Abstract
Tularemia is a zoonosis caused by the bacterium Francisella tularensis. Leporids are primary sources of human infections in the northern hemisphere. Africa is classically considered free of tularemia, but recent data indicate that this dogma might be wrong. We assessed the presence of this disease in wild leporids in Algeria. Between 2014 and 2018, we collected 74 leporids carcasses from spontaneously dead or hunted animals. Francisella tularensis DNA was detected by specific real-time PCR tests in 7/36 (19.44%) Cape hares (Lepus capensis) and 5/38 (13.15%) wild rabbits (Oryctolagus cuniculus). Known tularemia arthropod vectors infested half of the PCR-positive animals. At necropsy, F. tularensis-infected animals presented with an enlarged spleen (n = 12), enlarged adrenal glands (12), liver discoloration (12), hemorrhages (11), and pneumonia (11). Immunohistological examination of liver tissue from one animal was compatible with the presence of F. tularensis. Our study demonstrates the existence of tularemia in lagomorphs in Algeria. It should encourage investigations to detect this disease among the human population of this country.
Similar content being viewed by others
Introduction
Since antiquity, wildlife has been an essential source of infectious diseases transmissible to humans1,2. These infections, referred to as zoonoses, constitute a significant public health problem on all continents2. The importance of such diseases in humans and animals is increasingly recognized, and more attention in this area is needed2. Most emerging infectious diseases in humans are zoonoses3. The COVID-19 pandemic is the most recent example4,5. Wildlife represents a significant and often unknown reservoir for emerging pathogens and a source for the re-emergence of previously controlled zoonoses (e.g., brucellosis)6,7. In addition, such infectious agents also have harmful effects on domestic animals and wildlife, with substantial economic consequences and a threat to species preservation7,8. Zoonotic pathogens that infect both domestic and wildlife animals are more likely to emerge as human pathogens7.
An excellent example of an emerging zoonotic agent with many different modes of transmission to humans is Francisella tularensis, the causative agent of tularemia2,9. This Gram-negative, facultative intracellular bacterium is highly infectious and considered a biological threat agent10. It is mainly distributed in Northern hemisphere11. Two distinct subspecies that cause tularemia are recognized. Francisella tularensis subsp. tularensis (type A strains), the most virulent subspecies, is almost exclusively found in North America12. A few type A strains have been isolated from environmental sources and arthropods in Slovakia and Austria13. Francisella tularensis subsp. holarctica (type B strains) is endemic throughout the Northern hemisphere14. Recently, this subspecies has been isolated from tularemia patients and ringtail possums in Australia, especially in Tasmania and Sydney15,16.
Many wild animal species can be infected or are carriers of F. tularensis, but their epidemiological role remains unclear11. Francisella tularensis has been isolated from more than 250 species, including lagomorphs, rodents, insectivores, carnivores, ungulates, marsupials, birds, amphibians, fishes, and invertebrates11,17. However, lagomorphs and small rodents are considered as the primary sources of human infections18. Moreover, lagomorphs (especially hares) often die from F. tularensis infection and thus are deemed suitable sentinels for tularemia surveillance17. In the Northern hemisphere, human tularemia cases frequently occur through contact with infected animals19,20. However, other common modes of infection include arthropod bites (mainly ticks), inhalation of contaminated aerosols, ingestion of contaminated water or food, and contact with contaminated water or soil21.
Africa has been considered for decades as free of tularemia21,22. A suspected F. tularensis bacteremia was reported from Sudan, but the isolated bacterial strain was not fully characterized23. Besides, positive F. tularensis serological tests have been detected in febrile patients from Kenya24. However, there was no confirmation of F. tularensis infection by the isolation of bacteria or molecular tests. Finally, Francisella sp. DNA was detected in ticks collected on camels in Egypt, but F. tularensis was not identified among Francisella sp. positive ticks20. Altogether, we can conclude that, while strongly suspected, there is currently no definite confirmation of the presence of F. tularensis in Africa.
In Northern Algeria, from December 2012 to March 2013, much higher mortality rates than usual were observed in hares and wild rabbits, suggesting an epizootic. Although Algeria is currently not considered endemic for tularemia, this study was conducted to investigate the presence of this disease in wild leporids. Our investigations were performed in Northern Algeria for 4 years following the epizootic, in areas where an excess of mortality in lagomorphs was observed.
Results
Study area
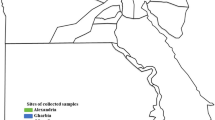
This study was conducted in the central part of Northern Algeria, which is part of the Tell Atlas mountain range and located on the shores of the Mediterranean Sea. It covers five wilayas (provinces): Tipaza (36° 35′ 31″ N, 2° 26′ 58″ E), Ain El Defla (36° 15′ 55″ N, 1° 58′ 13″ E), Medea (36° 16′ 03″ N, 2° 45′ 00″ E), Blida (36° 29′ 00″ N, 2° 50′ 00″ E), and Algiers (36° 46′ 34″ N, 3° 03′ 36″ E) (Fig. 1). These provinces include mountains, hills, and plains. The climate is of Mediterranean type with an average rainfall of 600–800 mm per year and an average annual temperature of 18 °C near the coast and 25 °C in the inner wilayas. A diverse fauna, including sedentary terrestrial birds (e.g., rapacious, sparrow, partridge), migratory birds, reptiles, and mammals (e.g., wild boars, carnivores, leporids, and rodents), characterizes this region.
Map of five localities of wild leporids collected in the study area. (The map was generated in paint standard software from a virgin map figure. The map of Algeria is freely accessible at this internet link https://d-maps.com/m/africa/algeria/algerienord_fr/algerienord_fr19.gif.
Animal specimens and real-time PCR (qPCR) testing
During the period 2014–2016 and in 2018, we collected 74 wild leporids (36 Cape hares and 38 wild rabbits), which were shot by hunters (n = 60) or found dead (n = 14). The characteristics of the animals (age, sex, location and period of collection) are summarized in Table 1. In total, 74 spleen and 20 liver specimens were tested for the presence of Francisella DNA using three qPCRs assays.
The ISFtu2-qPCR targets the multi-copy ISFtu2 insertion sequence. It is highly sensitive but lacks specificity because this ISFtu2 is found in several Francisella species. The Tul4-qPCR targets the gene encoding the membrane protein Tul4, which is more specific to F. tularensis but also detects F. novicida25. The Type B-qPCR targets a particular DNA fragment specific to F. tularensis subsp. holarctica, the only subspecies causing tularemia in Eurasia26.
Overall, 60 samples were negative for the three qPCR tests, 22 were positive only for ISFtu2-qPCR (level 1), ten were positive for both ISFtu2-qPCR and Tul4-qPCR (level 2), and two were positive for all three qPCR tests (level 3). Among the 74 spleen samples, 48 were negative, 15 graded level 1, nine level 2, and two level 3. For the 20 liver samples, 12 were negative, seven level 1, one level 2, and none level 3 (Table 2).
Tularemia infected animals
Since no F. tularensis strain could be isolated from animal samples, the animals were classified (see “Materials and methods” section) as “probable” tularemia case (at least one level 3 sample), “possible” case (at least one level 2 but no level 3 samples), and “uncertain” case (at least one level 1 but no level 2 or 3 samples).
Overall, two probable and ten possible tularemia case animals were considered infected with F. tularensis, including 7/36 (19.44%) Cape hares and 5/38 (13.15%) wild rabbits (Table 1). The uncertain cases corresponded to 8/36 (22.22%) Cape hares and 11/38 (28.94%) wild rabbits. The geographic distribution of probable and possible cases included the provinces Tipaza (5 hares and 5 wild rabbits), Medea (1 hare), and Ain El Defla (1 hare)). These animals included 7 males and 5 females, and 11 mature animals and 1 juvenile. They were collected in March (n = 1), August (2), September (1), October (2), November (4), and December (2). The uncertain cases were detected in Tipaza (7 hares and 9 wild rabbits), Blida (2 wild rabbits), and Algiers (1 hare). These animals included 12 males, 7 females, 16 mature animals, and 3 juveniles. They were collected in January (1), March (4), April (1), July (1), August (1), October (3), November (2), and December (6).
Pathological findings
Of the 12 probable and possible tularemia cases, only one hare was found dead, while 11 animals were hunted (the two probable cases were hunted animals). The body condition of these 12 animals was good. Two were found dead for the 19 uncertain cases (one from a traffic accident), while 17 were hunted. The body condition of the 19 uncertain cases was good in 18 cases and weak for one hunted rabbit. Of the 43 negative leporids, most (i.e., 20 hares and 20 wild rabbits) were in good body condition, and only one dead hare and two hunted wild rabbits were in weak condition. The main pathological findings were retrieved from the hares and wild rabbits diagnosed with probable or possible tularemia. Details of organ lesions detected at necropsy in all collected animals are presented in Tables S2 and S3. Figures 2 and 3 show the main lesions observed in tularemic animals. In addition, Table S4 indicates the degree of organ involvement (severe or moderate).
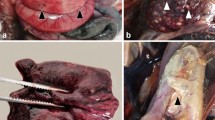
Organ lesions in Cape hare (cadaver). (A): Splenomegaly. The spleen is markedly enlarged (6 cm in length) and has prominent rounded edges with some discoloration areas. (B): Heterogeneous discoloration in the liver lobes with hemorrhage areas and an enlarged and full gallbladder. (C): Congestion of the kidney with capsule congestion. (D): The enlargement of adrenal glands.
Organ lesions in wild rabbit (Hunted animal). (A): Splenomegaly. The spleen is markedly enlarged (5 cm in length) and has prominent rounded edges. (B): Heterogeneous discoloration in the liver lobes with hemorrhage areas and a few small white necrotic foci (arrow). (C): Congestion of the kidney with a white focus of necrosis foci in the capsule (arrow). (D): Pneumonia with multifocal hemorrhages and dark hyperemic areas are scattered throughout the lungs, also few small white necrotic foci (arrow). (E): The enlargement of adrenal glands. (F): Numerous necrotic white foci in the ovary.
Immunohistochemistry (IHC) examination
IHC was conducted on four specimens graded level 3 (two spleens and two livers) from two animals. It was also performed on four specimens graded level 2 (two spleens and two livers) from two other animals. IHC was negative for all the spleen samples. In contrast, for one level 2 liver sample, bacteria were revealed by the anti-F. tularensis antibody within the hepatocytes' cytosol (Fig. 4).
Immunohistochemistry results. (A) and (C) positive control corresponding to Francisella tularensis in amoebae labeled with FB11 monoclonal antibody 1/1000 in Leica dilution buffer. (B) and (D) negative control corresponding to Francisella philomiragia in amoebae labeled with FB11 monoclonal antibody. (E) and (G) show Francisella tularensis labeled in the cytosol of liver cells. (F) and (H) show the background generated by the Leica revelation system without antibodies. A, B, E and F: objective × 20. C, D, G, and H: objective × 40. Added insert: objective × 100.
Ectoparasites
Potential arthropod vectors of tularemia were found for two probable tularemia cases. One hare was infested by nymphs of Rhipicephalus spp. and Hyalomma spp. One wild rabbit was infested by Ixodes ricinus and Spilopsyllus cuniculi. Concerning the ten possible cases, five animals were infested by known tularemia vectors (4 cases by I. ricinus and one case each by Rh. sanguineus, S. cuniculi, or lice). In addition, three animals were infested by Rhipicephalus spp, Hyalomma spp, and fleas (Ctenocephalides canis and Ctenocephalides felis). Finally, 13 of the uncertain cases were infested by tularemia vectors (I. ricinus, nine cases; S. cuniculi, five cases; Rh. sanguineus, one case; and Haemodipsus lepori and Haemodipsus setoni, one case). Other ectoparasites were detected, including Rhipicephalus pusilus, Rhipicephalus spp, Hyalomma spp, C. felis, C. canis, and mites. The qPCR-negative animals were also infested by some tularemia vectors (I. ricinus, 11 cases; S. cuniculi, 5 cases; and Rh. sanguineus, 3 cases) (Table S5).
Discussion
Tularemia affects animal welfare, human health, and the environment and is thus better approached from a one-health perspective27. Several studies in the Northern hemisphere28, and more recently in Australia15,16, have provided a vital research track in the epidemiology of this disease. In contrast, studies in Africa are too limited and scarce. The aim of this study was to investigate the presence of tularemia in wild leporids collected in Northern Algeria. These animals are highly susceptible to F. tularensis infection and considered sentinel hosts for surveillance of tularemia. The strategy we used to detect F. tularensis in leporids mainly used molecular, histological and immunohistochemical analyzes of tissues taken from animals found dead or hunted. To the best of our knowledge, detection of F. tularensis by PCR or culture has not been previously reported in wild leporidae in Algeria or other African countries.
Animal tissue samples were tested using three qPCR assays of variable sensitivity and specificity. The Type B-qPCR test targets a specific junction between ISFtu2 and a flanking 3′ region, which is considered specific for F. tularensis subsp. holarctica26, the only tularemia agent found in Europe and Asia. The Tul4-qPCR assay targets a simple copy gene encoding a surface protein, which can be found in the genome of all F. tularensis subspecies causing tularemia and that of the aquatic bacterium F. novicida. Because F. novicida has never been isolated from lagomorphs or other animal species, and very rarely from human29, a positive Tul4 qPCR for the studied tissue samples likely indicated the presence of F. tularensis DNA. The ISFtu2 qPCR is considered highly sensitive because multiple copies of this insertion sequence are found in the F. tularensis genome. However, it lacks specificity because ISFtu2 is also found in many other Francisella species25.
Two animals were considered “probable” tularemia cases because some of their samples were positive for the three qPCR tests. Ten animals were considered “possible” tularemia cases because their samples were positive for the ISFtu2 and Tul4 qPCRs but not the Type B qPCR. Finally 19 leporids were “uncertain” cases because only samples positive for the ISFtu2 qPCR were found. For the remaining 43 animals, all the tested samples were negative for the three qPCRs. Overall, we detected F. tularensis DNA-positive samples in 12/74 (16.21%) leporids, which strongly suggest that tularemia is present in the lagomorph population of the study area. The positive Type B qPCR tests in two animals suggested that F. tularensis subsp. holarctica could be the involved subspecies. We did not confirm these data by isolating F. tularensis from the studied leporids. However, the isolation of this pathogen from human or animal samples is tedious and has a low sensitivity13. Moreover, most of our samples were not appropriate for F. tularensis culture because of their long-term preservation in ethanol 70° or 10% formalin. Further study using fresh (non-fixed) tissue samples from dead leporids collected in the same study area is needed to definitively confirm the presence of tularemia in these animals and characterize the F. tularensis subspecies and genotypes involved.
Although PCR is usually more sensitive than culture for detecting F. tularensis, it also has some limitations. Firstly, the DNA extraction from organs preserved in ethanol for several months was difficult although easier for spleen than for liver samples. Some tissue samples could be lysed only after overnight incubation with proteinase K. Secondly, tissue samples contained PCR inhibitors as demonstrated by better DNA amplification from some samples after their dilution in PCR grade water. To reduce the effect of PCR inhibitors, organ samples with negative qPCR were retested using Bovine Serum Albumin (BSA) and the Real-time PCR system TaqMan (Applied Biosystems, Munich, Germany)30. Finally, DNA regions to be amplified were optimized to obtain high sensitivity and specificity of qPCR tests.
IHC detection of F. tularensis in formalin-fixed tissue can be helpful for tularemia diagnosis31,32. For one possible tularemia case, F. tularensis could be detected on immunohistochemical (IHC) examination of a liver sample using a specific anti-F. tularensis antibody. The intensity and localization of positive staining were comparable to those previously recorded for other animals32,33. IHC did not provide interpretable findings for four other tested specimens. Such negative results might be explained by an inhomogeneous distribution of infectious foci in the involved organs as well as a low bacterial inoculum in infected tissues. This has been previously demonstrated in tularemia granulomatous lesions in cell types like epithelial cells of the kidney, testis, and epididymis, hepatocytes, and bronchiolar epithelial cells31. Besides, IHC is a delicate technology whose results are highly dependent on the quality and fixation time of the organ tissues34. IHC analysis of dead animal tissues remains challenging, especially in case of tissue necrosis34.
In our limited case series we found a F. tularensis infection prevalence in leporids of 2.7% (2/74) for probable tularemia cases and 16.2% (12/74) when considering both probable and posible cases. We cannot make a guess about the prevalence of tularemia because our series is not representative of the general lagomorph population in the study area. In Germany, F. tularensis DNA was detected in 1.1% of European Brown hares and 2.4% of wild rabbits collected between 2009 and 201435. Higher infection rates were reported in the same country, including 11.8% (100/848 animals) in hares collcted in the North Rhine-Westphalia region36 and 30% (55/179) in brown hares collected between 2010 and 2016 in Baden-Wuerttemberg37. In Hungary, the prevalence of tularemia in hares was evaluated at 4.9–5.3%38. In Portugal, prevalences of 4.3% and 6.3% were reported in brown hares and wild rabbits, respectively39. However, the comparison of the reported tularemia prevalences in leporids is irrelevant because studies involved different animal species and geographic areas, and used different methods for F. tularensis detection.
Two possibilities could explain the lack of detection of tularemia in Algeria before this study. The first hypothesis is that this disease was not searched for in previous years, while it could have been present in this country for decades. The second hypothesis is that tularemia was recently imported in Algeria. Migratory birds may have been involved in the long-distance spread of F. tularensis40. These hosts can be infested by ectoparasites such as ticks which are the primary vectors of tularemia41,42. They can also spread the bacteria in the hydro-telluric environment through their secretions and feces18,43,44. An alternative possibility is that F. tularensis-infected animals (especially game animals) have been imported in Algeria from endemic countries. Whatever the mode of introduction of tularemia in Algeria, the dissemination of this disease over time might have been facilitated by the ability of F. tularensis to infect multiple hosts and its better survival in a cool environment45, which characterizes Northern Algeria climate. The emergence or re-emergence of tularemia in other countries has been related to climate change, human-mediated movement of infected animals, and wartime resulting in a significant rise of F. tularensis infections in the rodent populations39,46.
In our study, infected animals were collected throughout 4 years, although more frequently in autumn. Probable and possible tularemia cases were mainly collected during the hunting season (i.e., September, October, November, and December). Animals could not be collected in February because of heavy rains and in May and June because it corresponds to female leporids' lactation period. In most endemic countries, tularemia cases are typically more frequent in late spring, the summer months, and early autumn37,47,48,49,50. Occasionally, fatal tularemia cases in hares have been predominantly reported during the cold season11,51. The climatic conditions can affect tularemia outbreaks in animals, depending on the reservoir involved and the predominant modes of infection52.
We detected tularemia more frequently in female than in male hares, and the reverse was true for wild rabbits. The prevalence of tularemia in male or female lagomorphs varies between studies. In Sweden, Morener et al.50 reported a tularemia case series only involving male hares. In the same country, Borg et al.50 observed an overrepresentation of females in the epizootic of 1967. They suggested that, compared to males, females had a higher risk of exposure to infected mosquitoes or were more vulnerable to tularemia because they were pregnant or had just given birth to a litter50. Tularemia was found in a few juveline leporids, which might be explained by a shorter exposure time to F. tularensis, a higher death rates due to higher susceptibility to F. tularensis infection or easier predation by their natural enemies, or more frequent hunting of adults compared to the juveniles53.
Tularemia is usually more frequently detected in leporids found dead than in hunted animals. As an example, a German study reported a higher prevalence of tularemia in hares found dead (2.9%) than in hunted ones (0.7%)35. In our study, most qPCR-positive animals were hunted. Our study might not be representative of the prevalence of tularemia in either population because most collected animals had been hunted.
The incubation period and clinical presentation of tularemia in leporids vary according to the species considered. Tularemia is typically an acute disease in mountain hares (Lepus timidus) in Scandinavia and has a chronic pattern in European brown hares (Lepus europaeus) in Central Europe50. The incubation time and clinical presentation of tularemia can be different in Cape hares (Lepus capensis). Wild rabbits are less sensitive to F. tularensis infection than hares31,39,54. An extended incubation period and chronic evolution of tularemia would facilitate the detection of F. tularensis in infected animals. In our study, a similar tularemia prevalence was found in the Cape hares and wild rabbits, which might reflect exposure to a same biotope area and environmental reservoirs of F. tularensis.
The pathological lesions of tularemiia in leporids can vary according to the F. tularensis strain involved, the mode and route of infection, and the susceptibility and immune status of the host32,50. In the European brown hares, granulomas with central necrosis have been reported in the lungs and kidneys and occasionally in the liver, spleen, bone marrow, and lymph nodes50. In contrast, only acute necrosis in the liver, spleen, bone marrow, and lymph nodes have been found in Lepus timudus hares in Sweden50. The lesions in the Japanese hare (Lepus brachyurus angustidens) are comparable to those of Lepus timidus, except for cutaneous, lung, brain, and adrenal gland lesions32. In the European rabbit, Oryctolagus cuniculus, tularemia is not associated with identifiable macroscopic tissue lesions39,55. To our knowledge, no reports describing post-mortem lesions in Cape hares with tularemia are available. In this study, similar lesions were found in hares and wild rabbits except necrotic foci only observed in some wild rabbit organs (such as liver, lungs, kidney, ovary). Most animals had pathological lesions of pneumonia, gastritis and enteritis. Kidney lesions and adrenal glands enlargment were oberved. Necrotic lesions were occasionally found in the lungs, liver, spleen and ovary and hemorrhages in the lungs, liver, and intestines.
Tularemia is an arthropod-born disease in most endemic areas14,22,28. In our study, 50% of positive leporids were infested by known tularemia vectors such as ticks (Ixodes ricinus56,57, Rhipicephalus sanguineus39), fleas (Spillopsylus cuniculi58), and lice of lagomorphs (Haemodipsus lepori and Haemodipsus setoni59,60). Ticks are the most significant arthropod vectors of tularemia61. Ticks are frequently involved in the transmission of tularemia in North America, including Dermacentor andersoni, D. variabilis, and Amblyomma americanum57,62,63. In Europe, tick-borne tularemia represents 13% to 26% of human cases57,64. The involved species include D. marginatus, D. reticulatus, I. ricinus, R. sanguineus, and Haemaphysalis concinna65,66. Further research on wild leporid sucking arthropods is needed to confirm the presence and clarify the ecology of F. tularensis in Algeria.
Our study reports for the first time the detection of F. tularensis DNA in leporids from Northern Algeria. The markers most in favor of tularemia in the animals studied are the positivity of qPCR tests, in particular, the "type B" qPCR test which amplifies a specific DNA sequence of F. tularensis subsp. holarctica, and a positive immunohistological examination in one animal. Further investigation is needed to confirm our results by the isolation of this pathogen from animal samples and determine the F. tularensis subspecies and genotypes involved. This would allow the characterization of the F. tularensis subspecies and genotypes present in Algeria. Furthermore, our findings push us in future studies to seek tularemia in the Algerian human population. To achieve this, interdisciplinary or trans-disciplinary collaborative efforts underpinned by the One Health concept will be necessary.
Materials and methods
Animal samples
During the period 2014–2016 and in 2018, we collected 74 wild leporids (36 Cape hares and 38 wild rabbits) shot by hunters (n = 60) or found dead (n = 14) for post-mortem examination. A questionnaire was completed for each animal with information on the age (adult/juveniles), sex, region of collection, and the hunter or person who discovered the animal. Tissue samples (spleen, liver) were collected and stored in ethanol 70° for PCR testing or 10% buffered formalin for histology and immunohistochemistry analyses.
Necropsy examination of animals
The skin was scanned for the presence of ectoparasites or the lesions they cause. The following lesions were sought in the organs: enlargement, hemorrhagic lesions, congestion, edema, necrosis, degeneration, and inflammation. These lesions were graded as mild, moderate, or severe.
Detection of Franscisella tularensis DNA
DNA extraction from animal tissues
A tissue sample of 25 mg was lysed by incubation in proteinase K overnight at 56 °C. Then, the DNA was extracted using the NucleoSpin Blood kit (Macherey Nagel, Hoerdt, France) according to the manufacturer's instructions.
Real-time PCR (qPCR)
Francisella tularensis DNA was detected in animal samples using three real-time PCR (qPCR) tests. The ISFtu2-qPCR targets the ISFtu2 insertion sequence, which is present in multi-copy in the genomes of all Francisella species and Francisella-like endosymbionts of arthropods. The Tul4-qPCR targets the gene encoding Tul4, a membrane protein of 17 kDa found in all subspecies of F. tularensis and the aquatic species F. novicida25. The Type B-qPCR targets a specific junction between ISFtu2 and a flanking 3' region, only found in F tularensis subsp. holarctica, the only subspecies causing tularemia in Eurasia26. The TaqMan Fast Advanced PCR Master Mix kit (ThermoFisher Scientific Walthman, US) was used. The qPCR mixes also contained 100 ng of DNA extract, 10 µM of primers, and 2 µM of the appropriate probe (Table S1). When PCR tests were negative, they were repeated with 0.4 µL BSA (1 ng/µL) to block the potential PCR inhibitors. qPCR tests were run on the Lightcycler 480 (Roche Diagnostics, Meylan, France), and the PCR program was 50 °C for 2 min, 95 °C for 2 min, and 45 cycles at 95 °C for 3 s and 60 °C for 30 s. All qPCR tests were performed in duplicate or triplicate for inconsistent results. A qPCR test was considered positive only when two replicate tests were positive and using a cycle threshold (Ct) value < 38 for ISFtu2-qPCR and < 40 for Tul4-qPCR and Type B-qPCR. The specimens were classified into four categories according to qPCR tests results: level 3 for a sample positive for the three qPCR tests; level 2 when only ISFtu2- and Tul4-qPCR were positive; level one when only ISFtu2-qPCR was positive, and negative when all qPCR tests were negative.
The qPCR controls were performed with DNA extracts from the following strains: F. philomiragia ATCC 25,015, F. noatunensis LMG 23,800, F. tularensis subsp. novicida U112, and the clinical strain of F. tularensis subsp. holarctica. Our F. tularensis collection has been approved by the Agence Nationale de Securité du Médicament et des Produits de santé (France) (ANSM, authorization number ADE-103892019-7).
Immunohistochemistry analysis
Immunohistochemistry (IHC) analysis was performed for qPCR-positive spleen and liver samples from four animals. These samples were classified as level 3 (n = 4, two spleens, two livers) or level 2 (n = 4, two spleens, and two livers). IHC was performed on 3 µm paraffin sections using Bond Polymer Refine Detection (#DS9800, Leica Biosystems, Newcastle, UK).
Deparaffinized slides were incubated in a citrate solution (pH = 6) for 20 min, at 100 °C for epitopes retrieval. Staining was performed using an F. tularensis LPS-directed mouse monoclonal antibody (FB11, used at 1:1000, ThermoFisher Scientific, Rockford USA) applied for one hour at room temperature. Peroxidase revelation was performed using the Bond Polymer Refine Detection Kit (Leica Biosystems, Newcastle, UK), according to the manufacturer's recommendations. The pictures were taken with a Leica DFC295 camera, using the Leica Application Suite 4.13.
The positive control was F. tularensis-infected amoebae fixed with 4% formaldehyde assembled in a cytobloc using Shandon reagent and paraffin-embedded. The negative control was Francisella philomiragia-infected amoebae treated at the same time and similarly to the positive control.
Tularemia case definition
According to the specificities of the qPCR assays for F. tularensis detection, the animals were considered as a: 1/probable tularemia case when at least one sample was level 3; 2/possible tularemia case when at least one sample was level 2 (but no sample was level 3); 3/uncertain tularemia case when at least one sample was level 1 (but no sample as level 3 or level 2); and 4/not infected by F. tularensis when all three qPCR were negative for all tested samples. Pathological and immunohistochemistry findings could not be used in this definition because the former lacked specificity, and the latter was only performed on a few samples.
Ethical statement
All animal experiments were approved by the Algerian ethics committee of the General Directorate of Forests of the Ministry of Agriculture, Rural Development and Fisheries (GDF-MARDF). All methods were carried out in accordance with relevant guidelines and regulations, and in accordance with ARRIVE guidelines. The animal corpses were obtained during legal hunting activities in accordance with the laws of the Algerian Republic and the DGF-MARDF.
References
Namusisi, S. et al. A descriptive study of zoonotic disease risk at the human-wildlife interface in a biodiversity hot spot in South Western Uganda. PLOS Negl. Trop. Dis. https://doi.org/10.1371/journal.pntd.0008633 (2021).
Kruse, H., Kirkemo, A.-M. & Handeland, K. Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 10, 2067–2072 (2004).
Belay, E. D. et al. Zoonotic disease programs for enhancing global health security. Emerg. Infect. Dis. https://doi.org/10.3201/eid2313.170544 (2017).
Delahay, R. J. et al. Assessing the risks of SARS-CoV-2 in wildlife. One Health Outlook https://doi.org/10.1186/s42522-021-00039-6 (2021).
Kiros, M. et al. COVID-19 pandemic: Current knowledge about the role of pets and other animals in disease transmission. Virol. J. https://doi.org/10.1186/s12985-020-01416-9 (2020).
Greatorex, Z. F. et al. Wildlife trade and human health in Lao PDR: An assessment of the zoonotic disease risk in markets. PLoS ONE https://doi.org/10.1371/journal.pone.0150666 (2016).
Chomel, B. B., Belotto, A. & Meslin, F.-X. Wildlife, exotic pets, and emerging zoonoses1. Emerg. Infect. Dis. 13, 6–11 (2007).
Bacigalupo, S. A., Dixon, L. K., Gubbins, S., Kucharski, A. J. & Drewe, J. A. Towards a unified generic framework to define and observe contacts between livestock and wildlife: A systematic review. PeerJ https://doi.org/10.7717/peerj.10221 (2020).
Blancou, J., Chomel, B. B., Belotto, A. & Meslin, F. X. Emerging or re-emerging bacterial zoonoses: Factors of emergence, surveillance and control. Vet. Res. 36, 507–522 (2005).
Dennis, D. T. et al. Tularemia as a biological weapon: Medical and public health management. JAMA 285, 2763 (2001).
Decors, A. et al. Outbreak of tularaemia in brown hares (Lepus europaeus) in France, January to March 2011. Eurosurveillance https://doi.org/10.2807/ese.16.28.19913-en (2011).
Müller, W. et al. German Francisella tularensis isolates from European brown hares (Lepus europaeus) reveal genetic and phenotypic diversity. BMC Microbiol. 13, 61 (2013).
Chaudhuri, R. R. et al. Genome sequencing shows that European Isolates of Francisella tularensis subspecies tularensis are almost identical to US Laboratory Strain Schu S4. PLoS ONE https://doi.org/10.1371/journal.pone.0000352 (2007).
Maurin, M. & Gyuranecz, M. Tularaemia: Clinical aspects in Europe. Lancet. Infect. Dis 16, 113–124 (2016).
Jackson, J. et al. Francisella tularensis subspecies holarctica, Tasmania, Australia, 2011. Emerg. Infect. Dis. 18, 1484–1486 (2012).
Eden, J.-S. et al. Francisella tularensis ssp. holarctica in Ringtail Possums Australia. Emerg. Infect. Dis. 23, 1198–1201 (2017).
Carvalho, C. L., Lopes de Carvalho, I., Zé-Zé, L., Núncio, M. S. & Duarte, E. L. Tularaemia: A challenging zoonosis. Comp. Immunol. Microbiol. Infect. Dis. 37, 85–96 (2014).
Esmaeili, S. et al. Epidemiological survey of tularemia in Ilam Province, west of Iran. BMC Infect. Dis. https://doi.org/10.1186/s12879-019-4121-1 (2019).
Böhm, S. et al. Epidemiological investigation of a tularaemia outbreak after a hare hunt in Bavaria, Germany, 2018. Zoonoses Public Health 69, 106–116. https://doi.org/10.1111/zph.12899 (2021).
Ghoneim, N. H., Abdel-Moein, K. A. & Zaher, H. M. Molecular detection of Francisella spp. among ticks attached to camels in Egypt. Vector-Borne Zoonotic Dis. 17, 384–387 (2017).
Gurcan, S. Epidemiology of Tularemia. Balkan Med. J. 33, 3–10 (2014).
Yeni, D. K., Büyük, F., Ashraf, A. & Shah, MSu. D. Tularemia: A re-emerging tick-borne infectious disease. Folia Microbiol. 66, 1–14 (2021).
Mohamed, S. E. R., Mubarak, A. I. & Alfarooq, L. O. Francisella tularensis bacteremia: A case report from Sudan. Case Rep. Infect. Dis. 2012, 1–2 (2012).
Njeru, J. et al. Serological evidence of Francisella tularensis in febrile patients seeking treatment at remote hospitals, northeastern Kenya, 2014–2015. New Microb. New Infect. 19, 62–66 (2017).
Versage, J. L., Severin, D. D. M., Chu, M. C. & Petersen, J. M. Development of a multitarget real-time TaqMan PCR assay for enhanced detection of Francisella tularensis in complex specimens. J. Clin. Microbiol. 41, 5492–5499 (2003).
Kugeler, K., Pappert, R., Zhou, Y. & Petersen, J. Real-time PCR for Francisella tularensis types A and B. Emerg. Infect. Dis. 12, 1799–1801 (2006).
Tomaso, H. & Hotzel, H. Tularemia from a one health perspective. Curr. Clin. Microbiol. Rep. 4, 36–42 (2017).
Sjostedt, A. Tularemia: History, epidemiology, pathogen physiology, and clinical manifestations. Ann. NY. Acad. Sci. 1105, 1–29 (2007).
Kingry, L. C. & Petersen, J. M. Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. https://doi.org/10.3389/fcimb.2014.00035 (2014).
Farell, E. M. & Alexandre, G. Bovine serum albumin further enhances the effects of organic solvents on increased yield of polymerase chain reaction of GC-rich templates. BMC Res. Notes 5, 257 (2012).
Gyuranecz, M. et al. Tularemia of European Brown Hare ( Lepus europaeus ): A pathological, histopathological, and immunohistochemical study. Vet. Pathol. 47, 958–963 (2010).
Park, C.-H. et al. Pathological and microbiological studies of Japanese Hare (Lepus brachyurus angustidens) naturally infected with Francisella tularensis subsp. holarctica. J. Vet. Med. Sci. 71, 1629–1635 (2009).
Origgi, F. C. & Pilo, P. Francisella tularensis clades B.FTN002–00 and B.13 are associated with distinct pathology in the European Brown Hare (Lepus europaeus). Vet. Pathol. 53, 1220–1232 (2016).
Grizzle, W. E. et al. Factors affecting lmmunohistochemical evaluation of biomarker expression in Neoplasia. In Tumor Marker Protocols (eds Hanausek, M. & Walaszek, Z.) 161–180 (Humana Press, 1998).
Runge, M. et al. Prevalence of Francisella tularensis in brown hare (Lepus europaeus) populations in Lower Saxony Germany. Eur. J. Wildlife Res. 57, 1085–1089 (2011).
Tomaso, H. et al. Francisella tularensis and other bacteria in hares and ticks in North Rhine-Westphalia (Germany). Ticks Tick-borne Dis. 9, 325–329 (2018).
Stalb, S. et al. Detection of tularemia in European brown hares (Lepus europaeus) and humans reveals endemic and seasonal occurrence in Baden-Wuerttemberg, Germany. Berl Münch Tierärztl Wochensch 130, 293–299. https://doi.org/10.2376/0005-9366-16079 (2017).
Gyuranecz, M. et al. Investigation of the Ecology of Francisella tularensis during an inter-epizootic period. Vector-Borne Zoonotic Dis. 11, 1031–1035 (2011).
Lopes de Carvalho, I. et al. Francisella species in ticks and animals, Iberian Peninsula. Ticks Tick-borne Dis. 7, 159–165 (2016).
Lopes de Carvalho, I. et al. Borrelia garinii and Francisella tularensis subsp. holarctica detected in migratory shorebirds in Portugal. Eur. J. Wildlife Res. 58, 857–861 (2012).
Padeshki, P. I., Ivanov, I. N., Popov, B. & Kantardjie, T. V. Short report: The role of birds in dissemination of Francisella Tularensis: First direct molecular evidence for bird-to-human transmission. Epidemiol. Infect 138, 5 (2010).
Mörner, T. & Krogh, G. An endemic case of tularemia in the mountain hare (Lepus timidus) on the island of Stora Karlsö. Nord Vet Med. 36, 310–313 (1984).
Gürcan, S. et al. Tularemia re-emerging in European part of Turkey after 60 years. Jpn. J. Infect. Dis. 59, 391–393 (2006).
Leblebicioglu, H. et al. Outbreak of tularemia: A case–control study and environmental investigation in Turkey. Int. J. Infect. Dis. 12, 265–269 (2008).
Rossow, H. et al. Experimental infection of voles with Francisella tularensis indicates their amplification role in tularemia outbreaks. PLoS ONE https://doi.org/10.1371/journal.pone.0108864 (2014).
Splettstoesser, W. D. et al. Tularemia in Germany: The tip of the iceberg? Epidemiol. Infect. 137, 736–743 (2009).
Centers for Disease Control and Prevention (CDC). Tularemia United States, 1990–2000. MMWR Morb. Mortal Wkly. Rep. 51, 181–184 (2002).
Tärnvik, A., Priebe, H. & Grunow, R. Tularaemia in Europe: An epidemiological overview. Scand. J. Infect. Dis. 36, 350–355 (2004).
World Health Organization. WHO Guidelines on tularaemia. 115 (2007).
Mörner, T., Sandström, G., Mattsson, R. & Nilsson, P. O. Infections with Francisella tularensis biovar palaearctica in hares (Lepus timidus, Lepus europaeus) from Sweden. J. Wildl. Dis. 24, 422–433 (1988).
Larssen, K. W. et al. Outbreak of tularaemia in central Norway, January to March 2011. Eurosurveillance https://doi.org/10.2807/ese.16.13.19828-en (2011).
Mörner, T. The ecology of tularaemia. Rev. Sci. Tech. 11, 1123–1130 (1992).
Seiwald, S., Simeon, A., Hofer, E., Weiss, G. & Bellmann-Weiler, R. Tularemia Goes west: Epidemiology of an emerging infection in Austria. Microorganisms 8, 1597 (2020).
Ellis, J., Oyston, P. C. F., Green, M. & Titball, R. W. Tularemia. Clin. Microbiol. Rev. 15, 631–646 (2002).
Pilo, P. Phylogenetic lineages of Francisella tularensis in animals. Front. Cell. Infect. Microbiol. https://doi.org/10.3389/fcimb.2018.00258 (2018).
Zellner, B. & Huntley, J. F. Ticks and Tularemia: Do we know what we don’t know? Front. Cell. Infect. Microbiol. https://doi.org/10.3389/fcimb.2019.00146 (2019).
Genchi, M. et al. Francisella tularensis: No evidence for transovarial transmission in the tularemia tick vectors dermacentor reticulatus and Ixodes ricinus. PLoS ONE 10, e0133593. https://doi.org/10.1371/journal.pone.0133593 (2015).
Frank, R. et al. Parasites of wild rabbits (Oryctolagus cuniculus) from an urban area in Germany, in relation to worldwide results. Parasitol. Res. 112, 4255–4266 (2013).
Beaucournu, J.-C. Les anoploures de lagomorphes, rongeurs et Insectivores dans la Région Paléarctique Occidentale et en particulier en France. Ann. Parasitol. Hum. Comp. 43, 201–271 (1968).
Dik, B. & Uslu, U. Ectoparasites of hares (Lepus europaeus Pallas) in Konya Province, Turkey. Turk. J. Vet. Anim. Sci. 42, 65–72 (2018).
Gehringer, H. et al. Presence of an emerging subclone of Francisella tularensis holarctica in Ixodes ricinus ticks from south-western Germany. Ticks Tick-borne Dis. 4, 93–100 (2013).
Petersen, J. M., Mead, P. S. & Schriefer, M. E. Francisella tularensis: An arthropod-borne pathogen. Vet. Res. 40, 07 (2009).
Brown, R. N., Lane, R. S. & Dennis, D. T. Geographic distributions of tick-borne diseases and their vectors. In Tick-Borne Diseases of Humans (eds Goodman, J. L. et al.) 361–391 (ASM Press, USA, 2014). https://doi.org/10.1128/9781555816490.ch21.
Gurycová, D., Tináková, K., Výrosteková, V. & Gacíková, E. The incidence of tularemia in Slovakia in 1997–2008. Epidemiol. Mikrobiol. Imunol. 59, 39–44 (2010).
Toledo, A. et al. Tick-borne zoonotic bacteria in ticks collected from central Spain. Am. J. Trop. Med. Hyg. 81, 67–74 (2009).
Franke, J. et al. Coexistence of pathogens in host-seeking and feeding ticks within a single natural habitat in central germany. Appl. Environ. Microbiol. 76, 6829–6836 (2010).
Acknowledgements
We wish to express our thanks to the forests conservation staff of Tipaza province for their valuable help with the fieldwork. We are grateful to all hunters for their participation in the specimen collections and the general Directorate of Forests at the Ministry of Agriculture, Rural Development, and Fisheries that made administrative facilities for capturing wild leporids in Algeria. Also, we are so grateful for Facilities supported by the French National Reference Center for Francisella, Institute of Biology and Pathology, Grenoble Alpes University Hospital Center, Grenoble, France, and the Institute of veterinary sciences, University of Saad Dahlab—Blida 1, Algeria. We also thank the Agence Innovation Défense of the Direction Générale de l'Armement of France for their support.
Funding
This research was funded by the Agence Innovation Defense of the Direction Generale of l'Armement of France; grant number ANR-17-ASTR-0024 and international joint laboratory, Mediterranean research in emerging and re-emerging infections (LMI REMIDIER), Algeria; code 2R25700: VITROME.
Author information
Authors and Affiliations
Contributions
I.A.: conceived the idea and collected animal samples. I.A., C.D.B., M.M.: designed the study and drafted the manuscript. I.A., C.D.B., J.P., S.B., J.B., N.B.F., I.B., M.M.: performed laboratory analyses and revised the manuscript. M.M., I.B.: guided and supervised all work. K.R., I.B.: Project administration. I.A., M.M and I.B.: Funding acquisition. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ammam, I., Brunet, C.D., Boukenaoui-Ferrouk, N. et al. Francisella tularensis PCR detection in Cape hares (Lepus capensis) and wild rabbits (Oryctolagus cuniculus) in Algeria. Sci Rep 12, 21451 (2022). https://doi.org/10.1038/s41598-022-25188-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25188-0
This article is cited by
-
Molecular and serological investigation of Francisella tularensis among wild animals in Yamaguchi prefecture
Veterinary Research Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.