Abstract
At our tertiary children’s hospital, infections with newly detected methicillin-resistant Staphylococcus aureus (MRSA) among children attending primary (age 6–12 years) and secondary school (age 13–16 years) nearly doubled in 2018 compared to previous years. This observation initiated an epidemiological outbreak investigation including phenotypic (susceptibility testing) and genotypic (whole genome sequencing) characterization of the isolates. In addition, a cross-sectional study was conducted to determine source of the outbreak, colonization frequency and to identify risk factors for transmission using a questionnaire. As a result, 49 individuals were detected with 57 corresponding isolates. Based on the case definition combined with whole genome sequencing, a core cluster was identified that shared common genetic features and a similar antimicrobial susceptibility pattern (efflux-mediated macrolide resistance, tetracycline susceptibility along with presence of Panton-Valentine leukocidin). Epidemiologic evaluation identified a distinct school as a common risk factor. However, the source of the clustered infections within that school could not be further specified. No further cases could be detected after decolonization of infected and colonized children.
Similar content being viewed by others
Introduction
Over the last 25 years, infections with community associated methicillin-resistant Staphylococcus aureus (CA-MRSA) have increased all over the world. In Switzerland, MRSA detection rates decreased in recent years in adults, but increased in the younger population based on data from the Swiss antimicrobial resistance surveillance network (ANRESIS, http://www.anresis.ch)1. However, MRSA frequency in children admitted to hospitals is still low2,3 and general screening is not recommended. Most children colonized with CA-MRSA remain asymptomatic. Symptomatic children mainly present with skin and soft tissue infections (SSTI) infections4 and severe invasive infections are rarely seen. CA-MRSA infections are epidemic in some countries which may suggest, that CA-MRSA are more virulent and transmissible than hospital-associated MRSA strains5. CA-MRSA frequently carry the cytotoxic toxin Panton-Valentin Leukocidin (PVL) as a virulence factor associated with more severe clinical manifestations, although conflicting data exist on the role of PVL in the pathogenesis of CA-MRSA infections6. Clusters of sequence type 5 (ST5) PVL producing CA-MRSA strains in inpatient neonates7, in school-children8 and of different CA-MRSA strains have been previously described in Switzerland9,10.
For many years, pulsed-field gel electrophoresis (PFGE), spa typing and SCCmec typing alongside with multi-locus sequence typing (MLST) represented the primary MRSA typing techniques11. Nowadays, because of next-generation sequencing (NGS) techniques, healthcare facilities are capable of tracing outbreaks with higher resolution than ever before. Whole-genome sequencing (WGS) as part of a surveillance system can be used to monitor pathogens of concern within a healthcare facility, a region or even countrywide. Furthermore, the use of core genome (cg) MLST allows a more in-depth view inside the evolution and spread of MRSA.
A cluster of CA-MRSA infections was detected in an 80-bed tertiary children’s hospital in Switzerland with the number of CA-MRSA cases doubling compared to previous years in the first half of 2018. Furthermore, susceptibility testing of most of the CA-MRSA cases revealed an efflux-mediated, non-inducible macrolide-streptogramin B antibiotic resistance pattern and positivity for PVL, further supporting our observation of a possible cluster. In consequence, epidemiological and genomic outbreak investigations were initiated with the aims of identifying the source and risk factors for the transmission of CA-MRSA, to implement measures to prevent transmissions and to end the outbreak.
Methods
Outbreak investigation
Outbreak investigation included (i) microbiological case definition by microbiological culture with susceptibility testing to detect CA-MRSA, followed by WGS and (ii) detection of additional cases by cross-sectional study. This allowed for epidemiological investigation with evaluation of risk factors by questionnaires, case and outbreak description, and finally implementation of measures i.e., decolonization of CA-MRSA cases.
Microbial culture and MRSA detection
Copan eSwabs™ (Copan, Brescia, Italy) were used for swabbing and sent to the laboratory immediately after sampling. Ten microliters of the 1 ml liquid amies medium included in a Copan eSwab™ and containing the swab sample were transferred into enrichment broth (Brain heart infusion (BHI) with 6% NaCl, manufactured in house) and incubated overnight. Of the enriched broth 10 μl was inoculated onto a chromID chromogenic MRSA plate (bioMérieux, Marcy l’Etoile, France) with a WASP instrument (Copan, Brescia, Italy). After incubating the plates for 24 h in the smart incubators of a WASPLab™ high-resolution images of media plates were inspected using the WASPLab™ WebApp software. Colonies, indicative for MRSA were identified by a software algorithm (“segregation”) and sent to reading and picking for further workup12. Identification was done with a MALDI-ToF instrument using the BDAL 9.0 database (MALDI Biotyper Smart System, Bruker Daltonics, Bremen, Germany) with the colony transfer method (“direct smear”). Colonies identified as S. aureus were further tested with a PBP2a (penicillin-binding protein 2a, which confers methicillin-resistance) lateral flow immunochromatography assay (Alere Clearview PBP2a Culture Colony Test, Alere Inc. Waltham MA, USA), which confirmed MRSA. A BD™ Phoenix instrument (Becton Dickinson, Sparks, MD, USA) with PMIC-88 cartridge (tailored for the susceptibility testing of staphylococci and other Gram-positive bacteria) was used for susceptibility testing, except vancomycin, which was tested with epsilometer (E-) test (bioMérieux, Marcy l’Etoile, France). Antimicrobial susceptibility testing (AST) data were interpreted according to respective EUCAST guidelines (version 8.0 or 9.0 in 2018 and 2019, AST data not shown). This procedure applies to all isolates processed by the Centre for Laboratory Medicine in St. Gallen. Five isolates were tested elsewhere (i.e. available as laboratory report, only) and the data could not be verified.
Genomic evaluation by whole genome sequencing (WGS)
DNA extraction was performed using the QIAsymphony DSP DNA Mini Kit (QIAGEN GmbH, Hilden, Germany). DNA was quantified using the Qubit dsDNA BR HS Assay Kit and Qubit fluorometers (Invitrogen, https://www.thermofisher.com). WGS was performed using Illumina MiSeq with the Nextera XTlibrary preparation kit (Illumina Inc., USA), according to the manufacturer’s procedure (paired-end sequencing). After mapping the raw data against the reference genome, we retained reads with at least 90% sequence identity, from areas with an average coverage (unassembled) of more than 57 reads, and reads longer than 123 base pairs. Trimming and assembly of raw reads was performed using the Velvet assembler of the SeqSphere software (Ridom, https://www.ridom.de, version 7.7.5 using S. aureus cgMLST v1.3 and S. aureus v1.1). The analysis included MLST, cgMLST (1861 targets), spa typing, clonal complex typing and detecting resistance/virulence genes (mecA, macrolide resistance genes (ermA, ermC, mphC, msrA) and PVL (LukS-PV and LukF-PV)) by using SeqSphere. Coverage was at least 50-fold. We defined core clusters as groups of isolates with ≤ 5 different SNPs between neighboring isolates based on the cgMLST. To generate phylogenetic SNP trees, we used SeqSphere (Ridom; Münster, Germany) in the pairwise ignore missing values mode and an unweighted pair group method. The WGS data has been submitted to NCBI under the following submission number PRJNA738326.
Case definition based on microbiological results
The following case definition was used to categorize MRSA cases. First, all culture positive MRSA cases were “suspected cases”. Second, presence of the specific efflux-mediated macrolide resistance (phenotype) was assessed. Cases with an alternative antimicrobial susceptibility testing result were categorized as “non-cluster cases”. “Suspected cases” having efflux-mediated macrolide resistance were “probable cases”. In “probable cases” where strains were available, WGS was performed. As there are no international standards defining the cut-off between closely related and unrelated MRSA strains, we used the following two thresholds to categorize our strains13. If strains were ST5 using cgMLST, cases were “confirmed cases” and considered either as “core cluster” (≤ 5 single nucleotide polymorphisms (SNPs)) or “cluster” (> 5 but < 15 SNPs). All cases not belonging to ST5 or ST5 with > 15 SNPs were again classified as “non-cluster cases” (see Fig. 1).
Case definitions based on microbiological results used in this study. AST antimicrobial susceptibility testing, cgMLST core genome multi-locus sequence typing, WGS whole-genome sequencing, MRSA methicillin-resistant Staphylococcus aureus, MSSA methicillin-susceptible Staphylococcus aureus, ST sequence type, SNP single nucleotide polymorphism.
Identification of risk factors by questionnaires
Potential risk factors for CA-MRSA acquisition were assessed by two questionnaires before (column “Questionnaire” and during (column “Modified questionnaire”) the cross-sectional study and are provided in the Supplementary Table S1. The first questionnaire (“Questionnaire” was sent in spring to summer 2019 by email and/or mail to the families of confirmed and probable cases. Questions covered schooling (nursery, primary and secondary school), contact with the health-care systems (e.g., inpatient care), current, previous, or recurrent infections with MRSA or skin infections of the case and household contacts, profession, workplace, pets, leisure activities including sports, events, school or holiday camps or trips and attendance at the local children’s festival held in June 2018. The second questionnaire (“Modified questionnaire”) was used during the cross-sectional study (see “Cross-sectional study” section) and was electronically filled-in on site.
Cross-sectional study
Given persistent transmission throughout 18 months (2018–2019), a cross-sectional study was initiated to identify additional cases. Results of the epidemiological investigation and the first questionnaire supported the hypothesis that the outbreak was linked to a particular primary and secondary school. Accordingly, all students and all staff at the affected school with proven direct contact to the students (i.e. teachers, pre-schooling teachers) were included in the study in November 2019. Clinical examination for skin lesions was performed in all students and staff. Swabs for MRSA (from axilla/inguinal region, nose/throat, wounds) were taken from all individuals with SSTI on clinical examination. These individuals also had to answer a second questionnaire, which was adapted from the previous one in summer 2018. Questions focused on school activities and leisure activities. Furthermore, every fifth student and staff without symptoms of CA-MRSA infection were randomly assigned to the cross-sectional study to receive swabbing and answer the questionnaire.
Decolonization of CA-MRSA positive individuals
Protocols for decolonization can lead to the reduction of colonization and CA-MRSA infections14. To decolonize skin and mucous membranes, disinfectant soaps (i.e. octenidine-containing wash lotion), nasal ointment (i.e. containing mupirocin) and oral gel (i.e. chlorhexidine gluconate) were applied to the skin, hair, nose and throat 1–2 times daily for five days. Furthermore clothes, bed sheets and towels had to be changed daily and washed at 40–60 °C in a washing machine.
Statistics
Continuous variables were assessed by t tests or Mann–Whitney U tests as appropriate. For categorical values, comparison was done by Fisher’s exact test. Missing data were not imputed. A two-sided p value < 0.05 was considered as statistically significant. Given the small number of each outcome, only the univariate but not the multivariable analyses are reported. Data analysis was performed with SAS (version 9.3, SAS Institute Inc., Cary, NC, USA).
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, mandated, and supported by the responsible cantonal authorities, i.e. the Division of Public Health, Department of Health, St. Gallen, the highest medical supervisory authority in the canton. In addition, formal informed consent was waived because this study involved anonymized health-related data and sequencing data of bacterial strains both of which were included in the general consent that all “cases” gave upon admission to the hospital. Therefore, additional approval by the local ethical committee was not applicable.
Results
Outbreak description and epidemiological investigation
First cases of CA-MRSA were initially identified and treated at the local tertiary care children’s hospital. A common denominator was quickly identified during clinical examination and case history taking: most cases were students attending a school within the same city. As a result of case accumulation within one school, the school medical service and the responsible health department were involved, and hygiene measures (intensified hand hygiene and disinfection of surfaces) initiated at the affected school in October 2018. To increase awareness of further CA-MRSA cases, pediatricians and family physicians in the city area were informed of an CA-MRSA outbreak and encouraged to take skin swabs if a typical clinical skin infections were observed and to decolonize in case CA-MRSA was detected and forward results to study team. Furthermore, parents of children from this school received an information letter in spring 2019 on CA-MRSA and the request to visit their family doctor if the child or themselves were symptomatic. Also, a media release was published in the regional newspaper. Physicians and parents were notified once more when the cross-sectional study took place (“awareness campaign”).
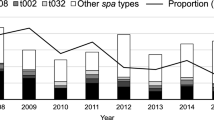
Although the number of cases decreased during the winter 2018/2019, new cases continued to occur (see Fig. 2). Hence, a first questionnaire was developed to identify other risk factors and determine venues in common and activities of cases. The questionnaire was distributed among 97% of confirmed and probable cases. Response rate was 65%, confirming that 23 (77%) cases were related to the school. There was no clustering regarding other risk factors of CA-MRSA than attendance of this school (see Supplementary Table S1). Affected children were not confined to a particular class, nor to any other shared venue or activity. As no additional risk factors were identified beyond attendance at this school, but additional infections were observed during the summer of 2019, a cross-sectional study was initiated (see below).
Clinical description of cases
CA-MRSA was identified in 49 individuals from January 2018 until June 2020. Eight cases had two isolates (total 57 different isolates). Two children were hospitalized. One adolescent was hospitalized because of osteomyelitis and in addition to intravenous antibiotic administration, surgical procedures and drainage were required. Based on the case definition, 27 cases were classified as confirmed (ST5 according to WGS), 6 as probable (not available for WGS) and 16 were non-cluster cases (9 cases were non-ST5 and 7 had a divergent antimicrobial susceptibility pattern). The non-cluster isolates either showed a MLSB phenotype with resistance to clindamycin (i.e., constitutive, or inducible), did not have macrolide resistance or had non-ST5 on whole genome sequencing.
Most cases of the outbreak were children and had mild clinical symptoms of MRSA infection. Clinical characteristics of the 33 probable and confirmed cases are provided in Table 1. Association with the affected school was observed in 79% of cases (see Fig. 1).
Cross-sectional study
The cross-sectional study was performed between November 27–29, 2019, and 129 participants from the affected school were screened (104 students and 25 staff including teachers). SSTI were observed in 17 students (13%) but in no staff. MRSA infection was detected in 1.6% (2 students and no staff [p = 0.64]) and MRSA colonisation in 4.7% (6 students and no staff [p = 0.26]). Three of these six students had been symptomatic and identified as part of the MRSA cluster before but were not symptomatic at the time of the cross-sectional study. Skin infection was present both in participants with positive and negative MRSA results (p = 0.35). WGS revealed ST5 in all but one case from the cross-sectional study (87.5%). There was one non-cluster case (ST72). Using the modified questionnaire, we were again unable to identify risk factors for CA-MRSA colonisation (Supplementary Table S1) on univariate analysis comparing those with MRSA-detection to those without. All CA-MRSA positive individuals identified in the cross-sectional study (colonized with or without infection) underwent skin decolonization following local recommendations. Ongoing surveillance did not reveal new cases of CA-MRSA. Three individuals already known to have been colonized with CA-MRSA were found persistently colonized with CA-MRSA (Fig. 2).
Results of WGS
As a result of the outbreak investigation and the cross-sectional study, 39/49 cases (79%) were available for WGS (including seven individuals with two isolates) and 27/49 individuals (55%) belonged to the core cluster (31 isolates whereof 4 had an additional follow-up isolate). Results from WGS included MLST typing (sequence type), detection of mecA (Methicillin resistance gene), spa typing, clonal complex (CC) typing, the macrolide resistance genes (ermA, ermC, mphC, msrA, vga, vgbB), PVL genes (LukS-PV/LukF-PV) and the case definition (Table 2). 35 CA-MRSA isolates belonged to ST5 (spa type t002, CC5) and differed by a maximum of 259 SNPs. The outbreak cluster consisted of 31 isolates with a maximum of 9 SNPs (Fig. 3). The number of SNPs increased over time (Fig. 4). Within the sequenced ST5 isolates were five pairs from the same individual but 2 to 23 months apart, which also showed differences of 0,1, 2, 4, and 8 SNPs compared to the closest ST5 strain. This suggests evolution over time during this event. The methicillin and macrolide resistance genes (mecA, ermA, ermC, mphC, msrA) matched to their respective phenotypes. The macrolide resistance genes of the core cluster were a combination of the mphC and msrA genes, except for one isolate (SGMRSA37) that harbored no macrolide resistance genes and belonged to spa type t045. Besides the core cluster, several isolates harboring the MLSB genes ermA and ermC were also discovered.
SNP tree generated by SeqSphere using the pairwise ignore missing values mode (46 samples with 28,657 columns) and an unweighted pair group method. Sample IDs refers to IDs provided in Table 2 and Fig. 4. Numbers close to connecting lines between the isolates represent the difference in the number of SNPs between isolates within the core cluster. The different connecting lines reflect the relatedness between the respective isolates. 27 confirmed cases plus 4 duplicates are shown; SGMRSA32_2 with 9 SNPs difference is not shown.
Isolates are stratified according to month of isolation. The number of SNPs observed with respect to the first isolate (SGMRSA14) is given. Note, that isolates obtained in November 2019 were from the cross-sectional study. Colors code the numbers of SNPs, with dark green for 0 SNPs. Increasing SNPs are colored from yellow to red.
Discussion
This epidemiological and microbiological investigation of 49 children and adults with a newly detected CA-MRSA identified 33 cases that belonged to the outbreak. Molecular typing revealed a cluster with ST5 and a common efflux-mediated, non-inducible macrolide-lincosamide-streptogramin B (MLSB) antibiotic resistance pattern. Epidemiologic investigation found that in 79% of cases, a local elementary and secondary school (one campus) were the main sites of transmission. Following the implementation of interventions in the school and surrounding community as well as the conduct of a cross-sectional study with subsequent decolonization of participants in whom CA-MRSA could be found and no further cases occurred.
Although several CA-MRSA clusters have been described in Switzerland before, this is—to the best of our knowledge—the largest cluster of symptomatic children reported to date. The cluster also included 5 adults which had direct contact to the affected children and teachers (2 teachers, 1 father, 1 teacher’s wife, 1 mother). A cluster of ST5, SCCmec type IV PVL producing CA-MRSA strains has been previously described in Switzerland7. However, this cluster included seven inpatient neonates with a phenotypically different MRSA with respect to the antimicrobial resistance. In a surveillance report on 58 CA-MRSA cases from Western Switzerland, 13 distinct transmission clusters within 26 cases were detected over 3-year period9. About one fifth were children under 10 years of age and about two thirds of the isolates produced the PVL toxin. Finally a PVL-producing cluster of methicillin-susceptible Staphylococcus aureus within 10 of 22 schoolchildren and one of two teachers has been reported in Western Switzerland8. Six children were symptomatic with skin infections but none of the strains involved were MRSA. Similar to the cluster described here, no further cases occurred after implementation of treatment and decolonization. Further outbreaks with a ST5 PVL producing CA-MRSA strains have been described internationally15. To illustrate this, spread of a ST516 has been linked to an increase of community onset staphylococcal diseases in children in Argentina and is a predominant clone in Australia15 and New Zealand17.
Molecular microbiological methods (i.e. WGS) enabled clear delimitation and description of this CA-MRSA ST5 cluster, providing important information for evaluating whether, where and for how long preventive measures are needed. This is highly useful information for managing an outbreak. Still, although we surveyed cases and families on risk factors by questionnaires (Supplementary Table S1) and conducted a cross-sectional study in the affected school, the key important insight on the common source of this CA-MRSA cluster remained unknown.
From the discussions during the investigations with the school representatives, the school medical service and the responsible health department, a hypothesis remains, which is supported by the WGS results. Initially, from February to November 2018, nine strains were identified by WGS with no difference in cgMLST (i.e. zero SNPs). These findings suggest an initial transmission event occurred in spring 2018.
We can only speculate about the exact timing and circumstances of this event. But it may have been in the context of a regional children’s festival in the spring of 2018, which takes place every 4 years. During rehearsals as part of the preparations, clothes worn directly on the skin were immediately changed between the children without being washed. Unfortunately, the questionnaire did not confirm this hypothesis since all children (with and without CA-MRSA) attended this event. However, given most children in the region participate, this is not unexpected. Further and less frequent transmissions during the following years must have taken part during school and leisure activities since CA-MRSA is mainly transmitted by direct contact.
Consistent with the epidemic curve (Fig. 2), the isolates had increasing numbers of SNPs over time compared to the first isolate in February 2018. While the actual magnitude of previous clusters reported from the literature remains unclear, we here provide evidence of a spatially and temporally restricted ST5 CA-MRSA PVL producing cluster.
Finally, we identified 4.7% CA-MRSA carriage on a school-based cross-sectional study during an outbreak and CA-MRSA infection in 1.6%. Data on the frequency of CA-MRSA carriage in the healthy population is not readily available since antimicrobial resistance testing according to the national antimicrobial resistance surveillance network ANRESIS is based on clinical isolates and not on swabs of healthy individuals. Thus, the rate of colonisation in healthy students with CA-MRSA in Switzerland is unknown. There are data from a Swiss single-center cross-sectional study in 340 hospitalized children with no MRSA detection3. Asymptomatic colonization was found in 0.1% of pre-clinical students in Warsaw, Poland which was distinctively lower than the CA-MRSA carriage rate in our population of students18. Higher rates have been observed in nursing homes (14.5–38%)19,20, intravenous drug users (8.7%)21 and in persons with contact with livestock (6.7%)22. In a worldwide study comprising of > 1000 surgeons only 2% were colonized with MRSA23 and 4.6% of health-care workers on meta-analysis24. We therefore consider the CA-MRSA carriage rate of 4.7% in healthy individuals to be above what would be expected in our setting. The identification of a single non-ST5 isolate on cross-sectional study (0.78%) that likely represents baseline carriage further strengthens our hypothesis. Overall, and although the ST5 strain produced PVL, the clinical phenotypes in the children were benign and non-invasive, except for one case with bacteremia and osteomyelitis.
Our study has several limitations: Since we have not systematically compared our data with a nationwide strain collection, we still have only a limited view of the “big picture”. However, we assume that for the majority of CA-MRSA-infections (i.e. SSTI) isolates are currently not characterized by cgMLST and nationwide analysis is likely to be biased towards isolates from the hospital setting. In addition, we did not conduct the cross-sectional study in a control school without a cluster of cases nor did we include a control group from the general population. Therefore, we are not able to compare the results with the MRSA carrier rate in asymptomatic school children. Moreover, the outbreak investigation was still taking a long time, so the actual additional benefit of rapidly available WGS results could not yet be demonstrated in this study. However, this may improve with faster availability of WGS in routine practice. Finally, while we could not determine the exact start of the cluster, transmission chains or independent predictors for the cluster. However, the observed microevolution (i.e. accumulation of SNPs) suggests an initial transmission event with accumulated SNPs over time, reflecting a molecular clock, driving genetic diversity.
There are several points that substantiate our findings. First, we did not only focus on the initial cluster but followed school classes longitudinally followed for over 2 years. Since single cases of CA-MRSA SSTI still occurred at that time, a rigid cross-sectional study was conducted. This allowed us not only to describe the cluster itself, but to analyze microevolution over time. Second, since the number of SNPs within the cluster was low as expected, we performed WGS of randomly chosen MRSA isolates over time to diversify the molecular data. This was done for three isolates, two of the cluster and one isolate of different ST and phenotype.
Conclusion
We identified and characterized a school outbreak of ST5 PVL CA-MRSA in 33 individuals by epidemiological investigation combined with WGS. As detailed risk assessment and a cross-sectional case–control study revealed no other source of transmission than a local school, therefore transmission likely occurred during school activities. Our data suggest an initial transmission event that could have been the local children’s festival. After implementing hygienic measures, informing parents and local physicians and treating and decolonizing all infected and colonized CA-MRSA carriers, no further cases were detected during continued surveillance. A combined approach of epidemiological and microbiological methods is successful to terminate an outbreak of CA-MRSA.
Data availability
The datasets generated and/or analyzed during the current study are currently not publicly available. Deidentified participant data might be available on reasonable request by email to the corresponding author. The WGS data has been submitted to NCBI under the following submission number PRJNA738326.
References
Olearo, F. et al. Staphylococcus aureus and methicillin resistance in Switzerland: Regional differences and trends from 2004 to 2014. Swiss Med. Wkly. 146, w14339 (2016).
Heininger, U. et al. Prevalence of nasal colonization with methicillin-resistant Staphylococcus aureus (MRSA) in children a multicenter cross-sectional study. Pediatr. Infect. Dis. J. 26, 544–546 (2007).
Johnson, K., Frei, R. & Heininger, U. Prospective, cross-sectional study on MSSA and MRSA colonisation in hospitalised children. Eur. J. Pediatr. 174, 1255–1262 (2015).
Herold, B. C. et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279, 593–598 (1998).
DeLeo, F. R., Otto, M., Kreiswirth, B. N. & Chambers, H. F. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568 (2010).
Saeed, K. et al. Panton-valentine leukocidin-positive Staphylococcus aureus: A position statement from the International Society of Chemotherapy. Int. J. Antimicrob. Agents 51, 16–25 (2018).
Sax, H. et al. Control of a cluster of community-associated, methicillin-resistant Staphylococcus aureus in neonatology. J. Hosp. Infect. 63, 93–100 (2006).
Boubaker, K. et al. Panton-valentine leukocidin and staphyloccoccal skin infections in school children. Emerg. Infect. Dis. J. 10, 121 (2004).
Aramburu, C. et al. Community-acquired methicillin-resistant Staphylococcus aureus in Switzerland: First surveillance report. Euro Surveill 11, 42–43 (2006).
Meyer Sauteur, P. M. et al. Methicillin-resistant Staphylococcus aureus eradication in young infants should include the diaper area and treat housing contacts. Acta Paediatr. 104, e581–e584 (2015).
Lakhundi, S. & Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31, e00020 (2018).
Faron, M. L. et al. Automated scoring of chromogenic media for detection of methicillin-resistant Staphylococcus aureus by Use of WASPLab image analysis software. J. Clin. Microbiol. 54, 620–624 (2016).
Cunningham, S. A. et al. Comparison of whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J. Clin. Microbiol. 55, 1946–1953 (2017).
Huang, S. S. et al. Decolonization to reduce postdischarge infection risk among MRSA carriers. N. Engl. J. Med. 380, 638–650 (2019).
McGuinness, S. L. et al. Clinical and molecular epidemiology of an emerging panton-valentine leukocidin-positive ST5 methicillin-resistant Staphylococcus aureus clone in Northern Australia. mSphere 6, e00651 (2021).
Sola, C. et al. Spread of epidemic MRSA-ST5-IV clone encoding PVL as a major cause of community onset Staphylococcal infections in Argentinean children. PLoS ONE 7, e30487 (2012).
Williamson, D. A. et al. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in New Zealand: Rapid emergence of sequence type 5 (ST5)-SCCmec-IV as the dominant community-associated MRSA clone. PLoS ONE 8, e62020 (2013).
Szymanek-Majchrzak, K. et al. Prevalence of methicillin resistant and mupirocin-resistant Staphylococcus aureus strains among medical students of Medical University in Warsaw. Przegl. Epidemiol. 73, 39–48 (2019).
Moschou, A. et al. Prevalence and molecular epidemiology of Staphylococcus aureus nasal colonization in four nursing home residents in Crete, Greece. J. Infect. Chemother. 26, 199–204 (2020).
March, A. et al. Colonization of residents and staff of a long-term-care facility and adjacent acute-care hospital geriatric unit by multiresistant bacteria. Clin. Microbiol. Infect. 16, 934–944 (2010).
Packer, S. et al. Clonal expansion of community-associated meticillin-resistant Staphylococcus aureus (MRSA) in people who inject drugs (PWID): Prevalence, risk factors and molecular epidemiology, Bristol, United Kingdom, 2012 to 2017. Eurosurveillance 24, 1800124. https://doi.org/10.2807/1560-7917.ES.2019.24.13.1800124 (2019).
Neradova, K., Jakubu, V., Pomorska, K. & Zemlickova, H. Methicillin-resistant Staphylococcus aureus in veterinary professionals in 2017 in the Czech Republic. BMC Vet. Res. 16, 4 (2020).
Morgenstern, M. et al. Antibiotic resistance of commensal Staphylococcus aureus and coagulase-negative staphylococci in an international cohort of surgeons: A prospective point-prevalence study. PLoS ONE 11, e0148437 (2016).
Albrich, W. C. & Harbarth, S. Health-care workers: Source, vector, or victim of MRSA? Lancet Infect. Dis. 8, 289–301 (2008).
Acknowledgements
The authors would like to thank the school medical service of the city of St. Gallen and the school affected for their great support, which enabled us to conduct the necessary epidemiological investigations.
Funding
Funding of WGS was provided by the institutions involved: Children’s Hospital of Eastern Switzerland, Health Department of the Canton of St. Gallen, Centre for Laboratory Medicine.
Author information
Authors and Affiliations
Contributions
C.R.K. and O.N. initiated and designed the study with the support of M.S., C.B., and D.Z. C.R.K. and O.N. supervised the study. F.W. and C.K. performed the epidemiological investigation including the development of questionnaires and conducted the cross-sectional study with support of M.S., S.M. and A.W. F.W. and C.K. drafted the first version of the manuscript with support of O.N., S.S. and W.A. O.N. and S.S. were responsible for the microbiology work and analysis of NGS data with the support of A.E. and T.R. All authors contributed to data interpretation and revised and approved the manuscript for intellectual content. All authors agreed to the final version including submission for publication and accept responsibility for this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Waldeck, F., Seiffert, S.N., Manser, S. et al. Outbreak investigation including molecular characterization of community associated methicillin-resistant Staphylococcus aureus in a primary and secondary school in Eastern Switzerland. Sci Rep 12, 19826 (2022). https://doi.org/10.1038/s41598-022-24363-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24363-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.