Abstract
Cardiac Magnetic Resonance Fingerprinting (cMRF) has been demonstrated to enable robust and accurate T1 and T2 mapping for the detection of myocardial fibrosis and edema. However, the relatively long acquisition window (250 ms) used in previous cMRF studies might leave it vulnerable to motion artifacts in patients with high heart rates. The goal of this study was therefore to compare cMRF with a short acquisition window (154 ms) and low-rank reconstruction to routine cardiac T1 and T2 mapping at 1.5 T. Phantom studies showed that the proposed cMRF had a high T1 and T2 accuracy over a wider range than routine mapping techniques. In 9 healthy volunteers, the proposed cMRF showed small but significant myocardial T1 and T2 differences compared to routine mapping (ΔT1 = 1.5%, P = 0.031 and ΔT2 = − 7.1%, P < 0.001). In 61 consecutive patients referred for CMR, the native T1 values were slightly lower (ΔT1 = 1.6%; P = 0.02), while T2 values did not show statistical difference (ΔT2 = 4.3%; P = 0.11). However, the difference was higher in post-contrast myocardial T1 values (ΔT1 = 12.3%; P < 0.001), which was reflected in the extracellular volume (ΔECV = 2.4%; P < 0.001). Across all subjects, the proposed cMRF had a lower precision when compared to routine techniques, although its higher spatial resolution enabled the visualization of smaller details.
Similar content being viewed by others
Introduction
Over the past decade, the field of cardiac magnetic resonance (CMR) saw an increasing interest in the measurement of myocardial T1 and T2 relaxation times1,2,3, and multiple studies have now shown the added diagnostic value of myocardial T1 and T2 values5. Native T1 mapping (i.e. without a gadolinium-based contrast agent—GBCA) has become a widely used technique to detect interstitial fibrosis6,5,8, while T1 mapping after the administration of GBCA provides unique additional prognostic information through the estimation of the extracellular volume (ECV) fraction9,8,11. Increased myocardial T2 values similarly provide a quantitative measure of myocardial edema in various pathologies12,13.
While these findings are highly encouraging, the clinical adoption of parametric mapping is hindered by relatively long acquisition times and ongoing discussions about accuracy, precision, robustness, and sensitivity5. These challenges have encouraged the exploration of new mapping techniques14,13,14,15,16,19. It would especially be desirable to develop a fast and reliable method capable of simultaneously quantifying both T1 and T2 relaxation times, in order to save time and to remove the bias that one parameter can have on the estimation of the other.
Magnetic Resonance Fingerprinting (MRF) has been proposed20 as an alternative approach to parameter mapping that enables the simultaneous quantification of multiple relaxation times. The idea behind MRF is to deliberately vary the pulse sequence parameters in order to induce a unique signal evolution that depends exclusively on the tissue properties under investigation (e.g. T1 and T2). After acquiring data with a highly undersampled k-space sampling trajectory, a pattern matching algorithm is used to match each voxel’s time course to an entry in a dictionary containing simulated MRF time courses. The best-matching dictionary entries are used to populate the parametric maps with the corresponding values. The accuracy and efficiency of this approach have been demonstrated for stationary tissues such as the brain20,21 and prostate22.
The use of MRF for parametric mapping in the heart, however, faces additional challenges due to respiratory and especially cardiac motion. In order to address these challenges, cardiac MRF (cMRF) makes use of breath holds and electrocardiographic (ECG) triggering. Similar to established myocardial mapping techniques such as MOLLI23, SASHA16 or T2-prepared bSSFP24, cMRF data are acquired during an acquisition window in the mid-diastolic phase of each heartbeat, over the span of a single breath hold. This results in a shorter and interrupted acquisition, which limits the amount of signal that can be matched to the dictionary as well as the variation in the signal, which is required for accurate pattern matching. At the same time, since the duration of the mid-diastolic quiescent period depends on the subject’s heart rate, a longer acquisition window is not always possible in the presence of pathologies. Most MRF applications in stationary organs (e.g. brain) use sequence lengths of 1000 TRs or more, and many prior cMRF studies have used a scan duration of 15–16 heartbeats with a 250 ms acquisition window, leading to approximately 750 TRs that can be used for fitting. Reducing the acquisition window to 150 ms would be especially desirable in patients with high heart rates. However, this would result in a sequence length of approximately 400 TRs, which limits the amount of cMRF data that can be collected and in turn may result in a loss of measurement precision. To counter the expected loss in precision caused by shortening the acquisition window, the intrinsic redundancy and similarity of the magnetization time courses can be exploited. Instead of using the full dictionary, the redundancy allows for the use of a low-rank approximation of the dictionary, which is a highly efficient regularization constraint for iterative reconstruction. Such a low-rank iterative reconstruction can thus be used to reduce noise and aliasing artifacts and to compensate for the precision loss25,24,27.
The feasibility of a cMRF sequence that combines such a short acquisition window and low-rank reconstruction has recently been presented in healthy volunteers28. Therefore, the goal of this study was to assess the robustness of the technique in a routine clinical setting by comparing its accuracy and precision to routine myocardial T1 and T2 mapping in a cohort of patients referred for clinical CMR for a wide range of indications.
Results
Phantom studies
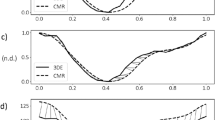
When considered over the entire tested range, the phantom mapping demonstrated similar or higher T1 and T2 accuracy for cMRF than the routine mapping techniques (Fig. 1A,B, Suppl. Fig. 1). In the T1 vials, the average bias and 95% limits of agreement obtained from the Bland–Altman analysis were equal to 12.81 ± 13.34% for cMRF with a 154 ms acquisition window and 13.03 ± 12.15% for the cMRF sequence with a 250 ms acquisition window (Fig. 1A). The two MOLLI variants had overall smaller biases and comparable limits of agreement, and the errors were equal to 5.38 ± 8.63% and 0.78 ± 12.39% for 5(3)3-MOLLI and 4(1)3(1)2-MOLLI, respectively (Fig. 1B). The same analysis was performed for T2 relaxation times and confirmed the overall good agreement with reference measurements. The biases and limits of agreement were respectively 1.55 ± 9.21% and 6.76 ± 11.93% for the two variants with shorter and longer acquisition window (Fig. 1C). T2-prepared bSSFP (Fig. 1D) showed a similar bias, but a considerably larger confidence interval: 8.76 ± 48.9%. The short-acquisition-window cMRF reconstruction time was 297 ± 20 s.
Accuracy of cMRF compared to reference techniques in the ISMRM-NIST phantom. (A) Both cMRF versions showed good agreement with one another and the bias was around 10% across a wide range of T1 values. (B) The two MOLLI sequences maintained a low bias, with 5(3)3-MOLLI performing slightly better at higher T1 values, as expected. (C) The cMRF T2 measurement were consistent between the two variations, with the shorter acquisition window showing lower bias overall. (D) T2-prepared bSSFP had higher error than cMRF, likely due to the high T1 values of the NIST phantom. The 95% confidence intervals are reported as matching semi-transparent colored bands in each plot, and the same color scheme was used to report the biases.
For both the T1 and T2 relaxation times, the heart-rate influence phantom study showed no significant variation or trend in the resulting T1 and T2 values as the R-R interval was increased (Suppl. Fig. 2). For the T1 relaxation times, the CoV for both cMRF variants was below 2%, which was comparable to that of 5(3)3-MOLLI and slightly better than that of 4(1)3(1)2-MOLLI. For the T2 relaxation times, the CoV was higher, and ranged between 2 and 8% across the relevant T2 values (Suppl. Fig. 2C,D). For both relaxation times, no significant difference was observed between the cMRF sequences with the shorter and longer acquisition windows. Moreover, for both cMRF variations, and both T1 and T2, the ANCOVA analysis found no significant difference between the linear regression slopes obtained at different heart-rates (all P values > 0.05, Suppl. Fig. 3A–D).
Healthy volunteers
In 9 healthy volunteers (age 25 ± 2 years, 66% female, heart rate 69 ± 17 bpm, ranging from 53 to 111), cMRF resulted in similar relaxation times with a slightly lower precision compared to routine techniques (Fig. 2). The inter-observer variability showed good agreement between two different observers: whole-heart T1 and T2 values were not significantly different for cMRF150ms (P = 0.148 and P = 0.535, respectively). A small but significant bias was found when analyzing the cMRF250ms T1 maps (P = 0.008, bias = 8.6 ms), while whole-heart T2 distributions did not differ significantly between the two observers (P = 0.177).
Circumferential polar plots of the myocardial relaxation times in 9 healthy volunteers. Myocardial areas were segmented according to the AHA guidelines29. Each segment reports the average relaxation time and its standard deviation across all volunteers in milliseconds. cMRF with a shorter acquisition window is shown in the first row (A and B); cMRF with a longer acquisition window is shown in the second row (B and C) routine T1 and T2 values are reported in the third row (E and F, respectively). No significant differences were found between the two variations of cMRF as well as between cMRF with a short acquisition window and routine techniques. Significant differences were found exclusively in the mid and apical slices between routine and cMRF with a longer acquisition window, as highlighted (*P < 0.05).
Averaged over the three slices and the nine volunteers, the myocardial T1 values were slightly higher and varied more for both versions of cMRF when compared to MOLLI (cMRF154ms: 1020 ± 45 ms; cMRF250ms: 1026 ± 34 ms; MOLLI: 1005 ± 20 ms). Both techniques also differed significantly from MOLLI (P = 0.031 and P = 0.002, respectively for cMRF154ms and cMRF250ms). No significant difference was found between the two cMRF variants (P > 0.999). When compared on a per-segment basis, the T1 difference was significant in the septum of the mid slice between cMRF250ms and MOLLI (segment 12; Fig. 2C–E). No significant difference was observed between the two cMRF variants and between cMRF154ms and MOLLI. Overall T2 values, on the contrary, were lower in cMRF compared to T2prep-bSSFP, and confirmed the trend observed in vitro (cMRF154ms: 42.4 ± 2.4 ms; cMRF250ms: 38.5 ± 2.6 ms; T2prep-bSSFP: 45.4 ± 2.0 ms). All whole-heart distributions were significantly different (P < 0.001 for all comparisons). When comparing the segments, significant difference was found between cMRF250ms and T2prep-bSSFP in the lateral part of all three slices (Fig. 2C–F). The per-segment precision was higher in routine techniques: cMRF with 154 ms acquisition window resulted in a T1 CoV of 7.9 ± 2.0% and T2 CoV of 7.7 ± 3.3%. cMRF with 250 ms acquisition window resulted in a T1 CoV of 6.8 ± 1.8% and T2 CoV of 8.1 ± 3.2%. MOLLI resulted in a T1 CoV of 5.3 ± 0.9% and T2prep-bSSFP in a T2 CoV of 6.5 ± 2.0%.
Patient scans
Scans were completed in n = 61 patients referred for CMR (Suppl. Fig. 4); in one patient, all mapping failed due to the presence of an implantable cardioverter-defibrillator (ICD). Native cMRF was completed in 59 patients; in one case, it failed due a sudden deep breath of the patient (not noticed until the maps were reconstructed). Native routine T1 and T2 mapping were completed in 58 and 13 subjects and failed in two and one cases respectively due to bSSFP banding artifacts. Post-contrast cMRF and routine T1 mapping were completed in 39 subjects, while for the latter, two acquisitions failed due to bSSFP banding artifacts. This resulted in n = 59 for native cMRF, n = 58 for native T1 mapping, n = 13 for native T2 mapping, n = 39 for post-contrast cMRF, and n = 37 for post-contrast T1 mapping.
Overall, we observed a robust performance of cMRF with a low number of failed acquisitions. We anecdotally observed that the higher spatial resolution of cMRF allowed for the easier identification of small and non-transmural lesions in several patients (Fig. 3, Suppl. Fig. 5). cMRF also still resulted in accurate maps in the presence of large respiratory drifts, albeit producing visibly noisier maps (Fig. 4).
T1 and T2 maps in a 40 y.o. patient with viral myocarditis and old sub-epicardial scar in the basal infero-lateral and inferior wall. (A,F) Short- and long-axis late-gadolinium-enhanced (LGE) images showing the presence of the subtle subepicardial scar. (B,C) Native T1 maps. The routine T1 map had slightly higher overall T1 myocardial values. (D,E) T1 maps 20–25 min after contrast agent injection. We can observe that the small non-transmural subepicardial lesion is easier to identify in the cMRF scan. (G,H) ECV maps obtained with cMRF and routine technique. The sub-epicardial scar is visible in the cMRF ECV map.
Respiratory drift resulted in noisier cMRF T1 and T2 maps in a 35 y.o. patient with hypertrophic CMP. The patient did not comply with the breath holding protocol and slowly breathed out during several acquisitions. (A,B) T1 maps before contrast agent injection. The patient complied with the very first breath hold (the routine T1 map), but not the subsequent cMRF: its T1 map appears noisy and its borders are poorly defined. The myocardial T1 values are 1079 ± 99 ms for cMRF and 1119 ± 69 ms for MOLLI. (C,D) T1 maps 20–25 min after contrast agent injection. Despite some respiratory drift, the routine T1 map source images were correctly aligned during registration and resulted in a precise map. Also note the higher T1 values in the myocardium of the routine map (404 ± 39 ms for cMRF and 447 ± 29 ms for MOLLI). (E,F) Native T2 maps. Slightly higher regional variability can be observed in the routine map, and the total myocardial area appears smaller due to poor registration of the source images. The averages across the myocardium were 46 ± 5 ms for cMRF and 51 ± 5 ms for T2prep-bSSFP.
The agreement between cMRF and routine techniques seen in the phantom and healthy volunteer studies was confirmed in the native myocardial T1 relaxation times (cMRF: 1015 ± 61 ms; MOLLI: 1029 ± 53 ms; ΔT1 = 1.6%; P = 0.02). A small but significant difference was found in the inferior and inferolateral segments as well as in the blood pool, where cMRF measured lower T1 values than MOLLI (Figs. 5A, 6A,B, 7). cMRF showed a substantial difference from the routine post-contrast myocardial T1 value (cMRF: 395 ± 50 ms; MOLLI: 444 ± 53 ms; ΔT1 = 12.3%; P < 0.001, Fig. 6C), which was consistent in all segments (Fig. 5B). Contrary to this, the post-contrast blood-pool T1 value had similar average values (cMRF: 279 ± 45 ms; MOLLI: 289 ± 54 ms; ΔT1 = 3.6%; P < 0.001, Fig. 6D). The different post-contrast myocardial (and the native blood pool) T1 values were reflected in a higher ECV than that established with MOLLI (cMRF: 28.4 ± 4.8%; MOLLI: 26.0 ± 4.0%; ΔECV = 8.4%; P < 0.001, Figs. 5C, 6F). In the myocardium, T2 values obtained with cMRF were marginally lower compared to those obtained with T2-prep bSSFP, but no significant difference was observed (cMRF: 44.5 ± 5.5 ms; T2-prep bSSFP: 46.4 ± 3.7 ms; ΔT2 = 6.1%; P = 0.11; Figs. 5D, 6E). The segmental precision of cMRF was again lower than that of the routine techniques for the native T1 (cMRF: 12.0 ± 3.7%; MOLLI: 6.8 ± 1.5%), post-contrast T1 (cMRF: 10.6 ± 3.1%; MOLLI: 7.4 ± 2.5%), and T2 values (cMRF: 12.3 ± 2.9%; T2prep-bSSFP: 9.1 ± 1.7%); all with P < 0.001. Finally, none of measured relaxation times (routine or cMRF) resulted in a significant linear regression with the individual patient heartrate (P > 0.14 for all, Suppl. Fig. 6).
Box and whiskers plots of segmental relaxation times in the myocardium of the patient group. Boxes are medians with interquartile ranges; whiskers indicate the minimum and maximum values. The segments that showed a statistically significant difference (P < 0.05) are outlined in red. (A–C) Native T1, post-contrast T1 and synthetic ECV measured with short-acquisition-window cMRF (left, blue) and MOLLI (right segment, orange and green). cMRF and routine techniques have similar ranges. (D) Native T2 measured with cMRF (left, blue) and T2prep bSSFP (right, red). The numbers indicate the AHA segment number and ‘avg’ is the average across the entire myocardium.
Bland–Altman analyses of short-acquisition-window cMRF and routine techniques for the entire visible left-ventricular myocardium and the left-ventricular blood pool in patients. (A,B) Native T1 relaxation times obtained with cMRF vs. 5(3)3-MOLLI in the myocardium and blood pool areas, respectively. (C,D) Post-contrast T1 relaxation times obtained with cMRF vs. 4(1)3(1)2-MOLLI. A significant bias was observed, while the difference in the post-contrast LV blood T1 was smaller between the two techniques. (E) Native T2 relaxation times obtained with cMRF vs. T2-prepared bSSFP. (F) Synthetic ECV calculated with cMRF vs. MOLLI. P-values indicate the significance of the unpaired t-test; P < 0.05 is highlighted in red. None of the measured parameters showed significant trends or changes with increasing value.
Myocardial relaxation times and ECV measured with short-acquisition-window cMRF and routine techniques. Similar native relaxation times were measured with all techniques in both the healthy volunteers and patients. However, the bias in the post-contrast myocardial T1 values was higher between cMRF and MOLLI, which together with similar post-contrast blood T1 values resulted in slightly different ECV estimations. All P values are reported on the plots.
Discussion
Cardiac MR fingerprinting with a short acquisition window and low-rank reconstruction28 was characterized in vitro, in healthy volunteers and as part of a clinical CMR protocol to assess its accuracy and precision compared to routine T1 and T2 mapping techniques.
In vitro, cMRF had slightly lower accuracy than routine techniques when tested against reference relaxation times spanning from very low (90 ms) to very high (1900 ms) T1 values. T1 values in the native myocardial range were higher for both cMRF sequences, while excellent agreement was observed for the entire T2 range. Despite the small size of these differences, they were interestingly opposite to what was found in vivo, where good agreement was found between the native T1 values (around 1000 ms) of both healthy volunteers and patients, and lower values compared to MOLLI were found after GBCA injection. We observed high consistency between the longer and shorter cMRF sequences both in vitro and in vivo, with the measured relaxation times often overlapping. The study of heart-rate variability influence in the phantom also showed consistent values for both techniques across a wide range of relaxation times and simulated heart-rates. The slightly higher heartrate CoVs for the cMRF techniques than the routine technique over the entire heartrate range are still lower than a single segmental myocardial T2 standard deviation observed in vivo, from which we estimate that they can be ignored.
The differences between the cMRF and the IR-TSE reference likely have several causes, such as using a TSE instead of an SE sequence, and not including the slice profile of the RF pulse30, B0 and B1+ inhomogeneities31, and inversion efficiency17 in the fitting model of IR-TSE reference, which might lower its accuracy. It should be noted that the RF pulse profile is included in the cMRF dictionary, and that the presented cMRF technique used adiabatic inversion and refocusing pulses, as opposed to the IR-TSE and SE techniques, making it less susceptible to B1 inhomogeneities. Moreover, the T1/T2 ratios in the phantom vary over a much broader range than in vivo. Given these sources for mismatching and discrepancies, it is encouraging to note that the overestimation in vitro still consistently remained below 10%.
Both in the healthy volunteer group and in the patients, cMRF showed a high agreement and comparable inter-patient standard deviations when compared to MOLLI and T2-prep bSSFP. In the patient group, the majority of the subjects was referred with suspected and not confirmed disease, and did not have significantly altered myocardial relaxation times with any of the used techniques. The global average across all patients was therefore close to that of the healthy volunteers, with virtually identical average T1 and T2 values measured in both groups. When a subgroup of patients with a specific disease was selected (not reported here), this also held true, although small sub-endocardial infarctions were more clearly delineated in the cMRF maps, most likely due to the higher spatial resolution.
The relaxation times observed here are in line with previous studies where the accuracy and precision of cMRF were established32,31,34. In particular, Hamilton et al. found moderately lower relaxation times and equivalent standard deviations (964 ± 71 ms and 41.2 ± 4.2 ms for T1 and T2, respectively) in fifty healthy volunteers at 1.5 T32. In twenty-four patients with suspected inflammatory cardiomyopathy33, cMRF resulted in very similar myocardial T1 = 1028 ± 64 ms and higher T2 = 52.8 ± 3.8 ms. The same comparison between cMRF and routine techniques was performed pre- and post-contrast in six hypertrophic cardiomyopathy patients and 12 healthy subjects34: small differences in myocardial relaxation times were found between routine techniques and cMRF, although both methods easily allowed for the distinction between patients and healthy subjects. Interestingly, larger differences were found in the ECV estimation, where cMRF measured consistently higher percentages in both groups.
The larger difference in the post-contrast myocardial T1 values is the main cause of difference in ECV estimation, although it should be noted that Treibel et al. obtained the empirical formula to calculate the hematocrit specifically from MOLLI sequences35. The timings of each post-contrast acquisition with respect to the GBCA injection were verified, as was the accuracy in the delineation of the myocardial area: both were not found to be significantly different between cMRF and routine techniques after verification by a second observer. To the best of our knowledge, neither of these factors alone could substantially influence the myocardial T1 estimation, and an adequate explanation for the mismatch remains to be found.
In both subject groups, the inter-subject standard deviations of cMRF and routine techniques were similar and between 5 and 12% of the measured value. The individual segmental precision of cMRF is in line with previous studies32,36,37. While it is lower than its routine counterparts, this is balanced with the 1.5-fold to 2.4-fold smaller pixel size of cMRF as well as the smoothing effects of the non-rigid motion registration techniques used for routine mapping techniques38. This suggests that it might be of interest to integrate a spatial regularization filter or a denoising step in the reconstruction pipeline of cMRF data27, while the cMRF spatial resolution could be lowered to that of the routine scans, if desired. This may result in a higher precision for cMRF, while leaving the other advantages of cMRF intact: acquisition of both maps at half the time, using the same protocol for all T1 (and T2) ranges, and intrinsic co-registration of the T1 and T2 maps5. If a larger spatial coverage is desired, the proposed shorter acquisition window could for example be combined with a simultaneous multislice (SMS) acquisition39, although the accuracy and precision of such a technique would need to be established. Similarly, a 3D free-breathing technique40 could be used if extra scan time can be allocated to obtain whole-heart coverage, which may especially be of use in diseases that manifest in unpredictable patchy patterns.
The main limitation of this study lies with the broad variety of scanned patients. While this variety is a strength of the study in that it enabled us to characterize cMRF in a cohort that represents a real heterogeneous clinical population, it did not result in a narrow range of relaxation times that was significantly different from healthy volunteers. This effect was further exacerbated by the patients only having suspected and not confirmed cardiac disease, and by the standardized segmental analysis that might average out any small regional T1 or T2 elevations. Therefore, it might be of interest to further characterize this technique in cohorts of patients with a specific and confirmed disease. In two of the aforementioned studies33,34, a blinded evaluation of cMRF against MOLLI and T2prep-bSSFP was performed, where cMRF compared favorably against both techniques. This might also be incorporated in future work to provide a better overall understanding of the diagnostic potential of cMRF.
In conclusion, the comparison of cMRF with a short acquisition window with routine myocardial T1 and T2 mapping techniques showed a robust performance in vitro, in healthy volunteers, and in a group of CMR patients covering a broad range of pathologies. The T1 and T2 maps generated from the single cMRF acquisition were generally comparable in accuracy, slightly less precise, and had a higher spatial resolution than those obtained with routine T1 and T2 mapping techniques. A notable exception was found in a lower estimation of the post-contrast myocardial T1 relaxation time, which led to a small but significantly higher ECV.
Materials and methods
The local ethical committee (Commission cantonale d'éthique de la recherche sur l'être humain du canton de Vaud CER-VD, Lausanne, Switzerland) approved all in vivo studies under protocol number 2018-00656 (principal investigator JS), all studies were performed in accordance with the relevant guidelines and regulations, and all participants provided written informed consent. All scans were performed on a clinical 1.5 T MR scanner (MAGNETOM Sola, Siemens Healthcare, Erlangen, Germany), using an 18-channel chest coil combined with a 12-channel spine coil.
Phantom study
The accuracy of the proposed cMRF technique was first assessed using the ISMRM-NIST phantom (QalibreMD, Boulder, CO, USA), a 200-mm spherical phantom with two layers comprised of 14 spheres, one with a wide range of T1 values and one with a wide range of T2 values. The T1 array was created by NiCl2-doped deionized water, while the T2 array spheres are filled with MnCl2-doped deionized water. A single slice was planned through the T2 layer of spheres (n = 14), since it contains a broad T1 and T2 range that mimics relevant physiological values. Because all sequences were designed to be accurate within physiological ranges, T1 values lower than 90 ms and higher than 1900 ms were discarded in the analysis. For the same reason, we considered T2 values ranging from 30 to 240 ms. For completeness, the measurements obtained in all 14 vials are reported as supplementary material.
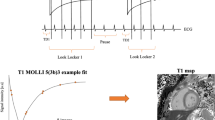
Parameters for the cMRF pulse sequence included: spiral readout, 15-heartbeat breathhold, 48-fold undersampling, field of view (FOV) = 300 × 300 mm2, pixel size = 1.6 × 1.6 mm2, slice thickness = 8 mm, 2.5 µs dwell time, and sampling bandwidth ± 200 kHz. The repetition time (TR) was fixed at 5.3 ms, while the flip angle was varied between 4° and 25°. To facilitate the distinction between different relaxation times, the cMRF sequence incorporates four different magnetization preparation modules: an adiabatic inversion recovery and three adiabatic T2 preparations41 with echo times of 30, 50 and 80 ms. Two variations of the cMRF sequence were considered and compared: one with a longer acquisition window = 250 ms and 48 undersampled image acquisitions per heartbeat and one with the shorter acquisition windows = 154 ms and 29 undersampled image acquisitions per heartbeat. We will differentiate between the two variants by indicating the acquisition window duration in a subscript.
A low-rank reconstruction was used to reduce the dimensionality of the generated dictionaries and reduce aliasing artifacts in the MRF images26,27,39. Similar to previous work, this approach compresses the dictionary along the time dimension by calculating the singular value decomposition of the dictionary and retaining the first five singular values. Images in this low-dimensional subspace are reconstructed using nonlinear conjugate gradient descent with both \({l}_{1}\) wavelet regularization and locally low-rank patch regularization with a patch size of 8 × 8. The dictionary took the slice profile and imperfections due to the efficiency of preparation pulses into account30. All reference and cMRF maps were reconstructed in MATLAB 2019a (The MathWorks, Natick, MA, USA; http://www.matlab.com) on a desktop PC with an i9-9900 CPU @3.10 GHz and integrated Intel UHD Graphics 630, and the reconstruction time was recorded.
As recommended in the phantom manual, reference T1 relaxation times were obtained with an inversion-recovery turbo spin echo (IR-TSE) pulse sequence with nine inversion times (35–3000 ms), TE = 7.9 ms, TR = 4500 ms, and spatial resolution = 1 × 1 mm2. A spin-echo (SE) pulse sequence was used to determine reference T2 values, with 25 echo times (TEs) ranging from 10 to 400 ms, pixel bandwidth = 279 Hz, TR = 5000 ms, spatial resolution = 1 × 1 mm2, and slice thickness = 6 mm.
Three clinical routine parametric mapping techniques were also characterized: 5(3)3-MOLLI (21) for T1 mapping in the native T1 range, 4(1)3(1)2-MOLLI4 for T1 mapping in the post-contrast T1 range (both variants used heartbeats for their timing), and T2-prepared bSSFP for T2 mapping24. 5(3)3-MOLLI and T2-prep bSSFP had FOV = 300 × 300 mm2, pixel size = 2.0 × 1.6 mm2 interpolated to 1.6 × 1.6 mm2, slice thickness = 8 mm. 4(1)3(1)2-MOLLI was performed with FOV = 306 × 360 mm2, pixel size = 2.1 × 1.4 mm2 interpolated to 1.4 × 1.4 mm2, slice thickness = 8 mm. Both MOLLI sequences and T2-prep bSSFP had TE = 1.1 ms. Acquisition window durations for cMRF, 5(5)3-MOLLI, 4(1)3(1)2-MOLLI and T2-prep bSSFP were 154 ms, 194 ms, 194 ms, and 144 ms, respectively. Routine maps were reconstructed on the scanner.
Multiple synthetic heart-rates were simulated in order to study the influence of heart-rate variability on the cMRF techniques. The R-R intervals varied from 600 to 1300 ms, at 100 ms intervals (8 in total).
Healthy volunteer study
We compared both cMRF versions to 5(3)3-MOLLI and T2-prepared bSSFP in 9 healthy volunteers. Three short-axis views were acquired with each technique (apex, middle, and base of the left ventricle) with breath holds in end-expiration. All three techniques were acquired with the same parameters used for the phantom study. After manual segmentation, the myocardium was subdivided according to the American Heart Association guidelines29.
Patient study
The cMRF with a shorter acquisition window was inserted in a clinical protocol for myocardial viability assessment in 61 consecutive patients referred for CMR at the Lausanne University Hospital (CHUV).
Patients (age 57 ± 14 years, 28% female, heart rate 66 ± 12 bpm ranging from 52 to 102) were referred for suspected myocarditis (n = 5), pericarditis (n = 4), ischemic cardiomyopathy (CMP, n = 27), dilated CMP (n = 4), hypertrophic CMP (n = 3), inflammatory CMP (n = 2), arrhythmia (n = 3), or other diseases (n = 13). A single cMRF slice was acquired in the same basal short-axis view as a 5(3)3-MOLLI T1 map and a T2-prepared bSSFP T2 map whenever either or both were deemed appropriate by the attending physician, and the clinical protocol allowed for the extra scan time. Similarly, cMRF and 4(1)3(1)2-MOLLI were acquired at the same location 20–25 min after the IV injection of 0.2 mmol/kg body weight GBCA (Gadovist, Bayer Healthcare, Leverkusen, Germany). Post-contrast cMRF T2 values were obtained but not reported due to low relevance and lack of comparison. The cMRF protocol with shorter acquisition window was kept the same as used for both healthy volunteers and the phantom; the two MOLLI maps were acquired with FOV = 306 × 360 mm2, pixel size = 2.0 × 1.4 mm2 interpolated to 1.4 × 1.4 mm2, and slice thickness = 8 mm, while T2prep-bSSFP had FOV = 288 × 360 mm2, pixel size = 2.5 × 1.9 mm2 interpolated to 1.9 × 1.9 mm2, and slice thickness = 8 mm.
In the patients where the native and post-contrast acquisitions were successful with both cMRF and routine techniques, the synthetic extracellular volume (ECV) was also computed:
where \(\Delta {R}_{1}={R}_{1}^{post{\text{-}}contrast}-{R}_{1}^{pre{\text{-}}contrast}\), and \({R}_{1}=1/{T}_{1}\). The value of the hematocrit (Hct) was estimated according to the empirical relationship found by Treibel et al. for MOLLI35:
The heartrate of each patient was recorded for linear regression with the pre- and post-GBCA relaxation times.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA; http://www.graphpad.com/scientific-software/prism/) and MATLAB. In vitro, we first computed the mean values and the standard deviation for each considered vial in the phantom. As a measure of accuracy, a Bland–Altman analysis and linear regression against reference values were performed for each technique, with a selected representative heart-rate of 60 bpm. In order to assess the influence of variable heart rates, we computed the coefficient of variation (CoV, standard deviation divided by the mean value) between scans acquired at different synthetic R-R intervals. Multiple linear regressions (one per heart-rate) were also computed for each technique, and Analysis of Covariance (ANCOVA) was used to test whether the slopes differed significantly.
In vivo, precision was defined per subject and segment and the coefficient of variation was again used as a measure of precision. When comparing the average measurements across multiple subjects, the standard deviations of the mean values were reported as absolute values (i.e. in milliseconds). Five datasets from healthy volunteers were segmented by a second observer, in order to assess the inter-observer variation that might be caused by differences in the segmentation. The resulting whole-heart distributions were compared by paired Student’s t-tests. Paired Student’s t-tests were also used to measure the agreement between cMRF and routine techniques. In healthy volunteers, since three slices from the same subject do not represent statistically independent variables, and three techniques were compared, a Bonferroni correction for multiple comparisons was used. When comparing global averages across the whole volunteer population, the three different slices were always averaged first, in order to generate one per-subject point. In all t-tests, P < 0.05 indicated statistical significance. Box and whiskers plots were reported to provide a graphical representation of the results from the multiple t-tests across the myocardial segments.
Data availability
The datasets used and analyzed during the current study available from the corresponding author on reasonable request.
References
Taylor, A. J., Salerno, M., Dharmakumar, R. & Jerosch-Herold, M. T1 mapping: Basic techniques and clinical applications. JACC Cardiovasc. Imaging. 9(1), 67–81 (2016).
Schofield, R., Bhuva, A., Manacho, K. & Moon, J. C. Cardiovascular magnetic resonance frontiers: Tissue characterisation with mapping. SA J. Radiol. 20(2), 1–6 (2016).
Messroghli, D. R. et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J. Cardiovasc. Magn. Reson. 19(1), 75 (2017).
Schelbert, E. B. et al. Myocardial extravascular extracellular volume fraction measurement by gadolinium cardiovascular magnetic resonance in humans: Slow infusion versus bolus. J. Cardiovasc. Magn. Reason. Off. J. Soc. Cardiovasc. Magn. Reson. 13, 16 (2011).
Ogier, A. C., Bustin, A., Cochet, H., Schwitter, J. & van Heeswijk, R. B. The road towards reproducibility of parametric mapping of the heart: a technical review. Front Cardiov Med. 9, 876475 https://doi.org/10.3389/fcvm.2022.876475 (2022).
Everett, R. J. et al. Assessment of myocardial fibrosis with T1 mapping MRI. Clin. Radiol. 71(8), 768–778 (2016).
Swoboda, P. P., McDiarmid, A. K., Page, S. P., Greenwood, J. P. & Plein, S. Role of T1 mapping in inherited cardiomyopathies. Eur. Cardiol. Rev. 11(2), 96–101 (2016).
Puntmann, V. O. et al. T1-mapping and outcome in nonischemic cardiomyopathy: All-cause mortality and heart failure. JACC Cardiovasc. Imaging. 9(1), 40–50 (2016).
Wong, T. C. et al. Myocardial extracellular volume fraction quantified by cardiovascular magnetic resonance is increased in diabetes and associated with mortality and incident heart failure admission. Eur. Heart J. 35(10), 657–664 (2014).
Piehler, K. et al. Association between extracellular matrix expansion quantified by cardiovascular magnetic resonance and short-term mortality. Circulation 126(10), 1206–1216 (2012).
Ugander, M. et al. Extracellular volume imaging by magnetic resonance imaging provides insights into overt and sub-clinical myocardial pathology. Eur. Heart J. 33(10), 1268–1278 (2012).
Paaladinesh, T. et al. Improved detection of myocardial involvement in acute inflammatory cardiomyopathies using T2 mapping. Circ. Cardiovasc. Imaging. 5(1), 102–110 (2012).
Spieker, M. et al. T2 mapping cardiovascular magnetic resonance identifies the presence of myocardial inflammation in patients with dilated cardiomyopathy as compared to endomyocardial biopsy. Eur. Heart J. Cardiovasc. Imaging. 19(5), 574–582 (2018).
van Heeswijk, R. B. et al. Free-breathing 3 T magnetic resonance T2-mapping of the heart. JACC Cardiovasc. Imaging. 5(12), 1231–1239 (2012).
Kvernby, S. et al. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. J. Cardiovasc. Magn. Reson. 16(1), 102 (2014).
Chow, K. et al. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn. Reason. Med. 71(6), 2082–2095 (2014).
Liu, Y., Hamilton, J., Rajagopalan, S. & Seiberlich, N. Cardiac magnetic resonance fingerprinting: Technical overview and initial results. JACC Cardiovasc. Imaging. 11(12), 1837–1853 (2018).
Christodoulou, A. G. et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat. Biomed. Eng. 2(4), 215–226 (2018).
Hashemi, S. S. et al. Comparison of three different cardiac T2-mapping techniques at 1.5 Tesla. Biomed. J. Sci. Tech. Res. 3(2), 3143–3150 (2018).
Ma, D. et al. Magnetic resonance fingerprinting. Nature 45(9), 4066–4084 (2013).
Jiang, Y., Ma, D., Seiberlich, N., Gulani, V. & Griswold, M. A. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn. Reason. Med. 74(6), 1621–1631 (2015).
Yu, A. C. et al. Development of a combined MR fingerprinting and diffusion examination for prostate cancer. Radiology 283(3), 729–738 (2017).
Messroghli, D. R. et al. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn. Reason. Med. 52(1), 141–146 (2004).
Giri, S. et al. T2 quantification for improved detection of myocardial edema. J. Cardiovasc. Magn. Reson. 11(1), 56 (2009).
McGivney, D. F. et al. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans. Med. Imaging. 33(12), 2311–2322 (2014).
Yang, M. et al. Low rank approximation methods for MR fingerprinting with large scale dictionaries. Magn Reson Med. 79(4), 2392–2400 (2018).
Lima da Cruz, G. et al. Sparsity and locally low rank regularization for MR fingerprinting. Magn. Reason. Med. 81(6), 3530–3543 (2019).
Eck, B., Seiberlich, N., Liu, Y. & Hamilton, J. I. Influence of scan window duration on parameter maps from cardiac magnetic resonance fingerprinting. In Proc Soc Cardiovasc Magn Reason. 325 (2019).
Cerqueira, M. D. et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. Circulation 105(4), 539–542 (2002).
Hamilton, J. I. et al. Investigating and reducing the effects of confounding factors for robust T1 and T2 mapping with cardiac MR fingerprinting. Magn. Reason. Imaging. 53, 40–51 (2018).
Cooper, M. A. et al. How accurate is MOLLI T1 mapping in vivo? Validation by spin echo methods. PLoS ONE 9(9), e107327 (2014).
Hamilton, J. I. et al. Simultaneous mapping of T1 and T2 using cardiac magnetic resonance fingerprinting in a cohort of healthy subjects at 1.5 T. J. Magn. Reason. Imaging. 52, 1044–1052 (2020).
Vincenti, G. et al. Cardiac magnetic resonance fingerprinting for the investigation of suspected inflammatory cardiomyopathy. In Proc Int Soc Magn Reson Med 27 (2019).
Patterson, A. et al. Initial utility of cardiac magnetic resonance fingerprinting for quantitative T1/T2 parametric mapping in hypertrophic cardiomyopathy. In Proc Int Soc Magn Reson Med 27 (2019).
Treibel, T. A. et al. Automatic measurement of the myocardial interstitium synthetic extracellular volume quantification without hematocrit sampling. JACC Cardiovasc. Imaging. 9(1), 54–63 (2016).
Jaubert, O. et al. Free-running cardiac magnetic resonance fingerprinting: Joint T1/T2 map and Cine imaging. Magn. Reason. Imaging. 68, 173–182 (2020).
Hamilton, J. I. et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn. Reason. Med. 77(4), 1446–1458 (2017).
Chefd’hotel, C., Hermosillo, G. & Faugeras, O. Flows of diffeomorphisms for multimodal image registration. In Proceedings IEEE International Symposium on Biomedical Imaging 753–756 (2002).
Hamilton, J. I. et al. Simultaneous multislice cardiac magnetic resonance fingerprinting using low rank reconstruction. NMR Biomed. 32(2), e4041 (2019).
Cruz, G. et al. 3D free-breathing cardiac magnetic resonance fingerprinting. NMR Biomed. 33(10), e4370 (2020).
Nezafat, R. et al. B1-insensitive T2 preparation for improved coronary magnetic resonance angiography at 3 T. Magn. Reason. Med. 55(4), 858–864 (2006).
Acknowledgements
The authors would like to thank Dr. Hélène Lajous, PhD (funded by SNSF grant 205321_182602) for providing the spin-echo T2 reference values for the phantom.
Funding
This study was funded by grants from the Swiss Heart Foundation and the Swiss National Science Foundation (grant number 32003B_182615) to RBvH.
Author information
Authors and Affiliations
Contributions
R.B.v.H., J.S. and N.S. designed the study. J.I.H. and N.S. developed and implemented the pulse sequence and image reconstruction algorithm. J.S. and A.P. recruited the patients and performed their diagnoses. R.B.v.H. supervised the analysis of MRI results and the manuscript draft. S.R. acquired the phantom and volunteer data. D.R. and A.P. acquired the patient data. S.R. analyzed the results, performed the statistical analysis and initially drafted the manuscript. All authors participated in manuscript drafting and editing and approved the final version of the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
JS receives research support by Bayer Healthcare, Switzerland. The other authors declare no potential competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rumac, S., Pavon, A.G., Hamilton, J.I. et al. Cardiac MR fingerprinting with a short acquisition window in consecutive patients referred for clinical CMR and healthy volunteers. Sci Rep 12, 18705 (2022). https://doi.org/10.1038/s41598-022-23573-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23573-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.