of the Association for Molecular Pathology, Clinical Pharmacogenetics Implementation Consortium, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, European Society for Pharmacogenomics and Personalized Therapy, and Pharmacogenomics Knowledgebase. J. Mol. Diagn. https://doi.org/10.1016/j.jmoldx.2022.06.007 (2022).
Moyer, A. M. NUDT15: A bench to bedside success story. Clin. Biochem. 92, 1–8. https://doi.org/10.1016/j.clinbiochem.2021.02.007 (2021).
Tsujimoto, S. et al. Diplotype analysis of NUDT15 variants and 6-mercaptopurine sensitivity in pediatric lymphoid neoplasms. Leukemia 32, 2710–2714. https://doi.org/10.1038/s41375-018-0190-1 (2018).
Scott, E. R. et al. Long-read HiFi sequencing of NUDT15: Phased full-gene haplotyping and pharmacogenomic allele discovery. Hum. Mutat. 43, 1557–1566. https://doi.org/10.1002/humu.24457 (2022).
Wahlund, M. et al. The role of TPMT, ITPA, and NUDT15 variants during mercaptopurine treatment of Swedish pediatric patients with acute lymphoblastic Leukemia. J Pediatr 216, 150-157.e151. https://doi.org/10.1016/j.jpeds.2019.09.024 (2020).
Tanaka, Y. et al. An international retrospective study for tolerability of 6-mercaptopurine on NUDT15 bi-allelic variants in children with acute lymphoblastic leukemia. Haematologica 106, 2026–2029. https://doi.org/10.3324/haematol.2020.266320 (2021).
Acknowledgements
The authors are grateful to all the patients who participated in this study and their parents. The authors also acknowledge the efforts of the TPOG (Taiwan Pediatric Oncology Group) and the Childhood Cancer Foundation in Taiwan.
Funding
Funding was provided by Ministry of Science and Technology, Taiwan (Grant No. 110-2314-B-002-091-MY3) and National Taiwan University Hospital (Grant No. 110-L1007).
Author information
Authors and Affiliations
Contributions
C.-H.Y. and Y.-L.Y. designed the study, analyzed the data, and wrote the manuscript. C.-H.Y., D.-S.W., Y.-L.N., P.-Y.X., and S.-W.L. acquired and processed patient specimens and performed experiments. C.-Y.L., Y-H.C., and H.-Y C. performed the analysis. K.-H.L., S.-T.J., M.-Y.L., K.-H.W., C.-N.C, H.-H.C., S.-W.C., M.-Y.S., and D.-T.L. provided clinical samples and data. The manuscript was written by C.-H.Y. and Y.-L.Y. and was reviewed and edited by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, CH., Chang, YH., Wang, DS. et al. Allele-specific polymerase chain reaction can determine the diplotype of NUDT15 variants in patients with childhood acute lymphoblastic Leukemia. Sci Rep 13, 490 (2023). https://doi.org/10.1038/s41598-023-27720-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-27720-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.