Abstract
This study revealed how Bacteria and Archaea communities and their metabolic functions differed between two groups of black deposits identified in gorge and cave environments. Scanning electron microscopy coupled with energy dispersive spectroscopy was used to analyse the presence of microbial biosignatures and the elemental composition of samples. Metabarcoding of the V3–V4 regions of 16S rRNA was used to investigate Bacteria and Archaea communities. Based on 16S rRNA sequencing results, PICRUSt software was used to predict metagenome functions. Micrographs showed that samples presented microbial biosignatures and microanalyses highlighted Mn concretions and layers on Al-Si surfaces. The 16S rRNA metabarcoding alpha-diversity metrics showed similar Simpson's and Shannon indices and different values of the Chao-1 index. The amplicon sequence variants (ASVs) analysis at the different taxonomic levels showed a diverse genera composition. However, the communities of all samples shared the presence of uncultured ASVs belonging to the Gemmatales family (Phylogenesis: Gemmataceae; Planctomycetes; Planctomycetota; Bacteria). The predicted metagenome functions analysis revealed diverse metabolic profiles of the Cave and Gorge groups. Genes coding for essential Mn metabolism were present in all samples. Overall, the findings on structure, microbiota, and predicted metagenome functions showed a similar microbial contribution to epigean and hypogean black deposits Mn metabolism.
Similar content being viewed by others
Introduction
Manganese is the second-most abundant transition metal available on the terrestrial surface and covers 0.1% of the Earth's total mass. It represents a crucial trace nutrient for the growth and survival of numerous living organisms, contributing, for instance, to oxygen production, redox reactions, or protecting cells from toxic metals, UV radiation and oxidative stress1. The manganese oxides control the availability of several elements because of their strong oxidizing capacity. The geochemical behaviour of Mn differs based on the oxygen profile2. Even if Mn occurs in seven different oxidation states (from 0 to + 7), manganese interacts as reduced soluble or adsorbed Mn2+, insoluble Mn3+ and Mn4+ oxides in the natural environment3. Direct and indirect evidence for the biogenic Mn coatings, such as crusts, and nodules, can occur in various environments on the Earth's surface and subsurface3. Typical natural ecosystems hosting nodules, black crusts, mineralized stromatolites, patinas, and rock varnishes of manganese are deserts, seawater, freshwaters, and caves3. The frequent occurrence of these intriguing formations in different environments has also been described by Darwin during his iconic voyage aboard the HMS Beagle. However, their origins are still partially unknown, with both the biogenic and abiotic models demonstrated in nature. When the conditions are favourable, different microbial groups can control the oxidation of Mn, competing with the abiotic process4.
Mn and other metals, play a major role in controlling both the redox balance and the carbon cycling via oxidation in the environments in which these elements are abundant5. Microorganisms, mainly bacteria and fungi, can perform oxidation/reduction of Mn for carbon oxidation and growth6. Mn microbial redox reactions of Mn(II) to Mn(IV) are most common, with Mn(III) as an occasional intermediate. In many cases, coupling Mn(IV) reduction to the complete oxidation of organic matter is reported5. Mn-oxidizing bacteria harbour a suite of enzymes that help scavenge Mn and other associated elements, while several Mn-reducing microorganisms, from highly aerobic to strictly anaerobic, have been described in the literature7. The microbial mechanisms of Mn reduction can be either an indirect process resulting from interactions with organic or inorganic compounds or comprising natural enzymatic processes based on an electron-transfer reactions6.
Oxide minerals associated with a biogenetic formation of black deposits are birnessite and todorokite, frequently found within rock varnish and ocean nodules and in black deposits from caves7. Specifically, Spilde et al. compared these two oxide minerals from cave deposits with those found in rock varnish and described a progression of increasing crystallinity from a filamentous to a fibrous form for those found underground8. From a morphologic point of view, Manganese oxides found in caves are different from the oxides in rock varnish. The manganese oxides from rock varnish are undulating, sometimes discontinuous, and laminated with clay and quartz, most likely dust particles. On the other hand, the cave coatings are thick and with a softer consistency9. Spilde suggests that the laminae that occur in rock varnish are wetted and dried because they were subjected to harsh conditions resulting in thin, flat layers, while in caves, the rock layers tend to preserve black crusts, shielded from the corrosion by weathering. Although there are these micro-and macroscopic differences, Spilde found similarities in the microbial communities between the cave and rock surface coatings8.
Studying microbial contribution to minerals formation can be challenging. Generally, deposits harbour microbial biosignatures that can help speculate about the microbial presence. However, preserving well-defined microbial structures is difficult because the cells collapse and degrade. In addition, the manganese oxides degrade the organic compounds left behind by living cells, and thus, they leave only a few traces of biological activity10. Mn oxidation/reduction mechanisms are still under debate, and more experimental research is necessary to understand the enzymatic pathways involved. The RNA-seq analysis is the most powerful tool to gather the differentially expressed genes within a community. However, few attempts to sequence ancient transcriptomes were carried out due to the high-degradative nature of RNA. The 16 s rRNA metabarcoding and bioinformatic tools may help obtain helpful information about the prokaryotic community (mainly Bacteria and Archaea) and metagenome functions involved11,12,13,14.
In our previous studies we investigated and described black deposits of gorge and cave environments, finding similar biological and structural traits12,15. For this reason, we hypothesized that black deposits of gorge and cave environments could present similar Bacteria and Archaea communities and common metabolic processes, especially concerning manganese metabolism. This work aims at investigating similarities and differences among the deposits of these two different environments, the similarities and differences among Bacteria and Archaea communities and metagenome functions that these hypogean and epigean environments share between two groups of black deposits found in gorge and cave environments. To evaluate the presence of microbial biosignatures and the element composition, samples were investigated by scanning electron microscopy coupled to energy dispersive spectroscopy (SEM–EDS). The Bacteria and Archaea communities were investigated by metabarcoding of V3-V4 regions of 16S rRNA. To predict the metagenome functions, sequencing data were analysed by PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved states, 2) software11.
Results
The black deposits were investigated by SEM–EDS. As presented in Figs. 1 and 2, both Gorge’s and Cave’s samples showed the presence of multi-layered structures and filaments attributable to microbial aggregation structures.
SEM–EDS investigations of gorge samples. Upper panels show the micrographs obtained by SEM observations. In the panels the putative microbial imprints are marked by yellow arrows, the microbial biofilms by blue arrows, and microbial mineralized structures by red arrows. The orange circle in sample G2 marks the presence of a diatom within the sample. Work distance (WD) = 8.5–11.5 mm; electron high tension (EHT) = 12–15 kV; Signal = BSD4 A (backscattered electrons); Magnitude = 5–8 K X (Scale bars are presented under each panel). Lower panels present the elemental maps obtained by EDS analyses. The microanalysis acquisitions were carried out at 12.00 kV (Scale bars are presented under each panel). Mn presence is underlined with light blue colour.
SEM–EDS investigations of cave samples. Upper panels show the micrographs obtained by SEM observations. In the panels the putative microbial imprints are marked by yellow arrows, the microbial biofilms by blue arrows, and microbial mineralized structures by red arrows. Work distance (WD) = 6.2–7.8 mm; electron high tension (EHT) = 5–7 kV; Signal = SE 2 (secondary electrons); Magnitude = 10 K X (Scale bars are presented under each panel). Lower panels present the elemental maps obtained by EDS analyses. The microanalysis acquisitions were carried out at 12.00 kV (Scale bars are presented under each panel). Mn presence is underlined with light blue colour.
The multi-layered structures were characterized by biofilms leaning on layers of mineral surfaces. Within these layers, many bores and cell-like shapes were found. Gorge's samples also showed the presence of many fossil diatoms embedded in mineral structures and concretions (Sample G2, Fig. 3). Figures 1 and 2 (Supplementary Figs. S1 and S2) also show the elemental composition of the samples. Mn layers were deposited or alternated with mineral fragments. The observations suggest the presence of a basic Si-Al mineral surface, with the same distribution and a distinct distribution to the other elements. This Si-Al mineral surface presented covering layers and concretions of Mn in all samples. The diffuse presence of other elements was also shown (i.e., Fe, K, Mg, and Na). In some cases, the distinct spatial distribution of Mn to S (Samples C3 and C4) and strict association with Fe (Sample C4) were observed. The Ca element was more diffuse in the cave's samples. Layers and concretions were instead observed in the gorge’s samples.
The black deposits of the Gorge and Cave groups were subjected to 16S sequencing. The ecological indices obtained for the analysed samples are shown in Table 1. Sample abundances ranged from 461 to 2045 (Chao-1), with samples G1 and C1 having the lowest values and C2 and C3 having the highest. The number of individuals displayed was comparable to the abundance recorded, implying a satisfactory sampling method. All the samples had substantial levels of diversity (H’ values higher than 4.5 and S 1-D near 1). The sample G2 had the most diversity, while the sample G1 had the least.
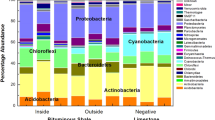
Sequencing results with a cutoff of 3% were processed and analysed at Domain, Phylum, Class, and Genus levels. At the Domain level, except for sample G1, which showed 27% Archaea and 73% Bacteria, the taxonomic data processing revealed that Bacteria accounted for most of the samples (97–100%) (Supplementary Table S1). The study of the total number of different ASVs belonging to each phylum (Supplementary Table S2) showed that the highest number of different ASVs was found in C2 for Acidobacteriota and Pseudomonadota and in G2 for Chloroflexota. The lowest values were recorded for G1. No marked differences were found among samples based on environment type. Except for sample G1, which included only Crenarchaeota, Micrarchaeota, and Thermoplasmatota, the Gorges samples were quite similar in their composition at the phylum level (Fig. 3A; Supplementary Table S2). For Cave samples, the situation was similar, with Gemmatimonadota and Patescibacteria being only present in samples C2 and C3, respectively (Fig. 3B; Supplementary Table S3). Nitrospirota, Actinobacteriota, Pseudomonadota, Chloroflexi, Patescibacteria, Acidobacteriota, Planctomycetota, Bacteroidota, and Gemmatimonadota were among the nine phyla that the Gorge and Cave groups shared (Fig. 3C; Supplementary Table S3). The Pseudomonadota phylum had a comparable abundance in all the samples, as shown in Fig. 3D. The other common phyla were present with different abundances. Therefore, at the class and genus levels, the differences between Gorge and Cave groups were more evident, underlying different compositions and abundances. However, some common traits were still found.
At the Class level, Alphaproteobacteria and Gammaproteobacteria were present in all samples of Gorge and Caves (Fig. 4A,B, respectively; Supplementary Table S4). Beyond these two classes, Vicinamibacteria, Bacteroidia, Gemmatimonadetes, Actinobacteria, Acidobacteriae, Blastocatellia, and Nitrospiria were found common between the two groups (Fig. 4C; Supplementary Table S4). Alphaproteobacteria and Gammaproteobacteria accounted for similar abundances within the bacterial communities of the samples (Fig. 4D).
Only the uncultured group was common at the Genus level in Gorge and Caves samples, respectively (Fig. 5A,B; Supplementary Table S5). The uncultured group mainly belonged to the Gemmatimonadaceae family (Phylogenesis: Gemmatales; Planctomycetes; Planctomycetota; Bacteria) but was not representative of all the samples (only G1 and G4). Except for sample G1, the unknown group was also common. The ASVs of this group mainly belonged to the Hydrogenedensaceae family (also present in G1 with abundance < 3%). The Nitrospira genus was found common in both groups (Fig. 5C; Supplementary Table S5). Except for C1, the uncultured and unknown groups accounted for a relevant abundance in all the samples (Fig. 5D). The other relevant genera were Acidothermus (for G1), Nitrospira (for G2, G3, C3, and C4), Crenothrix (for G3), Bacillus (for C1, C2, and C4), and wb1-P19 (for C2, C3, and C4).
To predict the microbial community functional profiles, the samples were also processed by using PICRUSt software. The EC, KO, and PWY outputs were firstly analysed to investigate similarities among the samples. Supplementary Fig. S3 reports the cluster outputs obtained for EC, KO, and PWY. All the evaluations underlined two distinct clusters based on sample origin. The Cave samples clustered within the first group, while Gorge samples clustered within the second group. To evaluate the differences among the groups, data were further investigated statistically. Figure 6A shows the differences among the EC predicted for each group. The statistical comparison showed that many enzymes were predicted differently based on Gorge and Cave samples. The enzymes with a significant statistical difference belonged to oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases families (statistical differences shown in Supplementary Table S6). Figure 6B depicts the differences among the KO predicted for each group. The evaluation showed that many predicted KO were statistically different between Gorge and Cave groups. Among them of relevance was the significant abundance within the gorge group of K16080, the functional ortholog of high-affinity Mn2+ porin (mnoP) (statistical differences shown in Supplementary Table S7). Figure 6C presents the statistical comparison of the PWY predicted for gorge and cave groups.
The investigation showed that only P621-PWY, nylon-6 oligomer degradation (a) and PWY-6876 (b), isopropanol biosynthesis, were different (Supplementary Table S8). This aspect suggested that even if the samples were constituted by different genera and had different predicted EC and KO, the pathways involved were mostly similar. To find out more about the potential redox biotransformation, metal resistance and respiratory processes, the EC outputs were also checked for the presence of enzymes involved in these processes. Figure 7 shows the heatmap generated for the most relevant abundances. Among the common enzymes, all samples with high abundances showed cytochrome-c oxidase, peroxiredoxin, and superoxide dismutase. The different distribution and abundance of other enzymes, such as Cd2+ exporting ATPase and sirohydrochlorin ferrochelatase, allowed clustering of the samples C1 and C2 in a different group. Sample G1 showed the abundance of most of the EC investigated and was the only sample found with cobaltochelase with good abundances.
Heat map of the enzymes involved in Mn metabolism predicted by PICURUSt software. The upper clusterization shows the sample distribution according to enzymes presence/absence and abundances. Heat map was realized based on resemblance matrix (Analyse between variables, measuring the index of association) and hierarchical cluster analysis (Group average cluster mode).
Data on alpha-diversity indices, phylum, Mn-involved EC, and environmental variables were also used to conduct a principal component analysis (Supplementary Fig. S4 and Supplementary Table S9). The results showed that the samples of gorge and cave groups occupied separated clusters, with temperature accounting more for gorge samples and pH and relative humidity for cave ones. For gorge site the temperature is more variable during the days and the seasons.
Discussion
Black deposits of gorge and cave environments have been investigated for many years3,5,8. However, despite the similarities they present, as far as we know, no studies dealt with the comparison of the Bacteria and Archaea communities for the identification of differences and similarities. The SEM–EDS investigation allowed to underline the presence of microbial imprints within the samples with a marked association of biological elements with the mineral matrices and the presence of Mn oxides layered upon a Si-Al mineral surface. These strict associations of the bacteria signatures within mineralized structures allow speculating on the contribution of microorganisms to the formation of these deposits. Microbial contribution is usually described by the mineralization/lithification of microbial cells and their aggregation structures, such as biofilms12, which can occur internally and externally. Many authors have described the presence of these biosignatures for several speleothems15,16,17,18. Our previous investigations described already the presence of these imprints within “Grotta Grande dei Cervi” black deposits12,19. This cave is one of the main caves present in the karst system of Pietrasecca together with “Ovito di Pietrasecca”. However, for the latter, microscopic observations of black deposits have not yet been reported in literature. Even with similar microbial imprints, the samples from “Ovito di Pietrasecca” (C3 and C4) showed a more prominent presence of N and P elements than those from “Grotta Grande dei Cervi”. These aspects should be further investigated by lithological investigations. However, we could speculate that this difference can be associated with different speleogenesis, which is still active in “Ovito di Pietrasecca” and inactive in “Grotta Grande dei Cervi”. These dynamics promote different environmental conditions, with a critical role in nitrogen and phosphate participation in the formation and degradation of primary and secondary deposits.
For gorge samples, the presence of black deposits is within ravine walls at low heights. This suggests that their formation dates to ages ago. As previously presented, this gorge develops primarily on volcanic substrates covering a sedimentary origin. Less than 2 million years ago, the waters of the Pliocene Sea covered all this emerged area. Following the marine regression, the young watercourses' volcanic genesis and erosive action gave rise to the deep incisions. Given the substantial presence of interactions between microbial and mineral imprints, the contribution of bacteria seems to have been influential in the formation of these deposits. Knowledge of these aspects and the observation of sampling sites lead us to associate these deposits with marine crusts-like deposits20, for which microbial activity is essential21. This speculation was supported by numerous fossils/traces of diatoms highlighted with SEM observations.
The 16S rRNA metabarcoding showed common ASVs at phylum and class levels. At the phylum level, Pseudomonadota was ubiquitous and represented important abundances. Pseudomonadota is one of the phyla that play a crucial role in the biogeochemical cycle of Mn6,22. It is represented by many Mn-oxidizing bacteria and is found in various settings (e.g., groundwaters, caves, soil)23. Alphaproteobacteria and Gammaproteobacteria are the classes with a joint distribution. For these classes, Mn oxidation is usually described by animal heme peroxidases (AHPs) and the multicopper oxidases (MCOs) enzymes, also found in the predicted metagenome functions of our study24. All the samples shared uncultured and unknown groups at the genus level. These groups are present in microbial communities of all types of environments. Uncultured bacteria are a key element of microbial communities. They represent a hidden population that provides an unexploited genetic resource encoding unique and valuable catalysts and enzymes. These aspects are important both for environmental and industrial application aspects25.
Among unknown ASVs, the representative taxon was assigned to the Hydrogenedensaceae family, common in all the samples. Limited information is available on the ecological role of this lineage. Their description in different types of wastewater treatments26,27 indicates anaerobic metabolism. Their distribution has been described in different environments. Lusa and Bomberg found this taxon in ombrotrophic Boreal bog communities, describing a distribution positively correlated to the presence of mineral elements (i.e., Mg, Al, Fe, Co, Cu, Se, Cs, Th, U) and negatively correlated to the organic matter content28. Cohen and collaborators described this family in permanently redox-stratified Fayetteville Green Lake as a part of the particle-associated bacterial community29.
Among the significant genera, Acidothermus (Actinobacteria phylum) is a thermophilic, acidophilic, and cellulolytic bacterium that can also break down chitin. It was initially isolated from an acidic hot spring in Yellowstone National Park30 and has also been described in several ecosystems, including as a part of the oxide sediment bacterial community of the eastern Mediterranean Sea31 and of an old Indian pond32. Crenothrix, are Mn oxidizer bacteria already used in biotechnologies for Mn biomining33. Nitrospira is the most varied and ubiquitous nitrifying in natural ecosystems and biological wastewater treatment34. Bacillus is one of the Mn-oxidizing bacteria used as a model due to its extraordinary biomining abilities35. Less is known on wb1-P19. However, phylogenetic studies suggested sulphur or nitrite-oxidizing autotrophic metabolism36 and its presence inside the cave has been already reported36,37,38,39.
Even with different microbial community structures and compositions, the samples presented similar predicted metagenome functions with some habitat-specific separations. The statistical approaches highlighted differences mainly in the presence and abundance of oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases. These differences are in line with the type of study conducted, based on the comparison of habitats with different environmental conditions. Within KO, MnoP was present with higher abundances in gorge samples. The MnoP is an outer membrane protein expressed only when there are low manganese levels. The mnoP gene is coordinated with the inner membrane transporter gene mntH. The Mur-binding site in the promoters of the mntH and mnoP genes allows occupancy in the presence of manganese and depletion when the metal is lacking40. Thanks to MnoP, Mn2+ can be translocated into proteoliposomes. Thus, it is essential for Mn2+ transport into cells and boosts the metals’ apparent affinity for cells40. Beyond these distinctions, PWY differences were mostly not significant. Differences were showed in PWY related to carbon metabolism and anthropogenic contaminants. The contribution of Mn to these PWY can be associated to the process of metals release induced by anthropogenic pollutants presence (e.g., organic contaminants) from aquifer sediments into groundwater41.
The more in-depth study of the predicted EC linked to Mn metabolism showed intriguing similarities. One of the most prevalent EC predicted with similar abundances in all the samples was cytochrome-c-oxidase, linked to Mn(II) oxidation14. Among the others, superoxide dismutase and peroxiredoxin were also shared by all samples. Superoxide dismutase and peroxiredoxins are linked to the oxidative stress response, mitigating superoxide and hydrogen peroxide excess, respectively, formed by Mn(IV) oxides bioreactions42. Moreover, superoxide dismutase isoenzymes can be overexpressed in different Mn reactions. Ercole et al., for example, reported that two superoxide dismutase isoenzymes are over expressed by Arthrobacter when cells are actively reducing MnO2 under aerobic conditions. Nevertheless, the same proteins are also expressed when reducing MnO4- anaerobically43. Cd2+ exporting ATPase and sirohydrochlorin ferrochelatase, only present in “Grotta Grande dei Cervi” samples, are instead involved in metal stress tolerance44 and transport and cobalamin biosynthesis45. The differences among these enzymes highlighted different capabilities of the communities of the samples to be involved in carbon, sulphur, and nitrogen cycling and metals resistance and transport, without a clear separation between Gorge and Cave samples. These differences are strictly related to the adaptable and diverse nature of the microbes under variable temperature, nutrition availability, and pH within diverse ecological microbial niches, mediated by a spectrum of processes (e.g., DNA damage repair, recombination, conjugation, transformation, and transduction)46.
The PICRUSt 2 software is only a predictive tool of metagenome functions, and some rare environment-specific functions may not be identified. In our case, Mn metabolism functions were identified in line with other studies that described other deposits from extreme environments. Gonzalez-Pimentel et al., for example, compared four lava tubes of caves, identifying biogeochemical methane cycle gene copies16. Moreover, Chung et al. compared eleven sediment samples from the tailings drill cores in a tungsten mine, identifying cellular, environmental, and cellular processes among the others47. Further studies should be carried out to characterize the lithological and geological characteristics of the samples. Datation analyses and mineral characterizations would clarify the microbiological role in the deposition of these deposits and their evolutionary aspects. Comparing other black crusts belonging to different habitats and integrating the analysis with environmental conditions and geological surveys data will help to understand the common and specific Mn deposit formation based on ecosystem type. Only a multidisciplinary approach could unveil some aspects that contributed to the formation of these deposits. All the findings obtained with 16 s RNA gene metabarcoding (and bioinformatic tools) are associated with the moment of sampling and not to the evolution. Few attempts to sequence ancient transcriptomes have been done. However, future studies may be also directed toward RNA-seq analysis to evaluate the actual differentially expressed genes within the active community and validate the obtained findings.
Methods
Sampling
Samplings were carried out in two different major areas, the “Infernaccio” gorge (Viterbo, Italy) and the two cave systems (“Grotta Grande dei Cervi”—inactive speleogenesis—and “Ovito di Pietrasecca”—active speleogenesis) of the central portion of the Monti Carseolani ridge (Central Apennines, Italy). The complete description and photos of the sampling sites are provided in Supplementary information S1. Along the walls of gorge, four different samples were taken (G1-G4), collecting many replicates possible in the same sterile container. At sampling time, the temperature of the “Infernaccio” gorge (Supplementary Fig. S5) was 18 °C with a relative humidity of 60% (samples pH = 6.7). Same procedure was carried out for samples of “Grotta Grande dei Cervi” (sample C1 and C2) and “Ovito di Pietrasecca” (sample C3 and C4) caves (Supplementary Fig. S6), physicochemical parameters of the sampling sites were respectively 8 °C and 98% of relative humidity (sample pH of 7.1). Except for samples G1 and G3, black corroded sediments, all samples had solid structures (details summarized in Supplementary Table S10). All the samples were collected using a geologist hammer for rock samples from the gorge and sterile tools. All specimens were transported into a portable refrigerator from the sampling areas to the laboratory. Aliquots for molecular analyses were processed before storage. For each sample, five aliquots were selected, pooled together, and transferred into a solution of RNAlater (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Aliquots for microscopic observations were transferred in sterile bags without treatments. All the samples were stored at − 80 °C until analysed.
Sample characterization by SEM–EDS
Scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDS) were used to examine black deposits' morphological and geochemical properties from hypogean and epigean samples. Samples were examined by a Gemini500 scanning electron microscope (SEM) (Zeiss, Germany), and EDS microanalysis was carried out by an INCA X-ACT PELTIER COOLED detector (Aztec Energy, Oxford). SEM–EDS acquisitions were performed at different values of Electron High Tension (EHT), and Work Distance (WD) based on the sample observed (details are shown in the figures’ captions).
DNA extraction and 16S rRNA analysis
Genomic DNA extraction was carried out, taking 500 mg of homogeneous samples, and extracting DNA with bead beating techniques according to the manufacturer’s protocol of the kit NucleoSpin®Soil (Macherey Nagel, Germany). Extracted samples were spectrophotometric and fluorometrically checked to evaluate DNA content and purity using a Nanodrop spectrophotometer (Thermo ScientificTM) and a Qubit fluorometer (Thermo ScientificTM). The different replicates were pooled together in an equimolar mixture for each sample. A specific 16S rRNA protocol was followed to amplify bacteria and archaea, using paired-end 16S rRNA community sequencing on the MiSeq Illumina platform (Bio-Fab Research, Italy). We focused on the V3 and V4 regions of 16S rRNA using the methodological approach previously described12. The reads were initially checked for quality and counted after filtering. QIIME2 (qiime2-2020.2 version; https://qiime2.org/)13 was used for ASV (Amplicon Sequence Variant) assembly with the DADA2 plugin version 1.18 (https://benjjneb.github.io/dada2/). From the 16S file obtained from the SILVA 132 database (https://www.arb-silva.de/ accessed on October 2021), the V3-V4 specific region was extracted and used for classifier training by the fit-classifier-naive-bayes plugin. Species accumulation curve and number of non-chimeric ASVs are presented in Supplementary Fig. S7 and Supplementary Table S9.
Metagenome functions prediction
Based on 16S rRNA gene sequencing data, functional abundances of microbial communities were predicted by PICRUSt2 software (version 2.0; https://huttenhower.sph.harvard.edu/picrust/)11. PICRUSt2 predictions were based on ASVs sequence profiles/abundances (BIOM file format obtained from qiime2 with a frequency filter of 100). Three gene family databases were used, Kyoto Encyclopedia of Genes and Genomes (KEGG), orthologs (KO), Enzyme Commission numbers (EC) and MetaCyc pathways abundances48,49,50. PICRUSt2 outputs (KO, EC, PWY) were analysed by QIIME2 software. The presence of enzymes involved in redox biotransformation, metal resistance, and respiratory processes was also verified in the EC outputs to learn more about these activities following (Supplementary Table S11)14.
Statistical analysis
Primer statistical software (version 7; https://www.primer-e.com/our-software/primer-version-7/) was used to calculate alpha-diversity metrics (i.e., Simpson, Shannon and Chao1 indices) and build taxonomy barplots of ASVs at Phylum, Class, and Genus levels. To underline the common ASVs within the different samples, Venn diagrams were built by Bioinformatics & Evolutionary Genomics tool (https://bioinformatics.psb.ugent.be/webtools/Venn/). PICRUSt outputs were analysed by the ALDEx2 package (version 1.29.2; https://bioconductor.unipi.it/packages/3.15/bioc/html/ALDEx2.html)51,52,53 to perform comparison tests between gorge and cave samples (generalized linear model—glm) and to plot the results into effect (MW) plot for each PICRUSt2 output. We underlined the most representative metabolic pathways and their correlation between the different samples, generating a heatmap elaborated with Primer 7 software.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. The nucleotide sequences of the partial 16S rRNA gene segments determined in this study have been deposited in the Sequence Read Archive of NCBI database repository, BioProject: PRJNA838991 (http://www.ncbi.nlm.nih.gov/bioproject/838991).
References
Spiro, T. G., Bargar, J. R., Sposito, G. & Tebo, B. M. Bacteriogenic manganese oxides. Acc. Chem. Res. 43, 2–9. https://doi.org/10.1021/ar800232a (2010).
Roitz, J. S., Flegal, A. R. & Bruland, K. W. The biogeochemical cycling of manganese in San Francisco Bay: Temporal and spatial variations in surface water concentrations. Estuar. Coast. Shelf Sci. 54, 227–239. https://doi.org/10.1006/ecss.2000.0839 (2002).
Tebo, B. M. et al. Biogenic manganese oxides: Properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 32, 287–328. https://doi.org/10.1146/annurev.earth.32.101802.120213 (2004).
Sujith, P. P. & Bharathi, P. A. L. Manganese oxidation by bacteria: Biogeochemical aspects. Prog. Mol. Subcell Biol. 52, 49–76. https://doi.org/10.1007/978-3-642-21230-7_3 (2011).
Lovley, D. R. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55, 259–287. https://doi.org/10.1016/S0065-2911(04)49005-5 (1991).
Tebo, B. M., Johnson, H. A., McCarthy, J. K. & Templeton, A. S. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 13, 421–428. https://doi.org/10.1016/j.tim.2005.07.009 (2005).
Carmichael, S. K. & Bräuer, S. L. 7. Microbial Diversity and Manganese Cycling: A Review of Manganese-oxidizing Microbial Cave Communities. in Microbial Life of Cave Systems (ed. Wagner, D.) 137–160 (DE GRUYTER, 2015). https://doi.org/10.1515/9783110339888-009.
Spilde, M. N. et al. Geomicrobiology of cave ferromanganese deposits: A field and laboratory investigation. Geomicrobiol. J. 22, 99–116. https://doi.org/10.1080/01490450590945889 (2005).
Northup, D. E. et al. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ. Microbiol. 5, 1071–1086. https://doi.org/10.1046/j.1462-2920.2003.00500.x (2003).
Northup, D. E. et al. Lava cave microbial communities within mats and secondary mineral deposits: Implications for life detection on other planets. Astrobiology 11, 601–618. https://doi.org/10.1089/ast.2010.0562 (2011).
Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. https://doi.org/10.1038/s41587-020-0548-6 (2020).
Vaccarelli, I., Matteucci, F., Pellegrini, M., Bellatreccia, F. & del Gallo, M. Exploring microbial biosignatures in Mn-deposits of deep biosphere: A preliminary cross-disciplinary approach to investigate geomicrobiological interactions in a cave in Central Italy. Front. Earth Sci. 9, 590257. https://doi.org/10.3389/feart.2021.590257 (2021).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. https://doi.org/10.1038/s41587-019-0209-9 (2019).
Newsome, L. et al. Natural attenuation of lead by microbial manganese oxides in a karst aquifer. Sci. Total Environ. 754, 142312. https://doi.org/10.1016/j.scitotenv.2020.142312 (2021).
Farda, B. et al. The “Infernaccio” Gorges: Microbial diversity of black deposits and isolation of manganese-solubilizing bacteria. Biology 11, 1204. https://doi.org/10.3390/biology11081204 (2022).
Gonzalez-Pimentel, J. L. et al. Prokaryotic communities from a lava tube cave in La Palma Island (Spain) are involved in the biogeochemical cycle of major elements. PeerJ 9, e11386. https://doi.org/10.7717/peerj.11386 (2021).
Kulkarni, H. V., Ford, J., Blank, J. G., Park, M. & Datta, S. Geochemical interactions among water, minerals, microbes, and organic matter in formation of speleothems in volcanic (lava tube) caves. Chem. Geol. 594, 120759. https://doi.org/10.1016/j.chemgeo.2022.120759 (2022).
Maciejewska, M. et al. Assessment of the potential role of streptomyces in cave moonmilk formation. Front. Microbiol. 8, 1181. https://doi.org/10.3389/fmicb.2017.01181 (2017).
Suchý, V. et al. Microbial signatures from speleothems: A petrographic and scanning electron microscopy study of coralloids from the Koněprusy Caves (the Bohemian Karst, Czech Republic). Sedimentology 68, 1198–1226. https://doi.org/10.1111/sed.12826 (2021).
Bernardini, S. et al. Morpho-mineralogical and bio-geochemical description of cave manganese stromatolite-like patinas (Grotta del Cervo, Central Italy) and hints on their paleohydrological-driven genesis. Front. Earth Sci. 9, 642667. https://doi.org/10.3389/feart.2021.642667 (2021).
McMurtry, G. M. Deep-Sea Sediment: Authigenic Deposits. in Encyclopedia of Ocean Sciences (eds. Cochran J. K.; Bokuniewicz, H. J. & Yager P. L.)121–132 (Elsevier, 2019). https://doi.org/10.1016/B978-0-12-409548-9.11645-8.
Jiang, X.-D. et al. Biomineralisation of the ferromanganese crusts in the Western Pacific Ocean. J. Asian Earth Sci. 136, 58–67. https://doi.org/10.1016/j.jseaes.2017.01.025 (2017).
Piazza, A. et al. Environmental bacteria involved in manganese(II) oxidation and removal from groundwater. Front. Microbiol. 10, 119. https://doi.org/10.3389/fmicb.2019.00119 (2019).
Zhang, G. Y., He, J. Z., Liu, F. & Zhang, L. M. Iron-manganese nodules harbor lower bacterial diversity and greater proportions of proteobacteria compared to bulk soils in four locations spanning from North to South China. Geomicrobiol. J. 31, 562–577. https://doi.org/10.1080/01490451.2013.854428 (2014).
Romano, C. A. et al. Biogenic manganese oxide nanoparticle formation by a multimeric multicopper oxidase Mnx. Nat. Commun. 8, 1–8. https://doi.org/10.1038/s41467-017-00896-8 (2017).
Wang, Y. et al. A culture-independent approach to unravel uncultured bacteria and functional genes in a complex microbial community. PLoS ONE 7, e47530. https://doi.org/10.1371/journal.pone.0047530 (2012).
Puig-Castellví, F. et al. Assessment of the microbial interplay during anaerobic co-digestion of wastewater sludge using common components analysis. PLoS ONE 15, e0232324. https://doi.org/10.1371/journal.pone.0232324 (2020).
Li, Y. et al. High-efficiency methanogenesis via kitchen wastes served as ethanol source to establish direct interspecies electron transfer during anaerobic Co-digestion with waste activated sludge. Water Res. 176, 115763. https://doi.org/10.1016/j.watres.2020.115763 (2020).
Lusa, M. & Bomberg, M. Microbial community composition correlates with metal sorption in an ombrotrophic boreal bog: Implications for radionuclide retention. Soil Syst. 5, 19. https://doi.org/10.3390/soilsystems5010019 (2021).
Cohen, A. B. et al. Particle-associated and free-living microbial assemblages are distinct in a permanently redox-stratified freshwater lake. bioRxiv 2021.11.24.469905. https://doi.org/10.1101/2021.11.24.469905 (2021) .
Mohagheghi, A., Grohmann, K., Himmel, M., Leighton, L. & Updegraff, D. M. Isolation and Characterization of Acidothermus cellulolyticus gen. nov., sp. nov., a New Genus of Thermophilic, Acidophilic, Cellulolytic Bacteria. Int. J. Syst. Bacteriol. 36, 435–443. https://doi.org/10.1099/00207713-36-3-435 (1986).
Polymenakou, P. N., Lampadariou, N., Mandalakis, M. & Tselepides, A. Phylogenetic diversity of sediment bacteria from the southern Cretan margin, Eastern Mediterranean Sea. Syst. Appl. Microbiol. 32, 17–26. https://doi.org/10.1016/j.syapm.2008.09.006 (2009).
Manikkam, R. et al. Metagenomic insights unveil the dominance of undescribed Actinobacteria in pond ecosystem of an Indian shrine. Meta Gene 23, 100639. https://doi.org/10.1016/j.mgene.2019.100639 (2020).
Cheng, Q., Huang, Y., Nengzi, L. & Zhang, J. Performance and microbial community profiles in pilot-scale biofilter for the simultaneous removal of ammonia, iron and manganese at different manganese concentrations. Bioprocess Biosyst. Eng. 42, 741–752. https://doi.org/10.1007/s00449-019-02077-x (2019).
Lücker, S. et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. 107, 13479–13484. https://doi.org/10.1073/pnas.1003860107 (2010).
Zhenggang, X. et al. Biosorption characteristics of Mn (II) by Bacillus cereus strain HM-5 isolated from soil contaminated by manganese ore. Pol. J. Environ. Stud. 28, 463–472. https://doi.org/10.15244/pjoes/84838 (2018).
Holmes, A. J. et al. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves Australia. Environ. Microbiol. 3, 256–264. https://doi.org/10.1046/j.1462-2920.2001.00187.x (2001).
Zhu, H.-Z. et al. Diversity, distribution and co-occurrence patterns of bacterial communities in a Karst cave system. Front. Microbiol. 10, 1726. https://doi.org/10.3389/fmicb.2019.01726 (2019).
Jurado, V. et al. Microbial communities in vermiculation deposits from an Alpine Cave. Front. Earth Sci. 8, 586248. https://doi.org/10.3389/feart.2020.586248 (2020).
Schabereiter-Gurtner, C., Saiz-Jimenez, C., Pinar, G., Lubitz, W. & Rolleke, S. Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ. Microbiol. 4, 392–400. https://doi.org/10.1046/j.1462-2920.2002.00303.x (2002).
Hohle, T. H., Franck, W. L., Stacey, G. & O’Brian, M. R. Bacterial outer membrane channel for divalent metal ion acquisition. Proc. Natl. Acad. Sci. 108, 15390–15395. https://doi.org/10.1073/pnas.1110137108 (2011).
Zhai, Y. et al. Anthropogenic organic pollutants in groundwater increase releases of Fe and Mn from aquifer sediments: Impacts of pollution degree, mineral content, and pH. Water 13, 1920. https://doi.org/10.3390/w13141920 (2021).
Learman, D. R., Voelker, B. M., Madden, A. S. & Hansel, C. M. Constraints on superoxide mediated formation of manganese oxides. Front. Microbiol. 4, 262. https://doi.org/10.3389/fmicb.2013.00262 (2013).
Ercole, C., Altieri, F. & Piccone, C. Influence of manganese dioxide and manganic ions on the production of two proteins in Arthrobacter sp. Geomicrobiol. J. 16, 95–103. https://doi.org/10.1080/014904599270785 (1999).
Solioz, M. & Vulpe, C. CPx-type ATPases: A class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21, 237–241. https://doi.org/10.1016/S0968-0004(96)20016-7 (1996).
Leech, H. K., Raux-Deery, E., Heathcote, P. & Warren, M. J. Production of cobalamin and sirohaem in Bacillus megaterium: an investigation into the role of the branchpoint chelatases sirohydrochlorin ferrochelatase (SirB) and sirohydrochlorin cobalt chelatase (CbiX). Biochem. Soc. Trans. 30, 610–613. https://doi.org/10.1042/bst0300610 (2002).
Chung, A. P. et al. Tailings microbial community profile and prediction of its functionality in basins of tungsten mine. Sci. Rep. 9, 19596. https://doi.org/10.1038/s41598-019-55706-6 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. https://doi.org/10.1093/nar/gkaa970 (2021).
Fernandes, A. D., Macklaim, J. M., Linn, T. G., Reid, G. & Gloor, G. B. ANOVA-like differential gene expression analysis of single-organism and meta-RNA-Seq. PLoS ONE 7, e67019. https://doi.org/10.1371/journal.pone.0067019 (2013).
Fernandes, D. A. et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2, 15. https://doi.org/10.1186/2049-2618-2-15 (2014).
Gloor, G. B., Macklaim, J. M. & Fernandes, A. D. Displaying variation in large datasets: A visual summary of effect sizes. J. Comput. Graph. Stat. 25, 971–979. https://doi.org/10.1080/10618600.2015.1131161 (2016).
Acknowledgements
The authors wish to thank Lorenzo Arrizza for SEM-EDS analysis, the “uPIX—uNDER PIXel Fotografia Ipogea” for the cave pictures, and Simone Chiani for the gorge pictures.
Author information
Authors and Affiliations
Contributions
M.P. conceived the experiment. All authors contributed to the study design. Material preparation and data collection were performed by B.F., I.V. and R.D. Data analysis was performed by B.F. and M.P. The first draft of the manuscript was written by B.F., I.V. and C.E. All authors commented on previous versions of the manuscript. Financial support was provided by M.D.G. The final version of the manuscript was revised by M.P. and M.D.G. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Farda, B., Vaccarelli, I., Ercole, C. et al. Exploring structure, microbiota, and metagenome functions of epigean and hypogean black deposits by microscopic, molecular and bioinformatic approaches. Sci Rep 12, 19405 (2022). https://doi.org/10.1038/s41598-022-24159-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24159-9
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.