Abstract
Pullulanase, a starch debranching enzyme, is required for the preparation of high glucose/maltose syrup from starch. In order to expand its narrow reaction conditions and improve its application value, Bacillus naganoensis pullulanase (PulA) was mutated by site-directed mutagenesis and the biochemical characteristics of the mutants were studied. The mutant PulA-N3 with mutations at asparagine 467, 492 and 709 residues was obtained. It displayed the activity maximum at 60 °C and pH 4.5 and exceeded 90% activities between 45 and 60 °C and from pH 4.0 to pH 5.5, which was improved greatly compared with wild-type PulA. Its thermostability and acidic pH stability were also remarkably improved. Its catalytic rate (kcat/Vmax) was 2.76 times that of PulA. In the preparation of high glucose syrup, the DX (glucose content, %) values of glucose mediated by PulA-N3 and glucoamylase reached 96.08%, which were 0.82% higher than that of PulA. In conclusion, a new pullulanase mutant PulA-N3 was successfully developed, which has high debranching activity in a wide range of temperature and pH, thereby paving the way for highly efficient starch saccharification.
Similar content being viewed by others
Introduction
Pullulanase is a kind of enzyme that specifically hydrolyzes α-1,6-glycosidic bonds at branch point of amylopectin. It plays an essential role in enzymatic processing of starch to either high glucose syrup or high maltose syrup1,2,3,4. Many different pullulanases have been biochemically identified and divided into five types (pullulanase type I, pullulanase type II, pullulan-hydrolysing enzyme type I or new pullulanase, pullulan-hydrolysing enzyme type II or isopullulanase and pullulan-hydrolysing enzyme type III) according to the differences in substrates and hydrolysis methods5 Currently, only type I pullulanases from Bacillus acidopullulyticus6 and Bacillus naganoensis7 are available on the market for industrial purposes.
When pullulanase is applied in the preparation of high glucose syrup, it often works with glucoamylase to promote starch saccharification efficiency. In order to meet the criteria necessary for starch saccharification, pulllulanase that has a temperature optimum of 60 °C or higher and pH optimum of 4.5 or lower is required (it is recommended by Novozymes that to operate at 61 °C and pH 4.3 for preparation of high-glucose syrup from starch). However, the wild type pullulanase from B. naganoensis has a temperature optimum of 55 °C and pH optimum of 5.0 with poor thermostability8 and its activity is limited in a narrow pH range (over 70% of its activity at pH 4.5 to 5.2)9, which are not well compatibility matching with that of glucoamylase. Therefore, it is valuable to obtain a pullulanase with the highest activity under more acidic pH and higher temperature with sufficient thermostability (temperature up to 60 °C) and pH stability (pH 4.0 or lower). For this purpose, several successful endeavors have been made. For example, the residue D787 lining the catalytic pocket of B. naganoensis pullulanase were identified and mutated, and the resulted mutant D787C maintained higher activity at temperatures over 60 °C and 90% residual activity at pH 4.0 and its enzymatic activity and specific activity were increased by 0.5-fold10. Using the evolutionary coupling saturation mutagenesis approach, seven residue pairs as evolutionary mutational hotspots far from the catalytic pocket of B. naganoensis pullulanase were predicted and the subsequent saturation mutagenesis yielded variants with enhanced catalytic activity11.
In the present studies, B. naganoensis pullulanase (PulA) was artificially evolved by site-directed mutagenesis technology. The mutants with improved starch debranching activities at higher temperature and under wider acidic pH range were developed. The values applied for starch saccharification to glucose were illustrated. To the best understanding of our knowledge, this is the first report that the debranching conditions (temperature and pH) of B. naganoensis pullulanase have been successfully extended and optimized.
Materials and methods
Strains, plasmids and primers
Escherichia coli JM109 and Bacillus licheniformis CBBD30212 were used as the hosts for the gene cloning and expression, respectively. The strains were grown at 37 °C and 220 rpm for 12–16 h in Luria–Bertani (LB) medium (tryptone 10.0 g/L, yeast extract 5.0 g/L, and NaCl 10.0 g/L) or at 37 °C on LB plates. When necessary, 100 µg/mL ampicillin or 20 µg/mL kanamycin was supplemented. pHY-WZX13 is an expressed vector in host strains from Bacillus genus and used to mediate the secretion and expression of pullulanase and its mutant genes. pWB-PulA carrying the pullulanase-encoded gene from B. naganoensis ATCC 53,909 constructed in the previous work7 was used as template for DNA manipulation. The sequences of oligonucleotides containing the appropriate base changes for site-directed mutagenesis are listed in Table 1 and chemically synthesized.
Prediction and structure analysis of pullulanase mutation sites
Based on the difference in the free energy of folding of the amino acid sequence, the mutation sites in PulA were predicted according to the method introduced in the literature14. The basic process for prediction of the putative mutation sites in PulA was to set the pH to 4.0 and the temperature to 60 °C in I-Mutant 2.014 as the selective parameters. Each of 926 amino acid residues in PulA was inputted and the outputs were selected and grouped according to the credibility index generated by the software. According to the difference in the free energy of protein unfolding (ΔΔG) formed by the amino acid residue changes in the amino acid sequence of pullulanase, the possible amino acid positions for mutation and mutated residues were further analyzed by homology modeling, structure matching and molecular docking as described below.
Pullulanase 2WAN (PDB: 2WAN)3 was selected as the template for PulA (GenBank: AEV53626.1) homology modeling using software SWISS-MODEL Protein-modeling Server15,16,17. Structure comparison and analysis between PulA and its mutants were performed by the program of PyMOL18. Maltotriose, a repeated unit joined by α-1,6 linkages in pullulan, the natural substrate of pullulanase19, was used as a pullulanase ligand for molecular docking by Auto Dock software20,21. The protein structure derived from the above homology modeling was used as the receptor structure.
Site-directed mutagenesis of PulA
The site-directed mutagenesis of pullulanase PulA encoding gene was adopted using primer-mediated overlapping PCR method22. In general, partial fragments of the pullulanase-encoding gene were amplified using pWB-PulA (Wang et al., 2014) as template and two pairs of primers PULA-F and 467-R, 467-F and PULA-R (Table 1). The full-length mutant pullulanase encoding gene was then overlapped and amplified under the mediation of primers PULA-F and PULA-R (Table 1) using the mixture of the above amplified fragments as template. Other site mutations were done in the similar procedure except different pairs of primers used (Table 1). The PCR amplification condition was set as follows: Initial denaturization at 95 °C for 5 min, cycling for 30 cycles at 94 °C for 10 s, 56 °C for 1 min, and 68 °C for 3 min. The mutated pullulanase-encoding gene was cloned into pHY-WZX13 and transformed into E. coli JM109 by heat shock transformation23, and then into B. licheniformis CBBD302 according to the method as described12 for preparation of pullulanase mutants. The mutated sites were verified by Sanger sequencing24.
Expression and purification of mutants
The transformants of B. licheniformis CBBD302 were grown at 37 °C and 200 rpm for 12–16 h in 50 mL LB liquid medium containing 20 μg/mL kanamycin. The culture was then inoculated at a 10% dose in 50 mL LB liquid medium in a 250 mL Erlenmeyer flask and incubated for 72 h at 37 °C and 220 rpm12. The supernatant was harvested by centrifugation at 8,000 rpm and 4 ℃ for 10 min and was precipitated with a saturated ammonium sulfate solution of 40–70% saturation when necessary. The enzyme was then purified by an ÄKTA Pure system using a Sephadex G-100 (10 × 500 mm) (Cytiva, Sweden) by eluting with 50 mM phosphate buffer (pH 7) at a flow rate of 0.5 mL/min. The chromatographic elution peaks were detected by the following method for the determination of the enzymatic activity of pullulanase, and the pullulanase activity parts were collected. The molecular weight and purity of the enzyme was estimated based on SDS-PAGE, selecting 10% (w/v) running gel and 5% (w/v) stacking gel25 using the unstained protein molecular weight marker #26,610 (Thermo Fisher Scientific, China). Protein concentration was determined by Bradford method with bovine serum albumin fraction V (Roche Diagnostics GmbH, Germany) as a reference standard26. The activities of PulA and the mutants were determined according to the method as described previously7 using pullulan (Sinopharm Chemical Reagent Co. Ltd, China) as substrate. One unit of the enzyme activity is defined as the amount of enzyme that produced one μmole of glucose reducing-sugars equivalents per minute under 60 °C and pH 4.5.
Effects of pH and temperature on enzyme activity and kinetic parameters
The temperature optimum was determined by measuring the enzyme activity across the temperature range from 40 to 70 °C. The pH optimum was determined by assaying the enzyme activity in 20 mM of acetate-sodium acetate buffer (pH 3.5–5.8) or phosphate buffer (pH 5.8–6.5) of different pH value ranging from 3.5 to 6.525. The results are shown as relative activity and the activity obtained at optimal condition was set as 100%. The thermostability was examined by measuring the enzyme activity after incubation at 45, 50, 55 and 60 °C for 0.5, 1, 1.5, 2, 2.5, and 3.0 h, respectively. The pH stability was examined by measuring the enzyme activity after adding 20 mM acetate or phosphate buffer of different pH value ranging from 3.5 to 6.525 and incubated at room temperature for 1 h. The activity of untreated enzyme is taken as 100% and the percentage of relative activity is calculated relative to the highest activity.
The kinetic parameters (Km, Vmax, and κcat values) of the enzymes were determined based on the method described previously27, where different concentrations (0.25, 0.5, 1, 2, 4 and 8 mg/mL) of pullulan (Sinopharm Chemical Reagent Co. Ltd, China) were adopted. The values of Vmax and Km were estimated by fitting the initial rate data to the Michaelis–Menten equation using nonlinear regression.
All experiments were carried out in triplicate. The results data were represented as mean ± standard deviation of triplicate experiments.
Preparation of glucose from starch
Preparation of glucose syrup from starch was carried out in a 100-mL reaction vessel by simulating the industrialized preparation process of starch glucose syrup. The enzyme solution comprising glucoamylase (Shandong Longkete Enzyme Co. Ltd., China) and pullulanase (PulA or PulA-N3) was added in a 100-mL reaction vessel containing 30% (w/v) maltodextrin from liquefied starch (DE value: 8, gifted by Roquette Americas). The enzyme dosage was equal to 150 U of glucoamylase and one unit of pullulanase per gram of dry maltodextrin. The saccharification process was carried out at 61 °C and pH 4.3 with 120 rpm stirring by a magnetic stirrer. During the saccharification process, samples were taken at certain time intervals and then boiled for 10 min to inactivate enzymes and the ingredients were quantitatively measured by an HPLC system (Agilent 1200 Series HPLC System) with a refractive index detector (Shodex RI-201H, Shoko Science Co. Ltd., Japan) using a Alltech Prevail Carbohydrate Es 5u column (4.6 mm IDx250 mm, 5 μm) and 65% (v/v) acetonitrile as mobile phase at a flow rate of 1 mL/min at 35°C28. Glucose, maltooligosaccharides and isomaltose as standards were purchased from Jiangsu Ruiyang Biotech Co., Ltd., China. The glucose contents (%, g/g) were expressed as DX value, which were calculated as the percentage of glucose content to dry matter29. The contents of other sugars were also measured by HPLC and calculated as the percentage of glucose content to dry matter (%, g/g).
Results and discussion
Selection of putative amino acid residues for mutagenesis
The possible mutation of amino acid residues that may affect stabilities or activity in lower pH and higher temperature of PulA were first predicted using I-Mutant 2.014 at pH 4.0 and 60 °C based on the difference in the free energy of protein unfolding (ΔΔG) formed by the change of amino acid residues in the amino acid sequence. Total 28 amino acid residues in the PulA molecular, including S99, E100, Q108, S112, N317, N322, K327, N342, S357, N450, K453, K460, K463, N467, K469, K476, N492, N521, N523, N587, K626, S630, K665, K669, N709, N734, E758 and G776, were finally selected and might play the roles in maintaining the stability against the pH and temperature environment. It was found that asparagine residues (39.3%, 11/28) in PulA molecule were the major predicted residues. It is found that the amide group of asparagine is important in forming the side chain-backbone hydrogen bonds and fulfills an important role in stabilizing local folded structures in proteins30. Its deamidation is dominant pathway for protein denaturation and degradation (enzyme inactivation) and is a pH dependent process31. Therefore, in the present report the predicted 11 asparagine residues in PulA molecule were focused on for further investigation. By mimic 3D analysis, N467, N492 or N709 mutation might cause a significant change in the structure (Fig. 1). Alterations of N467, N492 and N709 could raise effects on the spatial structure of PulA and the putative changes produced by the mutations of N467G, N492A or N709R are shown (Fig. 1). If N467 is assumed to be changed to glycine, it extends in the peptide chain as neutral amino acid for the side chain of glycine is hydrogen bonded32. While N492 is assumed to be mutated to alanine, it maintains the stability of tertiary structure through hydrophobic interaction for the side chain consists of hydrophobic methyl group33. The mutation of N709 to arginine results in a guanidine side chain, which could facilitate the formation of more hydrogen bonds. Besides, the electric charge of arginine is more exposed to the surface of protein, which could maintain the stability of the structure by forming electrostatic interaction, and further improve the heat resistance34.
To further illustrate the rational sites selected for mutation, the total binding energy of maltotriose-enzyme complex was calculated using molecular docking20,21. A pullulanase (PDB ID: 2WAN) with the catalytic triad composed of Asp622, Glu651 and Asp7363 (Turkenburg et al., 2009) was used as template for homologous modeling of PulA to predict its catalytic triad. As shown in Fig. 2A, their overall structures did not overlap completely (65.93% homology identity generated by alignment of the amino acid sequences), but their substrate binding pockets with highly conserved amino acid residues were very similar. Therefore, the catalytic triad of PulA was predicted and composed of Asp619, Glu648 and Asp733 (Fig. 2A). The schematic diagram of pullulanase binding maltotriose was then predicted by Auto Dock and the results are shown in Fig. 2B. In the substrate binding pocket for wild-type PulA, the amino acid residues directly formed hydrogen bonds with maltotriose were Arg617, Asp619, Glu648, Trp650 and Asp733. In the substrate binding pocket for the candidate mutant mutated at N467, N492 and N709 residues, the amino acid residues were changed to Arg617, Asp619 and Asn734 (Fig. 2B). They both formed eight hydrogen bonds with the substrate, however, their average binding free energy was − 3.56 and − 1.91 kcal/mol, respectively. Mutations at N467, N492 and N709 residues of PulA might considerably reduce the binding free energy.
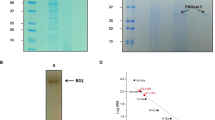
Structural schematic diagram of pullulanase. (A) Structural superposition of the predicted catalytic triads of PulA; B: the schematic diagram of PulA (a) and PulA-N3 (b) binding maltotriose. Pullulanase 2WAN (PDB ID: 2WAN) was used as template for homology modeling using software SWISS-MODEL Protein-modeling Server. Structure comparison and analysis of the wild-type enzyme and the mutants were performed by the program of PyMOL. Maltotriose was used as ligand for molecular docking by using Auto Dock software. The protein structure derived from the above homology modeling was used as the receptor structure. The number presented in the diagram represented the binding free energy.
Above analysis results strongly indicated that N467, N492 and N709 residues could play important roles in maintaining catalytic activity of PulA in lower pH and higher temperature. Therefore, they were selected for site-directed mutagenesis.
Mutation combination of N467, N492 and N709 resulted in improved activity of B. naganoensis pullulanase in higher temperature and more acidic pH ranges
N467, N492 and N709 residues in PulA as predicted above were mutated using primer-mediated site-directed mutagenesis22. The mutants were then expressed, prepared and purified to a single band on SDS-PAGE (Supplementary Fig. S1), and their biochemical properties were analyzed. Mutation of N467G, N492A or N709R caused meaningful changes in the specific activities. Of which, the specific activity of mutant N467G was increased about 11%, that of N492A and N709R were decreased (27% and 15%, respectively) (Table 2). Meanwhile, their temperature and pH optimum were changed, however, the change tendency was of variety (Fig. 3A). Mutant N467G had the maximal activity at 60 °C and pH 4.5, which was about 5 °C higher and 0.5 pH value lower than that of wild-type PulA. The activities of mutant N709R were improved under temperature from 55 ℃ to 65 ℃ and pH from 5.0 to 6.0, although its temperature and pH optima were not much changed. No remarkable changes in the temperature and pH optima occurred in mutant N492A comparison to PulA.
Effects of temperature (a) and pH (b) on the enzymatic activity of PulA mutants. (A) The temperature and pH optima of PulA (open diamond) and its single-site mutants N467G (solid diamond), N492A (solid triangle) and N709R (solid circular). (B) The temperature and pH optima of the multiple-site mutants PulA2-1 (N467G-N492A) (solid rectangle), PulA2-2 (N467G-709R) (solid triangle), PulA-N3 (N467G-N492A-N709R) (solid circular) and PulA (open diamond). (C) Thermostability and pH stability of PulA-N3 (circle) compared with PulA (diamond). The enzymes were incubated at 50 °C (solid icon) or 60 °C (open icon) for up to 3 h, or at different pH for 1 h. The residual activities were determined according to the Materials and Method section. The data were represented as average of triplicate experiments.
According to above results, mutation of N467 noticeably changed the temperature and pH activity profile of PulA, which encouraged us to further exploit mutants with better performance through mutation combination with residues N492 and N709. Three new mutants, PulA2-1 (mutations with N467G and N492A), PulA2-2 (mutations with N467G and N709R) and PulA-N3 (mutations with N467G, N492A and N709R), were developed, and their biochemical properties were determined. Obviously and interestingly, mutant PulA-N3 performed the highest activities from 45 ℃ to 60 ℃ and from pH 4.0 to pH 5.5, which dramatically broadened the enzymatic reaction condition (Fig. 3B). Its thermostability and pH stability were also improved (Fig. 3C). Under the optimal temperature and pH, PulA-N3 performed lower Km value and obviously increased κcat and κcat/Km values. The κcat/Km value of PulA-N3 was 2.76 folds of PulA (Table 3). Besides, the specific activity of PulA-N3 were increased about 20% (Table 2). The results indicated that PulA-N3 could have an enhanced catalytic rate on debranching of starch.
Glucose yield was improved in starch saccharification using PulA-N3
The pH and temperature optima of glucoamylase and pullulanase for starch saccharification were further comparatively examined and the results are summarized in Fig. 4. Glucoamylase displayed the highest activity at pH 4.0 and exhibited more than 90% of its maximum activity in the range of pH 4.0–4.5. Comparatively, mutant PulA-N3 did 96% and 90% of the highest enzyme activities at pH 4.3 and 4.0, respectively. The results clearly showed that PulA-N3 and glucoamylase shared well-matched pH optima, but PulA did not (Fig. 4A). Similar results were obtained for the optimal reaction temperature (Fig. 4B). Both Glucoamylase and PulA-N3 had the optimal temperature of 60 °C but PulA not (The optimal temperature of PulA was 55 °C and only 78% of its maximum activity at 60 °C) (Fig. 4B). The optimal reaction pH and temperature range of PulA-N3 should favor glucoamylase-led starch saccharification.
The time-course of corn starch saccharification with glucoamylase and PulA-N3. (A) Optimal pH for PulA-N3 and glucoamylase; (B) Optimal temperature for PulA-N3 and glucoamylase; (C) The time-course of corn starch saccharification. The saccharification experiments were carried out at 61 °C and pH 4.3 with 120 rpm stirring in 100-mL reaction vessels containing 30% (w/v) liquefied starch (DE value: 12) by adding 150 U/g (dried substrate) of glucoamylase and 1 U/g (dried substrate) of PulA-N3 or PulA. Samples were taken at certain time intervals and then boiled for 10 min to inactivate enzymes for measuring glucose content and the DX values (PulA and PulA-N3) were calculated to evaluate the degree of the saccharification by HPLC. The other sugars, including maltose (indicated as Maltose-PulA and Maltose-PulA-N3 in the figure), maltotriose (Maltotriose-PulA and Maltotriose-PulA-N3) and isomaltose (Isomaltose-PulA and Isomaltose-PulA-N3), were detected synchronously. The data were represented as average of triplicate experiments.
To obtain higher conversion rate of glucose on preparation of glucose syrup from starch, glucoamylase blended with pullulanase is required. The main function of glucoamylase is to hydrolyze α-1,4 glycosidic bonds to generate glucose, while pullulanase hydrolyzes α-1,6 glycosidic bonds to generate branched-chain polysaccharides, and further provides easy hydrolysis substrates for glucoamylase35. Since glucoamylase itself has a weak but obvious transglycosyl activity while the saccharification nearly completed in practice use, saccharification in more acidic condition is considered to reduce the amount of isomaltose formed by glucoamylase. Therefore, pullulanase mutants having the similar enzymatic properties to that of glucoamylase under more acidic condition is very likely to improve the yield from starch to glucose.
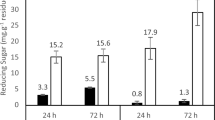
By simulating the industrial-scale process, the starch saccharification was carried out at 61 °C and pH 4.3 for 72 h using glucoamylase and PulA-N3 to hydrolyze 30% (w/v) maltodextrin. The results are summarized in Fig. 4C. In the early stages, the saccharification rate was very high. The DX values reached over 70% for 5 h incubation by glucoamylase blended with either PulA-N3 or PulA. The DX values reached 91.37% (with PulA-N3) and 89.82% (with PulA) for 12 h. After incubation for 24 h, the maximum glucose yields reached. A maximal DX value of 96.08% was achieved while PulA-N3 was used, which was increased by 0.82% comparison to that of PulA (95.26%). The content of glucose in glucose syrup is a key or even sole index for evaluating starch saccharification, which not only determines the quality and yield of high glucose syrup, but also contributes the main cost of down-stream purification. Therefore, in the starch sugar industry, whether the DX value of glucose syrup is higher than 96% or not is conventionally used as the main index for defining the qualities of both high glucose syrup and glucoamylase blends. Obviously, the use of more acidic pullulanase mutant will be more beneficial to the starch saccharification and glucose release by glucoamylase and of application potentials for formulation of high-efficiency glucoamylase blends.
Notably, the DX value of the glucose syrup showed a slow decline as the saccharification nears completion (Fig. 4C). Thus, the composition of the sugar spectrum formed during the preparation of glucose from starch was further analyzed (Fig. 4C). When the maximum DX value reached (for 24 h under this experiment conditions), there were still trace maltose and very trace maltotriose in the saccharification solution. The maltose and maltotriose were further hydrolyzed completely with the extension of saccharification time, meanwhile isomaltose was formed slowly and gradually increased over time (Fig. 4C). Since the commercially available pullulanase has no activity on isomalto-oligosaccharides36, optimization of saccharification endpoint and using optimized formula of glucoamylase blend may further improve the maximal DX value. Besides, other enzymes that can hydrolyze isomalto-oligosaccharides37 may involve in glucoamylase blend preparation.
In this report, we developed a new pullulanase mutant PulA-N3 from the wild pullulanase of B. naganoensis by rational site-directed mutagenesis of the predicted asparagine residues in PulA molecule. It displayed the improved activity and stability at higher temperature and under wider acidic pH ranges and the improved operating performance that well-matched with that of glucoamylase for starch saccharification (Tables 2, 3 and Figs. 3, 4). It is emphasized that the amide group of asparagine is important in forming the side chain-backbone hydrogen bonds and fulfills an important role in stabilizing local folded structures in proteins30. It is also well-illustrated that the deamidation of asparagine residues in proteins is dominant pathway for protein denaturation and degradation (enzyme inactivation) and is a pH dependent process31. Therefore, replacement of asparagine residues at specific sites of PulA with another specific amino acid may alter the side chain-backbone hydrogen forming and attenuate the deamidation. A combined mutation of N467G, N492A and N709R in PulA molecule contributed the improved activity and stability at higher temperature and low pH (Fig. 3), which indicated that asparagine residues at specific sites in pullulanases (and may also in other enzymes) were the suitable targets for artificially improving the enzyme’s activity, thermostability and acidic stability. Notably, with assistance of in silico experiments, more concise site-directed mutagenesis and less wet experiments for a desired mutant development can be obtained as the results presented in this report and many others10,11.
Interestingly, PulA-N3 displayed a broadened operating pH and temperature ranges, which is benefit for the process improvement of not only high glucose syrup preparation described in this report but also preparation of high maltose syrup, (iso-)malto-oligosaccharides and amylopectin as well as fruits and vegetables processing. As we know, most of natural enzymes, including pululanase, perform their maximal activity in a very narrow ranges of pH and temperature. However, the potential mechanism for these structure–function relationship should be more complex than we concerned above, which will be revealed in further studies.
Despite asparagine residues, other five amino acid residues in PulA molecule were also predicted, including the second dominant lysine residues (9/28) (K327, K453, K460, K463, K469, K476, K626, K665 and K669). It is known that lysine is the residue with the largest hydrophobic accessible surface in folded structures of proteins38. Site-directed mutagenesis of specific lysine residues on the enzyme surface improved the rigid β-sheet structure and thermostability of a Bacillus 1,3–1,4-β-glucanase39. Therefore, the predicted lysine residues in PulA molecule may also be the important mutation candidates to further improve its operating properties and catalytic activity under broadened pH and temperature ranges, which will be investigated in further research.
Conclusions
The mutant PulA-N3 of B. naganoensis pullulanase was successfully obtained through mutation site prediction, homologous modeling and structure matching, molecular docking and site-directed mutagenesis. It displayed the improved activity and stability at higher temperature and under wider acidic pH ranges. Notably, it was more suitable for starch saccharification to produce high glucose syrup from starch in combination of glucoamylase.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary information files.
References
Norman, B. E. A novel debranching enzyme for application in the glucose syrup industry. Starch/Stärke 34, 340–346. https://doi.org/10.1002/star.19820341005 (1982).
Bertoldo, C. & Antranikian, G. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 6, 151–160. https://doi.org/10.1016/S1367-5931(02)00311-3 (2002).
Turkenburg, J. P. et al. Structure of a pullulanase from Bacillus acidopullulyticus. Proteins 76, 516–519. https://doi.org/10.1002/prot.22416 (2009).
Wang, X., Nie, Y. & Xu, Y. Industrially produced pullulanases with thermostability: Discovery, engineering, and heterologous expression. Bioresour. Technol. 278, 360–371. https://doi.org/10.1016/j.biortech.2019.01.098 (2019).
Nisha, M. & Satyanarayana, T. Characteristics, protein engineering and applications of microbial thermostable pullulanases and pullulan hydrolases. Appl. Microbiol. Biotechnol. 100, 5661–5679. https://doi.org/10.1007/s00253-016-7572-y (2016).
Akassou, M. & Groleau, D. Advances and challenges in the production of extracellular thermoduric pullulanases by wild-type and recombinant microorganisms: A review. Crit. Rev. Biotechnol. 39, 337–350. https://doi.org/10.1080/07388551.2019.1566202 (2019).
Wang, Y., Liu, Y., Wang, Z.-X. & Lu, F. Influence of promoter and signal peptide on the expression of pullulanase in Bacillus subtilis. Biotechnol. Lett. 36, 1783–1789. https://doi.org/10.1007/s10529-014-1538-x (2014).
Nie, Y. et al. High-level expression of Bacillus naganoensis pullulanase from recombinant Escherichia coli with auto-induction: effect of lac operator. PLoS ONE 8, e78416. https://doi.org/10.1371/journal.pone.0078416 (2013).
Chang, M. et al. Improving the thermostability of acidic pullulanase from Bacillus naganoensis by rational design. PLoS ONE 11, e0165006. https://doi.org/10.1371/journal.pone.0165006 (2016).
Wang, X., Nie, Y. & Xu, Y. Improvement of the activity and stability of starch-debranching pullulanase from Bacillus naganoensis via tailoring of the active sites lining the catalytic pocket. J. Agric. Food Chem. 66, 13236–13242. https://doi.org/10.1021/acs.jafc.8b06002 (2018).
Wang, X. et al. Evolutionary coupling saturation mutagenesis: Coevolution-guided identification of distant sites influencing Bacillus naganoensis pullulanase activity. FEBS Lett. 594, 799–812. https://doi.org/10.1002/1873-3468.13652 (2020).
Niu, D., Zuo, Z., Shi, G. Y. & Wang, Z.-X. High yield recombinant thermostable α-amylase production using an improved Bacillus licheniformis system. Microb. Cell Fact. 8, 1–7. https://doi.org/10.1186/1475-2859-8-58 (2009).
Niu, D. & Wang, Z.-X. Development of a pair of bifunctional expression vectors for Escherichia coli and Bacillus licheniformis. J. Ind. Microbiol. Biotechnol. 34, 357–362. https://doi.org/10.1007/s10295-007-0204-x (2007).
Capriotti, E., Fariselli, P. & Casadio, R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 33, W306–W310. https://doi.org/10.1093/nar/gki375 (2005).
Xiang, Z. Advances in homology protein structure modeling. Curr. Protein Pept. Sci. 7, 217–227. https://doi.org/10.2174/138920306777452312 (2006).
Kryshtafovych, A. & Fidelis, K. Protein structure prediction and model quality assessment. Drug Discov. Today 14, 386–393. https://doi.org/10.1016/j.drudis.2008.11.010 (2009).
Zhang, L. et al. Homology modeling, molecular dynamic simulation and docking studies of cyclin dependent kinase 1. J. Mol. Model. 17, 219–226. https://doi.org/10.1007/s00894-010-0710-z (2011).
Yuan, S., Chan, H. S., Filipek, S. & Vogel, H. PyMOL and Inkscape bridge the data and the data visualization. Structure 24, 2041–2042. https://doi.org/10.1016/j.str.2016.11.012 (2016).
Leathers, T. D. Biotechnological production and applications of pullulan. Appl. Microbiol. Biotechnol. 62, 468–473. https://doi.org/10.1007/s00253-003-1386-4 (2003).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. https://doi.org/10.1002/jcc.21256 (2009).
Ferreira, L. G., Dos Santos, R. N., Oliva, G. & Andricopulo, A. D. Molecular docking and structure-based drug design strategies. Molecules 20, 13384–13421. https://doi.org/10.3390/molecules200713384 (2015).
Heckman, K. L. & Pease, L. R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932. https://doi.org/10.1038/nprot.2007.132 (2007).
Maniatis, T., Sambrook, J. & Fritsch, E. F. Molecular Cloning: A Laboratory Manual. (New York, 1989).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467. https://doi.org/10.1073/pnas.74.12.5463 (1977).
Zhuge, J. & Wang, Z.-X. A Lab Manual For Industrial Microbiology. (Beijing, 1994).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. https://doi.org/10.1016/0003-2697(76)90527-3 (1976).
Malle, D. et al. Overexpression, purification and preliminary X-ray analysis of pullulanase from Bacillus subtilis strain 168. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 381–384. https://doi.org/10.1107/s1744309106007901 (2006).
Li, S. et al. Gene cloning, heterologous expression, and characterization of a high maltose-producing α-amylase of Rhizopus oryzae. Appl. Biochem. Biotechnol. 164, 581–592. https://doi.org/10.1007/s12010-011-9159-5 (2011).
Linko, Y. Y. & Wu, X. Y. Improvement and estimation of enzymic starch saccharification process. Biotechnol. Tech. 7, 551–556. https://doi.org/10.1007/BF00156327 (1993).
Vasudev, P. G., Banerjee, M., Ramakrishnan, C. & Balaram, P. Asparagine and glutamine differ in their propensities to form specific side chain-backbone hydrogen bonded motifs in proteins. Proteins 80, 991–1002. https://doi.org/10.1002/prot.24001 (2012).
Kempkes, L. J., Martens, J. K., Grzetic, J., Berden, G. & Oomens, J. Deamidation reactions of protonated asparagine and glutamine investigated by ion spectroscopy. Rapid Commun. Mass Spectrom. 30, 483–490. https://doi.org/10.1002/rcm.7464 (2016).
Krieger, F., Möglich, A. & Kiefhaber, T. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 127, 3346–3352. https://doi.org/10.1021/ja042798i (2005).
Hacke, M., Gruber, T., Schulenburg, C., Balbach, J. & Arnold, U. Consequences of proline-to-alanine substitutions for the stability and refolding of onconase. FEBS J. 280, 4454–4462. https://doi.org/10.1111/febs.12406 (2013).
Sokalingam, S., Raghunathan, G., Soundrarajan, N. & Lee, S. G. A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS ONE 7, e40410. https://doi.org/10.1371/journal.pone.0040410 (2012).
Crabb, W. D. & Shetty, J. K. Commodity scale production of sugars from starches. Curr. Opin. Microbiol. 2, 252–256. https://doi.org/10.1016/S1369-5274(99)80044-7 (1999).
Niu, D. et al. Highly efficient enzymatic preparation of isomalto-oligosaccharides from starch using an enzyme cocktail. Electron. J. Biotechnol. 26, 46–51. https://doi.org/10.1016/j.ejbt.2016.12.002 (2017).
Dong, Z. et al. Molecular cloning and biochemical characterization of two novel oligo-1,6-glucosidases from Bacillus mycoides and Thermomyces lanuginosus. Starch/Stärke 70, 1700093. https://doi.org/10.1002/star.201700093 (2018).
Lins, L., Thomas, A. & Brasseur, R. Analysis of accessible surface of residues in proteins. Protein Sci. 12, 1406–1417. https://doi.org/10.1110/ps.0304803 (2003).
Niu, C., Zhu, L., Zhu, P. & Li, Q. Lysine-based site-directed mutagenesis increased rigid β-sheet structure and thermostability of mesophilic 1,3–1,4-β-glucanase. J. Agric. Food Chem. 63, 5249–5256. https://doi.org/10.1021/acs.jafc.5b00480 (2015).
Funding
This work was supported by the Tianjin Outstanding Talent Program (Grant No.: JC20200309) to Wang, Z.-X., the Intergovernmental International Scientific and Technological Innovation Cooperation Program, MOST, China (Grant No.:2021YFE0106200) to Niu, D., and the Intergovernmental International Scientific and Technological Innovation Cooperation Program, MOST, China (Grant No.: 2018YFE0100400) to Wang, Z.-X.
Author information
Authors and Affiliations
Contributions
D.N. designed and directed the experimental research and revised the manuscript, H.C. conducted the experiments and drawn the first version of the manuscript. Y.Z. conducted the experiments. N.M. conducted part of experiments and prepared the manuscript. W.Z.X. supervised the research and prepared the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Niu, D., Cong, H., Zhang, Y. et al. Pullulanase with high temperature and low pH optima improved starch saccharification efficiency. Sci Rep 12, 21942 (2022). https://doi.org/10.1038/s41598-022-26410-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26410-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.