Abstract
Biochemical correlates of stochastic single-cell fates have been elusive, even for the well-studied mammalian cell cycle. We monitored single-cell dynamics of the ERK and Akt pathways, critical cell cycle progression hubs and anti-cancer drug targets, and paired them to division events in the same single cells using the non-transformed MCF10A epithelial line. Following growth factor treatment, in cells that divide both ERK and Akt activities are significantly higher within the S-G2 time window (~ 8.5–40 h). Such differences were much smaller in the pre-S-phase, restriction point window which is traditionally associated with ERK and Akt activity dependence, suggesting unappreciated roles for ERK and Akt in S through G2. Simple metrics of central tendency in this time window are associated with subsequent cell division fates. ERK activity was more strongly associated with division fates than Akt activity, suggesting Akt activity dynamics may contribute less to the decision driving cell division in this context. We also find that ERK and Akt activities are less correlated with each other in cells that divide. Network reconstruction experiments demonstrated that this correlation behavior was likely not due to crosstalk, as ERK and Akt do not interact in this context, in contrast to other transformed cell types. Overall, our findings support roles for ERK and Akt activity throughout the cell cycle as opposed to just before the restriction point, and suggest ERK activity dynamics may be more important than Akt activity dynamics for driving cell division in this non-transformed context.
Similar content being viewed by others
Introduction
The mammalian cell cycle is in large part driven by growth factor activation of the Ras-ERK1,2,3,4,5 and the PI3K-Akt2,6,7,8,9,10,11 pathways. Growth factors cause auto-phosphorylation of receptor tyrosine kinases (RTKs). For the ERK pathway, RTK phosphorylation recruits the guanine exchange factor SOS to the membrane, catalyzing the exchange of GDP for GTP bound to Ras, initiating Raf activation4,5,6,12,13,14. This in turn activates the MEK-ERK phosphorylation cascade. When activated, the effector kinases ERK1/2 translocate from the cytoplasm to the nucleus and activate transcriptional regulators such as Elk-1 and CREB15,16,17. These transcriptional regulators induce immediate early genes (IEGs) like c-fos15,16 that then contribute to the expression of cyclin D14,6,8,17,18,19, a key step in S-phase entry20.
RTK activation can also initiate Akt pathway signaling. RTK autophosphorylation recruits adapter proteins like insulin receptor substrate (IRS-1) and GRB2-associated binder (GAB)2,21,22,23. These proteins in turn recruit Phosphatidylinositol (PtdIns) 3-kinase (PI3K) to the membrane2,19,24,25 where it phosphorylates the membrane phospholipid PtdIns (4,5) P2 (PIP2), generating PtdIns (3,4,5) P3 (PIP3). PIP3 recruits pleckstrin homology domain (PH)-containing proteins to the membrane such as phosphatidylinositol-dependent kinase-1 (PDK1)26 and the serine/threonine protein kinases Akt1/219,26. PDK1 phosphorylates Akt’s activation loop followed by mTORC2 phosphorylation of a second site on Akt for full activation6,7,19. This doubly phosphorylated, activated Akt promotes cell cycle progression by: (1) promoting protein translation via 4E-BP and p70S6K6,19, (2) promoting cyclin D17,18 CDK4/6, c-Myc, and E2F activity27 and (3) inhibiting p21 and p2728 (cyclin-dependent kinase inhibitors).
While ERK and Akt pathways have established roles prior to the restriction point marked by S-phase entry, the extent to which they are informative of cell cycle completion after S-phase is less clear. Beyond S-phase, studies suggest that Ras-ERK29,30,31,32,33,34,35,36 and PI3K-Akt26,37,38 may contribute towards regulating G2 progression. ERK activity was shown to play a role in the duration of DNA damage-induced G2 arrest39. Transient ERK activity maintains G2 arrest, whereas sustained ERK activity promotes escape by reducing p53 levels, and inducing the expression of pro-mitotic factors such as Plk1 and cyclin B36. Akt activity also contributes to G2-M progression as its inhibition is associated with reduced cyclin B levels, promoting Chk1 activity and G2 arrest37. These observations motivate a closer look at determining how ERK and Akt dynamics are informative of cell cycle completion after the canonical restriction point.
On a single cell level, both ERK and Akt activity dynamics have substantial cell-to-cell and dynamic variation, exhibiting complex pulses and more simple steady activity1,24,40,41,42,43,44,45,46,47,48. Such variation, when coupled with the observations that cell cycle progression is also heterogeneous1,49,50, have prompted investigations into the correlation between dynamics and cell cycle fate in single cells. What determines proliferation on a single cell level? What relative contributions do ERK and Akt activity have to the decision of individual cells to divide? Much prior work has focused on activity dynamics. Both Ras-ERK51,52,53 and PI3K-Akt54 exhibit biphasic growth factor-induced activation dynamics, with a transient peak followed by sustained activity hours later. The dynamics of each phase contributes differently towards driving progression to S-phase and is cell type dependent54,55,56. Live-cell imaging and analysis of recently divided sister cells reveal that time-integrated ERK activity has some predictive power of the timing to S-phase entry1. Time-integrated ERK dynamics were found to influence proliferation decisions in daughter cells45. Predicting PC12 cell differentiation/proliferation decisions required both ERK and Akt activity dynamics to best define the decision boundary between these two cell fate outcomes57. Yet, the extent to which both ERK and Akt activities throughout the cell cycle are associated with division in single cells remains unclear.
Here, we use live-cell imaging to pair measurements of growth factor-induced ERK and Akt activity to cell division outcomes in the same single cells. We aim to assess the extent to which these activities are associated with cell cycle progression beyond S-phase entry to single cell division responses, using the well-established non-transformed breast epithelial MCF10A cell line, a model system that is commonly used to study epithelial signaling biology and cell division control2,58,59,60,61,62,63. We found that following treatment of synchronized cells with growth factors EGF and insulin, both ERK and Akt activity are significantly higher within the S-G2 interval in dividing cells. Such differences were much smaller in the pre-S-phase window, which is traditionally associated with ERK and Akt activity dependence54,55,56, suggesting unappreciated roles for ERK and Akt in S through G2. ERK activity was more strongly associated with cell division events, suggesting Akt activity may play a smaller role in this context. Interestingly, we found that ERK and Akt activities are less correlated in cells that divide. Network reconstruction experiments demonstrated that this correlation behavior was not due to crosstalk, as ERK and Akt do not interact in this context, in contrast to other cell types64. Overall, our findings support roles for ERK and Akt activity throughout the cell cycle as opposed to just before the restriction point, and suggest ERK activity dynamics may be more important than Akt activity dynamics for driving cell division in this non-transformed context.
Results
Association of cell division events with univariate ERK and Akt dynamics
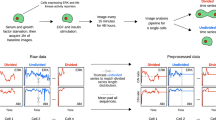
To evaluate associations between ERK or Akt signaling dynamics and cell division, we first conducted a series of live cell imaging experiments in MCF10A cells that express either ERK41,65 or Akt43 kinase translocation reporters (KTRs), but not yet both at once, and paired those single cell dynamics to cell division events (Fig. 1A). We first verified that cell cycle progression and division are related to ERK and Akt activity dynamics in MCF10A cells using small molecule inhibitor experiments (Fig. S1). KTR-expressing cells were G0-synchronized by serum and growth factor starvation for 24 h. After acquiring 1 h of baseline ERK or Akt activity, cells were treated with EGF and insulin, growth factors that promote cell division in MCF10A cells66. Images were acquired every 15 min for 48 h, and then single-cell data for kinase activity and division outcome were extracted using custom-built image processing pipelines (see “Methods”). Dynamic regimes of KTR specificity were determined using two independent (four total) MEK and Akt inhibitors (Fig. S2). ERK KTR was found to be specific in all regimes explored here, whereas the Akt KTR was found to be specific > ~ 1 h after EGF and insulin co-stimulation. Variability from cell-to-cell at a particular moment in time, and across a single cell over time, suggested that the probes had informative dynamic range outside of the acute time period following the ~ 4–8 h post-growth factor stimulation (Supplementary Figs. S9, S10).
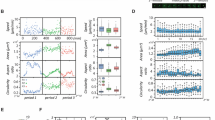
Evaluating the association of cell division with univariate single cell ERK or Akt dynamics. In these experiments, cells were either expressing the ERK or the Akt KTR. (A) Cell treatment workflow for pairing single cell KTR dynamics to cell division. ERK or Akt KTR expressing MCF10A cells were seeded, allowed to attach overnight, and then serum and growth factor starved. Following starvation, baseline images were acquired, cells were treated with EGF and insulin, and then imaged every 15 min for 48 h. Images were quantified using the analysis pipeline described in the “Methods”. (B, C) Quantified ERK or Akt KTR dynamics paired to division events for EGF and Insulin doses that match those used in culture medium (B) or tenfold less (C). Single cell traces of dividing (blue) and non-dividing (red) cells are shown with thin lines, and population median (per time point) is shown with thick lines. (D) Left, representative single cell trace of ERK KTR for a dividing (blue) or non-dividing (red) cell. Median ERK activity within the 8.5–40 h interval for each cell becomes a single dot in the boxplots. (E) notBoxplots for single cell median ERK or Akt activity within the 8.5–40 h interval for EGF and Insulin doses that match those used in culture medium (top) or tenfold less (bottom). p-values for the right-tailed rank-sum test were calculated at the 95% confidence interval. D, dividing; ND, non-dividing.
Single cell traces of ERK or Akt activity (thin lines) along with the population median (bold line) show rapid activation following growth factor treatment, which largely persists for the duration of the experiment (Fig. 1B, C). In (blue) dividing cells, population median ERK and Akt activity dynamics are higher throughout the cell cycle compared to non-dividing cells, with larger differences evident for ERK. In the pre-S-phase entry window (~ < 8 h after growth factor treatment), there are slight differences between dividing and non-dividing cells in terms of population median ERK and Akt dynamics. These differences grow larger in the subsequent 8.5–40 h interval post growth factor addition, which largely corresponds to S and G2 phases. These trends were also evident with ten-fold less concentration of growth factors (Fig. 1C). These results suggest that ERK and Akt activity may have importance after S through G2 phase.
To assess the statistical significance of this finding, we calculated the median ERK or Akt activity for individual single cells within the 8.5–40 h window post-growth factor treatment, and then compared median activity between dividing and non-dividing cells with the single tailed rank-sum test (Fig. 1D, E). Individual dots in the boxplot represent the median ERK or Akt activity calculated within the 8.5–40 h interval in a single cell. These median single cell activities were significantly different in dividing vs. non-dividing populations (Fig. 1E). ERK activity was more significantly different in dividing vs non-dividing cells than Akt activity. Nevertheless, there is substantial overlap in the two populations, indicating other factors influencing cell division events. We also cannot rule out that other dynamic features in addition to the 8.5–40 h median may be additionally informative.
Measurements of both ERK and Akt dynamics in the same single cells correlated with cell division fate
As shown above, ERK and Akt activity dynamics alone contain information about subsequent cell division. Do ERK and Akt activity covariations also correlate with cell division fate? Is one more strongly associated than the other? To answer these questions, we performed a similar experiment as described above using dual reporter expressing MCF10A cells (see “Methods”). For the duration of the time course, population median ERK and Akt dynamics are again elevated in dividing cells compared to non-dividing cells (Fig. 2A), with larger differences observed in the 8.5–40 h interval. Median ERK and Akt dynamics together had some ability to stratify dividing cells, but again, there is substantial overlap (Fig. 2B). Logistic regression indicated that median ERK activity was more strongly associated with cell division events as compared to median Akt activity (Fig. 2C). These results were confirmed in an independent experiment (Fig. S3). Moreover, imaging data acquired at a 5-min interval corroborated these findings (Fig. S8). We conclude that Akt dynamics add comparatively little new information to ERK dynamics for predicting single cell division events in this context. Although the median in the 8.5–40 h post-growth factor window was analyzed here, we cannot rule out the importance of other dynamic features such as pulsing, although in our datasets we did not observe significant pulsing1,24,40,41,42,43,44,45,46,47,48 (perhaps due to experimental differences such as subconfluency and serum-starvation—see “Discussion”). Moreover, because of strong growth factor stimulation and potential probe saturation in the 0–8.5 h window, we cannot rule out the importance of this earlier signaling window either.
Evaluating the association of cell division with paired, bivariate single-cell ERK and Akt dynamics. In these experiments, cells were expressing both the ERK and Akt KTR simultaneously. (A) Quantified ERK and Akt dynamics for EGF and Insulin doses that match those used in culture medium (top) or ten-fold less (bottom). Single cell traces of dividing (blue) and non-dividing (red) cells are shown with thin lines, and population median (per time point) is shown with thick lines. (B) Scatter plot of ERK vs. Akt KTR median activity in the 8.5–40 h window from 150 randomly sampled cells. Each dot is a single cell. Dividing cells are blue and non-dividing cells are red. Left and right are high and low growth factor concentrations, respectively. (C) Statistical significance of logistic regression coefficients for either ERK or Akt median activity (8.5–40 h), with regression coefficients shown in parentheses above the respective bars (uncertainty is standard error).
Inferring the topology of the ERK-Akt network
Information content is related to correlation, so we investigated the extent to which ERK and Akt dynamics in the same single cells were correlated, looking across every cell and every time point (Fig. 3A, replicate Fig. S3D). Interestingly, in dividing cells, single cell ERK and Akt dynamics within the 8.5–40 h window are significantly less correlated than in non-dividing cells, at both high and low growth factor doses. Network topology can strongly influence correlated behaviors. In different studies, ERK and Akt have been reported to exhibit very different network behavior, such as cross-pathway activation, inhibition6,18,19,23,64,67,68,69,70,71 and non-interaction21,64,72. Factors such as cell type and growth factor context can influence these discrepant network topologies3,64. Previous work conducted in panel of growth factors and cell lines show varying probabilities of forming an interaction network edge between ERK and Akt64. The differences in network edge formation can affect downstream signaling and cell fate decisions3. Could ERK and Akt network topology give insight into the division-related correlated behaviors observed above?
Investigating properties of the ERK and Akt network. (A) Single cell ERK and Akt activity plotted across all time points within the 8.5–40 h interval for dividing and non-dividing cells. These cells expressed both the ERK and Akt KTR. Pearson correlation coefficient, along with the number of cell-time datapoint combinations are indicated. Uncertainty in the correlation coefficients is calculated as described in “Methods”. (B) Cell treatment workflow for network reconstruction in the “acute” regime. Single ERK or Akt KTR expressing MCF10A cells were seeded, allowed to attach overnight, and then serum and growth factor starved. Following starvation, inhibitor was added (PD: PD0325091; MK: MK2206), baseline images were acquired, cells were treated with EGF and insulin, and then imaged every 15 min for 12 h. Images were quantified using the analysis pipeline described in the “Methods”. (C) Quantified ERK and Akt activity dynamics in the acute regime. Solid lines are population median (per time point), and shaded areas denote the standard deviation across cells. (D) Cell treatment workflow for network reconstruction in the “chronic” regime. Dual ERK and Akt KTR expressing MCF10A cells were seeded, allowed to attach overnight, and then serum and growth factor starved. Following starvation, EGF and insulin were added, baseline images were acquired, cells were treated with inhibitors, and then imaged every 6.5 min for the remaining ~ hour. Images were quantified using the analysis pipeline described in the “Methods”. (E) Quantified ERK and Akt activity dynamics in the chronic regime. Solid lines are population median (per time point), and shaded areas denote the standard deviation.
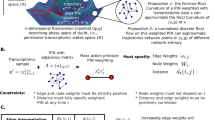
To reconstruct the ERK and Akt network in MCF10A cells, we implemented recent theory from our lab that specifies a sufficient experimental design for such tasks, based on perturbation time course data73. Specifically, for this 2-node network, three time course experiments should be done: response of ERK and Akt activity to EGF and Insulin co-treatment with (1) no inhibitor; (2) an ERK pathway inhibitor; and (3) an Akt pathway inhibitor. Additionally, we wanted to understand whether the network would be different in the acute phase of growth factor treatment from a serum starved state vs. the “chronic” condition where ERK and Akt activities are steady over time, particularly because these time regimes seem to have different biological information encoded for cell division decisions.
In the acute regime (Fig. 3B), MCF10A cells expressing either ERK or Akt KTR were seeded, serum and growth factor starved for 24 h, and then pretreated with either a MEK (PD0325901) or an Akt (MK2206) inhibitor for 30 min. The concentrations utilized were determined via titration experiments to ensure minimum possible doses were being used (Figs. S1, S2, S4). Following drug treatment, baseline KTR activity was acquired every 15 min for 1 h. Then, we treated cells with EGF and insulin and imaged. Single cell traces (thin) and population median activity (bold) were calculated for each condition, showing robust ERK and Akt activation (Fig. 3C). MEK inhibition ablates ERK activation and has a negligible effect on Akt activation (Fig. 3C). Akt inhibition ablates Akt activation and has a negligible effect on ERK activation (Fig. 3C). Although the Akt KTR may reflect kinase activity other than Akt in this acute stimulus regime, the fact that complete inhibition of the ERK pathway has negligible impact on the Akt KTR readout means that ERK does not impact Akt or the others. These results show that in the acute stimulus regime, ERK and Akt exhibit negligible crosstalk after treatment with EGF and insulin.
In the chronic regime, cells were pretreated with either EGF and insulin for 30 min followed by 30 min of baseline acquisition, leading to robust ERK and Akt activation (Fig. 3D, E, Replicates in S5). Akt inhibition reduces Akt activity, as expected, but negligibly affects ERK activity. MEK inhibition reduces ERK activity, as expected, but does not appreciably affect Akt activity. These conclusions are also consistent when a second set of MEK and Akt inhibitors are used (Fig. S4). Therefore, in the chronic regime ERK and Akt also do not exhibit appreciable cross pathway interactions after EGF and insulin co-treatment. We conclude it is unlikely that crosstalk interactions account for correlations that change in dividing vs. non-dividing cells.
Discussion
Binary single-cell responses, like division, to perturbations such as growth factor and drug treatments, are almost universally heterogeneous even in clonally derived populations. However, biochemical features associated with heterogeneous fates, present either before the perturbation, or from dynamics following the perturbation, are not well established. The ability to predict such binary responses would not only reflect a deep and fundamental understanding of the systems governing important cellular responses, but also have significant translational applications such as antibiotic resistance, tissue engineering, and anticancer therapy, where the fates of single cells can be of great importance. Here, we investigated growth-factor induced cell division fates in the well-studied, non-transformed mammalian epithelial cell line MCF10A, and how they may be associated with the dynamics of two central signaling pathways, PI-3 K/Akt and Ras/ERK. Answering such questions requires single-cell, non-destructive analysis of biochemical features, in this case ERK and Akt activities, that are paired to the eventual cell division outcome. They also must be carried out in a high-throughput manner to observe enough events to make statistically-supported conclusions. After setting up this experimental system and understanding its ranges of validity, we learned that (1) ERK and Akt activities are higher in the 8.5–40 h window after growth factor treatment in cells that divide, suggesting underappreciated roles post-restriction point, into S and G2 phases; (2) median activities in this time window are associated with cell division outcome, and ERK activity was more strongly associated than Akt activity; (3) ERK and Akt activities are less correlated in cells that divide; and (4) ERK and Akt do not exhibit crosstalk in this system, so division-related correlation is unlikely related to crosstalk.
We have performed these studies in the MCF10A cell line, a well-established model for non-transformed epithelial cells. An obvious next question is how the relationships between ERK, Akt and cell division found here translate to different cell lines, and transformation contexts. Many other cell lines are cancer-derived and genetically unstable, and/or contain multiple alterations to the systems that control cell cycle progression and division. A firm understanding of how ERK and Akt systems control the cell cycle in a system such as MCF10A is an important foundation for subsequently understanding how it may be altered in other cell lines, and also across different microenvironmental contexts, such as confluent settings. Indeed, there is a growing body of work that focuses on answering fundamental cell biological questions using studies on the MCF10A system alone (e.g.,74). It is appealing to consider MCF10A as an emerging model system for mammalian epithelial cells.
Nearly all the cells we observed had relatively simple dynamics for ERK and Akt activity, a rise then a somewhat constant higher than baseline steady-state. Other recent single cell studies have reported pulsatile ERK dynamics1,44,75,76 using a 5 min acquisition frequency. When we used a 5 min acquisition frequency, we still did not observe pulsatile activity, although this increased frequency of acquisition required shorter exposure times which increased noise in the images. Pulsatile vs non-pulsatile dynamics may be related to differences in growth factor concentration, the reporters used, being FRET-based77 or translocation based65. No live-cell imaging probe is perfect and of course has its drawbacks, some of which may be related to off-target responses, which may partly explain our Akt activity data in the “acute” phase first following growth factor treatment. For example, kinases other than Akt may recognize and phosphorylate the FOXO1-based Akt KTR docking site78,79,80,81,82. EGF and insulin stimulation may also promote activation of such kinases including PLK183, SGK and PKA84. Another aspect may have to do with cell–cell contact and density. In our study, cells were seeded at low density and serum/growth factor-starved prior to analysis, whereas pulsatile signaling was reported in high density environments in asynchronously cycling settings1,76. Yet others have found pulsatile dynamics can induce different sets of genes as compared to sustained dynamics41. However, phenotypic consequences, at least in terms of cell proliferation, still seem to be related to simple time-integrated signaling dynamics1,45, similar to what we found here.
ERK and Akt activity dynamics are only a subset of the potentially important variations that drive phenotypic variability in cell division responses. ERK activity was substantially more significant than Akt activity. This reinforces Akt activity as perhaps more relevant for cell maintenance and health, and more as a “checkpoint” for division but not a significant driver, at least in the studied system. As noted above, cell contacts and density are important. Such phenomena may potentially be controlled through micropatterning experiments, where cell shape and placement can be carefully controlled85,86. Cell “state”, corresponding to different epigenetic and/or metabolic states of cells prior to the experiment, has been reasonably well documented ubiquitously, and can contribute to variability, although is difficult to assess in the “track and follow” manner that can be done with live-cell kinase reporters. Metabolic or organelle abundance variability may also contribute87,88. Of course, there are other pathways and biochemical correlates that are likely important, such as a balance between p53 and p21 and/or CDK2 activity49,62,89. Given the multitude of fluorescent proteins, and improvements in cell tracking from non-labeled bright field images90,91, one may be able to measure more important biochemical readouts simultaneously for such purposes. There are also multiple checkpoints between growth factor treatment and cell division, such as the restriction point, and DNA damage checkpoints, that may contribute. Monitoring division with probes like the Fucci system that gives readouts of each cell cycle phase may help explore such phenomena92.
An interesting aspect of our study was differences between dividing and non-dividing cells in the time period that corresponds to S/G2 phases of the cell cycle. The roles of growth factor signaling through ERK and Akt pathways historically focused on passing the restriction point into S-phase93. Although we did not see large differences pre-S-phase, this may have been due to probe saturation in the acute period following growth factor stimulation. Nevertheless, our results suggest potential functional roles for ERK and Akt beyond this canonical understanding. Indeed, a recent study found time-integrated ERK activity in a mother cell’s G2 phase influenced the cell cycle progression in the subsequently daughter cells45. The mechanisms that may be driving such functional roles are a potentially interesting area of future study.
We also studied the ERK and Akt activity network, since we found that ERK and Akt activity are less correlated with each other in dividing cells compared to non-dividing cells. We found that the observed differences in correlation are likely not a result of network topology as ERK and Akt do not appreciably interact. This lack of interaction is surprising given that some prior studies describe these pathways as exhibiting cross pathway interactions21,23,64, albeit in other cell lines and in response to other growth factors. However, other studies in MCF10A cells across a panel of different growth factors show that ERK and Akt are insular, and do not interact64. Similar to MCF10A cells, 32D-EpoR; BaF3-EpoR; CFU-E cell lines exhibit minimal ERK and Akt cross pathway interaction under erythropoietin stimulation, a growth factor that activates both ERK and Akt72. These studies reveal that in non-interacting pathways, differences in protein expression influence the flow of erythropoietin signaling72. Therefore, in our model system, it is possible that the observed differences in ERK and Akt correlation may arise from differences in protein expression across dividing and non-dividing cells. It may also be that differences in upstream signaling capacity to ERK and Akt may be related. Characterizing the differences in protein expression level in single cells and following cell signaling and cell division can provide insight; but this becomes a quite challenging experiment given the number of probes to be measured simultaneously.
In conclusion, we have studied the relationship between ERK activity dynamics, Akt activity dynamics, and cell division, and found that simple measures of central tendency of these dynamics in a time coinciding with S/G2 phase are strongly associated with division in single cells. This implies unappreciated roles for ERK and Akt beyond the canonical restriction point. ERK activity is more strongly associated than Akt activity. Yet, neither ERK nor Akt activity perfectly segregate dividing from non-dividing cells, so it is clear other biochemical pathways are important factors for driving single cell division events. ERK and Akt do not interact with one another in the studied contexts, despite the fact that their activities are more correlated in cells that do not divide. These studies conducted in the non-transformed context provide a foundation to explore how cell transformation through oncogenic and/or loss-of-function mutations shape network topology, signaling dynamics, and cell division outcome in cancer, with the potential to identify and target pathway compensation behaviors that promote cell proliferation and survival94 in the diseased context. In addition, exploring the role of spatial temporal propagation of ERK and Akt signaling in a 3D tissue context, a model system that MCF10A cells are suited for, can provide insight into how these pathways regulate tissue homeostasis and how transformation disrupts this homeostasis.
Methods
Cell culture
MCF10A cells were gifted by Dr. Gordon Mills and cultured in complete sterile filtered (VWR 10040-436) media, consisting of DMEM F12 (Gibco #11330-032) supplemented with 2 mM l-Glutamine (Gibco # 25-005-CI), 20 ng/ml EGF (Peprotech AF-100-15), 10 μg/ml insulin (Sigma #I-1882), 0.5 μg/ml hydrocortisone (Sigma #H-0888), 100 ng/ml cholera toxin (Sigma #C-8052) and 5% horse serum (Invitrogen #16050-122). Cells were passaged with 0.25% trypsin (Gibco #25200056) to maintain sub confluency. Cells were maintained at 37 °C, 5% CO2. Starvation media and imaging media is phenol red free DMEM F12 (Fisher #11039-021) supplemented with 100 ng/ml cholera toxin.
HEK293T cells were gifted by the Dr. Dominguez and Dr. Pappapetrou labs and cultured in complete sterile filtered (VWR 10040-436) media, consisting of DMEM (Gibco #11965118) supplemented with 2 mM l-Glutamine (Gibco #25-005-CI) and 10% heat inactivated fetal bovine serum (Gibco #16140071). Cells were passaged with 0.05% trypsin (Gibco #25300054) to maintain sub confluency.
All inhibitors used for KTR validation were formulated as 10 mM stock solutions in DMSO (Sigma Aldrich D2650-5X0ML) and sterile filtered with a 0.22-micron syringe filter. PD0325901 (MEK inhibitor 1) was purchased from Sigma Aldrich (PZ0162-5MG). Trametinib (S2673) (MEK inhibitor 2) and Ipatasertib (S2808) (Akt inhibitor 2) were purchased from Selleck Chemicals. MK2206 (#CT-MK2206) (Akt inhibitor 1) was purchased from Chemietek.
Imaging
All live cell imaging experiments with a 15 min interval were acquired using an InCell 2200 microscope (GE Healthcare) under environmental control (37 °C, 5% CO2) with a Nikon 20X/0.75, Plan Apo, CFI/60 objective. For KTR imaging the following filter sets were utilized: FITC (Excitation: 475/28 nm Emission: 511/23 nm) (ERK-mClover, Akt-mClover KTR); Cy3 (Excitation: 542/27 nm Emission:597/45 nm) (H2B-mRuby2, mCherry-NLS); Cy5 (Excitation: 632/22 nm Emission: 679/34 nm) (ERK-iRFP); Brightfield.
KTR-expressing MCF10A cell lines (pool-see below) were seeded in separate rows of a 96 well plate (Corning #3603) at 5000 cells/well and treated as described. After growth factor and serum starvation, starvation media was aspirated, cells were washed with PBS and 100 μL imaging media was then placed in the wells. Following baseline imaging, cells were treated as indicated by adding 100 μL of 2× solutions in imaging media. Acquired images were processed as described in “Computational image analysis”.
Experiments with a 5-min sampling interval were acquired using the Leica DMI8 (Leica Microsystems) microscope equipped with a HC PL FLUOTAR L 20×/0.40 DRY objective, a pentacube filter (excitation: 458–490 nm, 524–584 nm, 624–754 nm, emission: 500–530 nm, 579–611 nm, 661–701 nm), a Leica DFC 9000GT camera and a CoolLED pE-4000 illumination system. Cells were maintained at 37 °C, 5% CO2 using a Pecon environmental control system. 12-bit images were acquired for each probe: Akt-mClover KTR; mCherry-NLS, ERK-iRFP and Brightfield. Raw images were not flatfield corrected or background subtracted due to potential signal loss due to the short acquisition times and exposure settings for this dataset.
Flow cytometry
EdU flow cytometry assays were performed using the Molecular Probes Click-iT Plus EdU flow cytometry assay kit (C10633 molecular probes). MCF10A cells were seeded in 6 well plates (Corning 353046) at a density of 127 cells/mm2 in complete DMEM F12 media. The following day, we serum and serum and growth factor starved cells in DMEM F12 media supplemented with 100 ng/ml cholera toxin for 24 h. Following starvation, cells were pretreated with a final concentration of 100 nM of MEK inhibitor 1 (PD0325901) or 10uM of Akt inhibitor 1 (MK2206) or DMSO control for 30 min. Cells that did not receive inhibitor pretreatment were treated with either MEK inhibitor 1 or Akt inhibitor 1 2, 4, 8, or 12 h post EGF and insulin addition (Final growth factor concentration: 20 ng/ml EGF, 10ug/mL insulin). 22 h post growth factor addition, a final concentration of 10 μM of EdU was added to each well and incubated for 2 h. 2 h post EdU addition, cells were washed with PBS and lifted with 0.25% Trypsin for 10 min. Trypsin was neutralized with complete DMEM F12 media. Cells were pelleted at 100×G for 5 min, resuspended in 100 μl of PBS, and processed as recommended by the manufacturer’s protocol (Molecular Probes Click-iT Plus EdU flow cytometry assay kit). During the last 5 min of permeabilization, 100 μl of diluted 1 μg/ml Hoechst 33342 (ThermoFisher H3570) stain was added. Cells were then washed with a 1% (g/100 ml) bovine serum albumin PBS solution and pelleted at 100 g. Cells were resuspended in permeabilization buffer and stained with EdU Click-iT reaction cocktail for 30 min at room temperature protected from light. Following EdU Click-iT labeling, cells were washed and resuspended in permeabilization buffer and analyzed using the BD Canto II flow cytometer configured with the following laser lines: excitation 640 nm, emission filter 660/20, excitation 405 nm, and emission filter 450/50. Data was gated and processed using FCS Express (Denovo Software).
Cloning
Akt KTR was modified from the transposase transfection system PSBbi-FoxO1_1R_10A_3D vector (Addgene # 106278)43 for lentiviral production (Fig. S6A). The lentiviral expression vector was developed using overlap PCR from fragments generated from the source vector PSBbi-FoxO1_1R_10A_3D: SV40NLS-mCherry-P2A and Gly-FT2DDD-KTR-mClover. Primer sequences are shown in Fig. S6C and were designed in SnapGene and ordered from Sigma Aldrich. Fragments for lentiviral vector construction were generated via PCR using Q5 polymerase (NEB M0491S) and primers specific to SV40NLS-mCherry-P2A and Gly-FT2DDD-KTR-mClover (Fig. S6C) regions of PSBbi-FoxO1_1R_10A_3D (Fig. S6, Fragment 1,2). Fragments were gel purified using the NEB Monarch gel extraction kit (NEB T1020S). Following gel extraction, a 10 cycle PCR reaction was performed using Q5 polymerase and equimolar SV40NLS-mCherry-P2A and Gly-FT2DDD-KTR-mClover fragments using an annealing temperature of 72 °C. 5 μl of the product was amplified using end primers (F: SV40NLS-mCherry-P2A, R: Gly-FT2DDD-KTR-mClover, Ta = 69 °C, Fig. S6C). Gateway ATTB sites were inserted at flanking ends using PCR and the ATTB primers (Fig. S6C). The Gateway cloning compatible fragment was inserted into donor vector pDONR221 (Invitrogen™ 12536017) using BP Clonase II (ThermoFisher# 11789020). High Efficiency NEB-5-alpha Competent E. coli (NEB C2987I) were transformed with pDONR221 containing the Akt KTR. Transformants were miniprepped with the PureYield Plasmid Miniprep System (Promega A1223) and Sanger sequence verified using GeneWiz. Akt KTR expression vector was generated by performing a LR reaction using pDONR 221-Akt, LR Clonase II (Thermofisher #11791020) and the lentiviral expression vector pLenti CMV Hygro DEST (Addgene #17454) generating the final product, a bi-cistronic hygromycin selectable lentiviral expression vector. The product was transformed into NEB® 5-alpha Competent E. coli. Transformations with the correct sequence were maxiprepped with PureYield Plasmid Maxiprep System (Promega #A2392) and used for lentiviral production.
We exchanged the antibiotic selectable marker on the lentiviral expression vector H2B-mRuby2 (Addgene #90236) from hygromycin to puromycin. Specifically, we transferred pLentiPGK Hygro DEST H2B-mRuby2 into pLentiCMV puromycin DEST (Addgene #17452) using BP Clonase II followed by LR Clonase II generating pLentiCMV puromycin DEST H2B-mRuby2 (H2B-mRuby2).
Lentiviral production
The lentiviral constructs for each cell line are shown in Fig. S6B. Lentiviral particles were generated by transfecting 5 million HEK293T cells seeded in a T75 flask allowed to attach overnight (Corning® T-75 flasks catalog #430641) using the TransIT-293 transfection reagent (Mirus Bio MIR2704) along with expression vector ERK KTR (mClover or iRFP), H2B-mRuby2, or Akt KTR along with packaging plasmid pPAX (Addgene #12260), and envelope protein pCMV-VSV-G (Addgene #8454) according to the manufacturer’s instructions. Two days post transfection, supernatant from was collected and concentrated using Amicon Ultra-15 100 kD centrifugation filters (Millipore #UFC910008). The concentrated lentiviral supernatant was aliquoted and stored at − 80 °C.
Lentiviral transduction
100,000 MCF10A cells were transduced in suspension in a 6 well plate (Corning 353046) containing complete DMEM F12 medium along with 100 μl lentiviral supernatant. Two days later, expression was validated by fluorescence imaging. ERK KTR MCF10A cell lines (ERK KTR-mClover Hygro, H2B-mRuby2 Puro) were selected in complete DMEM F12 media supplemented with hygromycin (35 μg/ml) and puromycin (2 μg/ml). Akt KTR expressing MCF10A cell lines (SV40nls-mCherry-Akt KTR-mClover Hygro) were selected with DMEM F12 media containing hygromycin (35 μg/ml). Cells were passaged every two to three days in selection media for about 2 weeks. Following selection, cells were expanded in complete DMEM F12 maintenance media containing either both hygromycin (1.5 μg/ml) and puromycin (0.1 μg/ml) (ERK KTR-mClover expressing cells) or hygromycin (1.5 μg/ml) (Akt-KTR-mClover expressing cells). Single clones were not selected, rather, analyses were done with pools. Live-cell imaging was conducted in the absence of selection antibiotics. Dual reporter expressing cells (ERK KTR-iRFP, Akt-KTR-mClover) lines were not selected, as the ERK KTR iRFP (Addgene #59150) lentiviral expression vector does not confer antibiotic resistance. For these, ERK KTR iRFP virus was added to cells for 24 h, and then subcultured as above prior to live-cell imaging analysis.
Computational image analysis
While many image analysis tools exist95,96,97, each application still requires much novel development tuned to the problem at hand. We developed an automated image analysis pipeline using both iLastik98 and CellProfiler97 software packages, along with MATLAB scripts (Fig. S2C). It is available at the Birtwistle Lab github repository (github.com/birtwistlelab/Predicting-Individual-Cell-Division-Events-from-Single-Cell-ERK-and-Akt-Dynamics), which includes some dockerized scripts. The analysis pipeline consists of (1) cell nuclei and cytoplasmic segmentation, (2) quantification of KTR fluorescence in both nuclei and cytoplasmic compartments, (3) tracking single cells across a time series, and (4) automatic detection of cell division.
-
1.
Prior to segmentation images were flatfield corrected and background subtracted using CellProfiler. Images of nuclear localized fluorescent protein H2B-mRuby2 (ERK KTR) and NLS-mCherry (Akt KTR) were input into iLastik. Nuclei were identified using a series of features—object intensity, edge detection and texture.
-
2.
The binary mask outputs from iLastik were input into CellProfiler to create a perinuclear ring known as the ‘Cytoring’65 which extends 10 pixels from the binary nuclei mask and into the cytoplasm (Fig. S2C). Calculating the cytoplasmic to nuclear KTR fluorescence ratio provides the relative activity of the pathway of interest for that particular cell at that time point.
-
3.
Segmented nuclei identified with iLastik were tracked using CellProfiler’s TrackObjects module96,97,99 and Follow Neighbors99. Each identified nucleus was assigned a numerical ID, which corresponds to the same cell across each timepoint. We filter tracks that are shorter than the duration of the time course to prevent quantification of cells that were transiently tracked.
-
4.
Cell division was detected using a feature of cytoplasmic to nuclear KTR fluorescence (C/N ratio) that is unique to dividing cells. As cells divide, there is a change in morphology resulting in a rapid decrease in C/N ratio (Fig. S7). MATLAB’s findpeaks function was used to detect when this steep decrease occurs. In dual reporter cells, the Akt KTR was used. We then truncated the time series 5 timepoints before the identified peak, which is attributed to actual kinase activity. For each processed dataset, the output of the cell division script was manually reviewed, and any misclassified cells were removed by directly observing ERK and Akt KTR time course plots for spurious peaks that erroneously resulted in a cell being marked as a dividing cell, or by directly observing the cell nucleus in time-lapse movies for a fusion event. These spurious peaks were more common in the 5-min dataset and could be the result of low dynamic range due to the acquisition parameters. The remaining cells that were not categorized as dividing were defined as non-dividing if the cell was present for 48 h (15-min interval), or greater than or equal to 41.67 h (5-min interval).
The CellProfiler pipeline exports CSV files first preprocessed in Microsoft Excel then analyzed in MATLAB. First, the csv are input into batchreader.m, which generates a cell array of tables containing each cell’s measured parameters. The data is then input into the script ktrTablePlotter.m, which plots KTR dynamics. Cell division events are detected using the script Div_detection.m. For single reporter experiments, ERK or Akt KTR dynamics were utilized for cell division detection. For experiments with cells containing both reporters under 15-min acquisition Akt KTR was utilized for division detection; for 5-min acquisition, ERK KTR was utilized. Division events were manually curated in addition as described above. Cells were separated by division status, and KTR dynamics were plotted for each class.
Statistics, classification, and visualization
Where box plots are shown, right tailed rank-sum tests were used to calculate p-values for differences between median dividing and non-dividing cells. For logistic regression analysis, individual cell median ERK and Akt activity were calculated across the 8.5–40 h interval. The MATLAB function fitglm, with the following parameters ('Distribution','binomial','link','logit') was applied to cell median activity and cell division response to generate the generalized linear regression model along with its coefficients, and p-values.
NotBoxPlot was retrieved from MATLAB Central File Exchange (Rob Campbell, 2021). Scatter plots of single cell ERK and Akt activity across all timepoints within the 8.5–40 h interval was generated using the MATLAB Central File Exchange script Scatplot.m—Alex Sanchez (2020). To assess statistical significance of the correlation coefficient, the mean (µ) and covariance (σ) between ERK and Akt were calculated across all biological replicates and used to sample matched numbers of data points from random multivariate normal distributions for dividing cells and non-dividing cells. This was repeated 1000 times to define the range of correlation coefficients between the 5th and 95th percentiles, which was reported and rounded up to the nearest 0.01.
Data availability
Analysis pipelines can be found on our lab’s GitHub, along with host links for sample datasets (github.com/birtwistlelab/Predicting-Individual-Cell-Division-Events-from-Single-Cell-ERK-and-Akt-Dynamics).
References
Albeck, J. G., Mills, G. B. & Brugge, J. S. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261 (2013).
Worster, D. T. et al. Akt and ERK control the proliferative response of mammary epithelial cells to the growth factors IGF-1 and EGF through the cell cycle inhibitor p57 Kip2. Sci. Signal. 5, 1–13 (2012).
Santos, S. D. M., Verveer, P. J. & Bastiaens, P. I. H. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 9, 324–330 (2007).
Wei, Z. & Liu, H. T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 (2002).
Roberts, P. J. & Der, C. J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26, 3291–3310 (2007).
De-Luca, A., Maiello, M. R., D’Alessio, A., Pergameno, M. & Normanno, N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther. Targets 16, 17–27 (2012).
Chang, F. et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: A target for cancer chemotherapy. Leukemia 17, 590–603 (2003).
Li, B. B. et al. Targeted profiling of RNA translation reveals mTOR- 4EBP1/2-independent translation regulation of mRNAs encoding ribosomal proteins. Proc. Natl. Acad. Sci. USA 115, E9325–E9332 (2018).
Morita, M. et al. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle Georget. Tex 14, 473–480 (2015).
Xu, N., Lao, Y., Zhang, Y. & Gillespie, D. A. Akt: A double-edged sword in cell proliferation and genome stability. J. Oncol. https://doi.org/10.1155/2012/951724 (2012).
Ripple, M. O., Kalmadi, S. & Eastman, A. Inhibition of either phosphatidylinositol 3-kinase/Akt or the mitogen/extracellular-regulated kinase, MEK/ERK, signaling pathways suppress growth of breast cancer cell lines, but MEK/ERK signaling is critical for cell survival. Breast Cancer Res. Treat. 93, 177–188 (2005).
McKay, M. M. & Morrison, D. K. Integrating signals from RTKs to ERK/MAPK. Oncogene 26, 3113–3121 (2007).
Cirit, M., Wang, C. C. & Haugh, J. M. Systematic quantification of negative feedback mechanisms in the extracellular signal-regulated kinase (ERK) signaling network. J. Biol. Chem. 285, 36736–36744 (2010).
Schaub, F. X. et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 6, 282-300.e2 (2018).
Murphy, L. O. & Blenis, J. MAPK signal specificity: The right place at the right time. Trends Biochem. Sci. 31, 268–275 (2006).
Murphy, L. O., MacKeigan, J. P. & Blenis, J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol. Cell. Biol. 24, 144–153 (2004).
Wee, P. & Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 9, 1–45 (2017).
Torii, S., Yamamoto, T., Tsuchiya, Y. & Nishida, E. ERK MAP kinase in G1 cell cycle progression and cancer. Cancer Sci. 97, 697–702 (2006).
Mendoza, M. C., Er, E. E. & Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 36, 320–328 (2011).
Dyson, N. The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 (1998).
Borisov, N. et al. Systems-level interactions between insulin–EGF networks amplify mitogenic signaling. Mol. Syst. Biol. 5, 256 (2009).
Gross, S. M. & Rotwein, P. Akt signaling dynamics in individual cells. J. Cell Sci. 128, 2509–2519 (2015).
Aksamitiene, E., Kiyatkin, A. & Kholodenko, B. N. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: A fine balance. Biochem. Soc. Trans. 40, 139–146 (2012).
Birtwistle, M. R. et al. Emergence of bimodal cell population responses from the interplay between analog single-cell signaling and protein expression noise. BMC Syst. Biol. 6, 109 (2012).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Xu, N., Lao, Y., Zhang, Y. & Gillespie, D. A. Akt: A double-edged sword in cell proliferation and genome stability. J. Oncol. 2012, 1–15 (2012).
Zhu, J., Blenis, J. & Yuan, J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. USA 105, 1–6 (2008).
Bretones, G., Delgado, M. D. & León, J. Myc and cell cycle control. Biochim. Biophys. Acta Gene Regul. Mech. 1849, 506–516 (2015).
Knauf, J. A. et al. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J. Biol. Chem. 281, 3800–3809 (2006).
Abbott, D. W. & Holt, J. T. Mitogen-activated protein kinase kinase 2 activation is essential for progression through the G2/M checkpoint arrest in cells exposed to ionizing radiation. J. Biol. Chem. 274, 2732–2742 (1999).
Wright, J. H. et al. Mitogen-activated protein kinase kinase activity is required for the G2/M transition of the cell cycle in mammalian fibroblasts. Proc. Natl. Acad. Sci. USA 96, 11335–11340 (1999).
Dumesic, P. A., Scholl, F. A., Barragan, D. I. & Khavari, P. A. Erk1/2 MAP kinases are required for epidermal G2/M progression. J. Cell Biol. 185, 409–422 (2009).
Liu, X., Yan, S., Zhou, T., Terada, Y. & Erikson, R. L. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene 23, 763–776 (2004).
Chambard, J. C., Lefloch, R., Pouysségur, J. & Lenormand, P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta Mol. Cell Res. 1773, 1299–1310 (2007).
Dangi, S., Chen, F. M. & Shapiro, P. Activation of extracellular signal-regulated kinase (ERK) in G2 phase delays mitotic entry through p21CIP1. Cell Prolif. 39, 261–279 (2006).
De, S., Campbell, C., Venkitaraman, A. R. & Esposito, A. Pulsatile MAPK signaling modulates p53 activity to control cell fate decisions at the G2 checkpoint for DNA Damage. Cell Rep. 30, 2083-2093.e5 (2020).
Shtivelman, E., Sussman, J. & Stokoe, D. A role for PI 3-Kinase and PKB activity in the G2/M phase of the cell cycle. Curr. Biol. 12, 919–924 (2002).
Lee, S. R. et al. Akt-induced promotion of cell-cycle progression at G2/M phase involves upregulation of NF-Y binding activity in PC12 cells. J. Cell. Physiol. 205, 270–277 (2005).
Jaiswal, H. et al. ATM /Wip1 activities at chromatin control Plk1 re-activation to determine G2 checkpoint duration. EMBO J. 36, 2161–2176 (2017).
Maryu, G., Matsuda, M. & Aoki, K. Multiplexed fluorescence imaging of ERK and Akt activities and cell-cycle progression. Cell Struct. Funct. 41, 81–92 (2016).
Goglia, A. G. et al. A live-cell screen for altered Erk dynamics reveals principles of proliferative control. Cell Syst. 10, 240-253.e6 (2020).
Regot, S., Hughey, J. J., Bajar, B. T., Carrasco, S. & Covert, M. W. High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 (2018).
Gross, S. M., Dane, M. A., Bucher, E. & Heiser, L. M. Individual cells can resolve variations in stimulus intensity along the IGF-PI3K-AKT signaling axis. Cell Syst. 9, 580-588.e4 (2019).
Shankaran, H. et al. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 5, 332 (2009).
Min, M., Rong, Y., Tian, C. & Spencer, S. Temporal integration of mitogen history in mother cells controls proliferation of daughter cells. Science 8241, 8241 (2020).
Meyer, R. et al. Heterogeneous kinetics of AKT signaling in individual cells are accounted for by variable protein concentration. Front. Physiol. 3, 5 (2012).
Blum, Y. et al. Temporal perturbation of ERK dynamics reveals network architecture of FGF2/MAPK signaling. Mol. Syst. Biol. 15, 11 (2019).
Uhlitz, F. et al. An immediate–late gene expression module decodes erk signal duration. Mol. Syst. Biol. 13, 928 (2017).
Wesley-Overton, K., Spencer, S. L., Noderer, W. L., Meyer, T. & Wang, C. L. Basal p21 controls population heterogeneity in cycling and quiescent cell cycle states. Proc. Natl. Acad. Sci. USA 111, E4386–E4393 (2014).
Bouhaddou, M. et al. A mechanistic pan-cancer pathway model informed by multi-omics data interprets stochastic cell fate responses to drugs and mitogens. PLOS Comput. Biol. 14, e1005985 (2018).
Gille, H. & Downward, J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J. Biol. Chem. 274, 22033–22040 (1999).
Taylor, S. J. & Shalloway, D. Cell cycle-dependent activation of Ras. Curr. Biol. 6, 1621–1627 (1996).
Sah, J. F., Eckert, R. L., Chandraratna, R. A. S. & Rorke, E. A. Retinoids suppress epidermal growth factor-associated cell proliferation by inhibiting epidermal growth factor receptor-dependent ERK1/2 activation. J. Biol. Chem. 277, 9728–9735 (2002).
Jones, S. M., Klinghoffer, R., Prestwich, G. D., Toker, A. & Kazlauskas, A. PDGF induces an early and a late wave of PI 3-kinase activity, and only the late wave is required for progression through G1. Curr. Biol. 9, 512–521 (1999).
Zwang, Y. et al. Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Mol. Cell 42, 524–535 (2011).
Jones, S. M. & Kazlauskas, A. Growth-factor-dependent mitogenesis requires two distinct phases of signalling. Nat. Cell Biol. 3, 165–172 (2001).
Chen, J. Y., Lin, J. R., Cimprich, K. A. & Meyer, T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol. Cell 45, 196–209 (2012).
Soule, H. D. et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 (1990).
Isakoff, S. J. et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 65, 10992–11000 (2005).
Flusberg, D. A., Roux, J., Spencer, S. L. & Sorger, P. K. Cells surviving fractional killing by TRAIL exhibit transient but sustainable resistance and inflammatory phenotypes. Mol. Biol. Cell 24, 2186–2200 (2013).
Heiser, L. M. et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA 109, 2724–2729 (2012).
Spencer, S. L. et al. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013).
Ng, P.K.-S. et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell 33, 450-462.e10 (2018).
Sampattavanich, S. et al. Encoding growth factor identity in the temporal dynamics of FOXO3 under the combinatorial control of ERK and AKT kinases. Cell Syst. 6, 664-678.e9 (2018).
Regot, S., Hughey, J. J., Bajar, B. T., Carrasco, S. & Covert, M. W. High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 (2014).
Debnath, J., Muthuswamy, S. K. & Brugge, J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003).
Arkun, Y. Dynamic modeling and analysis of the cross-talk between insulin/akt and mapk/erk signaling pathways. PLoS ONE 11, 1–22 (2016).
Hoeflich, K. P. et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. 15, 4649–4664 (2009).
Moelling, K., Schad, K., Bosse, M., Zimmermann, S. & Schweneker, M. Regulation of Raf-Akt cross-talk. J. Biol. Chem. 277, 31099–31106 (2002).
Sears, R. et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14, 2501–2514 (2000).
Massagué, J. G1 cell-cycle control and cancer. Nature 432, 298–306 (2004).
Adlung, L. et al. Protein abundance of AKT and ERK pathway components governs cell type-specific regulation of proliferation. Mol. Syst. Biol. 13, 904 (2017).
Smith, G. R. et al. Network reconstruction from perturbation time course data. bioRxiv 2020, 341008. https://doi.org/10.1101/341008 (2020).
Gookin, S. et al. A map of protein dynamics during cell-cycle progression and cell-cycle exit. PLOS Biol. 15, e2003268 (2017).
Hiratsuka, T., Bordeu, I., Pruessner, G. & Watt, F. M. Regulation of ERK basal and pulsatile activity control proliferation and exit from the stem cell compartment in mammalian epidermis. Proc. Natl. Acad. Sci. 117, 17796–17807 (2020).
Gagliardi, P. A. et al. Collective ERK/Akt activity waves orchestrate epithelial homeostasis by driving apoptosis-induced survival. Dev. Cell 56, 1712-1726.e6 (2021).
Komatsu, N. et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–4656 (2011).
Gross, S. M. & Rotwein, P. Quantification of growth factor signaling and pathway cross talk by live-cell imaging. Am. J. Physiol. Cell Physiol. 312, C328–C340 (2017).
Asada, S. et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell. Signal. 19, 519–527 (2007).
Huang, H., Regan, K. M., Lou, Z., Chen, J. & Tindall, D. J. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314, 294–297 (2006).
Ozcan, L. et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 15, 739–751 (2012).
Woods, Y. L. et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 355, 597–607 (2001).
Yuan, C., Wang, L., Zhou, L. & Fu, Z. The function of FOXO1 in the late phases of the cell cycle is suppressed by PLK1-mediated phosphorylation. Cell Cycle 13, 807–819 (2014).
Tzivion, G., Dobson, M. & Ramakrishnan, G. FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta BBA Mol. Cell Res. 1813, 1938–1945 (2011).
Camelliti, P., Gallagher, J. O., Kohl, P. & McCulloch, A. D. Micropatterned cell cultures on elastic membranes as an in vitro model of myocardium. Nat. Protoc. 1, 1379–1391 (2006).
Ron, A. et al. Cell shape information is transduced through tension-independent mechanisms. Nat. Commun. 8, 2145 (2017).
Snijder, B. et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461, 520–523 (2009).
Santos, L. C. et al. Mitochondrial origins of fractional control in regulated cell death. Nat. Commun. 10, 1313 (2019).
Yang, H. W., Chung, M., Kudo, T. & Meyer, T. Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature 549, 404–408 (2017).
Christiansen, E. M. et al. In silico labeling: Predicting fluorescent labels in unlabeled images. Cell 173, 792-803.e19 (2018).
Vicar, T. et al. Cell segmentation methods for label-free contrast microscopy: Review and comprehensive comparison. BMC Bioinform. 20, 360 (2019).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Vasjari, L., Bresan, S., Biskup, C., Pai, G. & Rubio, I. Ras signals principally via Erk in G1 but cooperates with PI3K/Akt for Cyclin D induction and S-phase entry. Cell Cycle Georget. Tex 18, 204–225 (2019).
Serra, V. et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 30, 2547–2557 (2011).
Sommer, C., Straehle, C., Köthe, U. & Hamprecht, F. A. Ilastik: Interactive learning and segmentation toolkit. In 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 230–233 (2011). https://doi.org/10.1109/ISBI.2011.5872394.
Carpenter, A. E. et al. Cell Profiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
McQuin, C. et al. Cell Profiler 3.0: Next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018).
Berg, S. et al. ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Delgado-Gonzalo, R., Denervaud, N., Maerkl, S. & Unser, M. Multi-target tracking of packed yeast cells. In 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 544–547 (IEEE, 2010). https://doi.org/10.1109/ISBI.2010.5490288.
Acknowledgements
We thank Sean Gross and Laura Heiser for sharing Akt KTR constructs, and John Albeck for helpful discussion. We thank NIH-NIGMS (R01GM104184 to MRB and F31GM129985 to ADS), Clemson University, and the Icahn School of Medicine at Mount Sinai for funding.
Author information
Authors and Affiliations
Contributions
A.D.S. generated the KTR expressing MCF10A cells lines, developed the image processing pipeline, acquired, and performed data analysis for KTR dynamics as well as flow cytometry data acquisition and processing. G.R.S. and D.S. developed the network analysis framework used for determining if interactions are present between ERK and Akt pathways. L.C.S. aided in collecting flow cytometry data for cell cycle progression. X.Z. F.L., helped with dockerization and code revision. X.L. helped with DNA preparation. G.P. helped with interpreting HCTSA results. R.I. and M.B. provided experimental guidance.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stern, A.D., Smith, G.R., Santos, L.C. et al. Relating individual cell division events to single-cell ERK and Akt activity time courses. Sci Rep 12, 18077 (2022). https://doi.org/10.1038/s41598-022-23071-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23071-6
This article is cited by
-
Low-frequency ERK and Akt activity dynamics are predictive of stochastic cell division events
npj Systems Biology and Applications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.