Abstract
Net-casting spiders (Deinopidae) comprise a charismatic family with an enigmatic evolutionary history. There are 67 described species of deinopids, placed among three genera, Deinopis, Menneus, and Asianopis, that are distributed globally throughout the tropics and subtropics. Deinopis and Asianopis, the ogre-faced spiders, are best known for their giant light-capturing posterior median eyes (PME), whereas Menneus does not have enlarged PMEs. Molecular phylogenetic studies have revealed discordance between morphology and molecular data. We employed a character-rich ultra-conserved element (UCE) dataset and a taxon-rich cytochrome-oxidase I (COI) dataset to reconstruct a genus-level phylogeny of Deinopidae, aiming to investigate the group’s historical biogeography, and examine PME size evolution. Although the phylogenetic results support the monophyly of Menneus and the single reduction of PME size in deinopids, these data also show that Deinopis is not monophyletic. Consequently, we formally transfer 24 Deinopis species to Asianopis; the transfers comprise all of the African, Australian, South Pacific, and a subset of Central American and Mexican species. Following the divergence of Eastern and Western deinopids in the Cretaceous, Deinopis/Asianopis dispersed from Africa, through Asia and into Australia with its biogeographic history reflecting separation of Western Gondwana as well as long-distance dispersal events.
Similar content being viewed by others
Introduction
Once characterized as rare1, the net-casting spiders (family: Deinopidae; C.L. Koch, 1850) are cryptic, challenging to collect, and historically under-sampled and understudied. Living deep in understory habitats throughout tropical and subtropical regions2, deinopids were seldom collected1 and their perceived rarity is likely due to inattention and their unusual habits. Therefore, despite being charismatic spiders admired by most arachnologists, the difficulty in sampling them has resulted in very poor knowledge of the group’s evolutionary history. Deinopids, like the majority of web-spinning spiders, are sit-and-wait predators; however, they have a unique hunting strategy. Instead of sitting in their web, these spiders employ a modified orb web3 that they manipulate using their two anterior leg pairs. Using a silken net spun of a fuzzy, mechanical-capture silk (cribellate silk) rather than gluey silk, deinopids remain motionless until a prospective prey comes into view. They lunge into a forward strike, extending their webs with their legs and then envelope their prey, rendering them immobile1,4,5,6,7. They are also capable of a backward strike detecting vibrations of flying prey, hence are not entirely reliant on their enlarged eyes for prey capture7.
Historically, phylogenetic analyses based on morphology have divided the family into two genera: Deinopis, the ogre-faced spiders, aptly named for their massive posterior median eyes (PMEs), and Menneus, the humpback spiders1. Both genera share the unique net-casting hunting strategy, yet only Deinopis species have distinctively enlarged PMEs. Eye size, function, orientation, and visual field overlap all contribute to how spiders perceive visual signals. While some spiders have visual fields that span 360°8, the spider optical system is highly diversified across taxa and typically aids in prey recognition, hunting, predator avoidance, mating, and courtship9,10,11. There are two eye types, the principal eyes (typically three pairs), which capture light, and a pair of eyes (the PMEs) which are resolution-based10,12. In deinopids, the PMEs are forward-facing with low visual acuity and are primarily responsible for detecting motion10. Remarkably, Deinopis PMEs are the largest simple eyes of any arthropod and are 2000 × more sensitive to light than human photoreceptors13. They are a particularly important feature that enables visual hunting at night in low-light conditions14. Although other characters differentiate the two nominal genera, including abdominal tubercles, genitalic features, and their geographic distributions, deinopids have been notoriously difficult to diagnose based on somatic characters alone, particularly to species level1.
Molecular phylogenetic treatments of Deinopidae have focused on species-level relationships within Deinopis15 and a newly described genus in Asia, Asianopis16, formerly Deinopis. Molecular data revealed that Eastern Hemisphere Deinopis are more closely related to Menneus from South Africa than to Western Hemisphere Deinopis15. Blest et al.17 postulated that Menneus PMEs were a plausible ancestral form of the enlarged Deinopis PMEs. Deinopis paraphyly contradicts a morphology-based phylogeny of deinopids1, and foments questions: did deinopids gain their large PMEs within the clade, or ancestrally followed by subsequent loss, and when did this shift in PME size occur? Furthermore, did this shift in PME size occur once or multiple times across the evolutionary history of deinopids? Eye size reductions and even complete eye loss are typical in troglomorphic organisms, including crustaceans18,19, fish20,21, blind mole rats22, beetles23, and spiders24,25,26,27, where, in eternal darkness, energetically costly eyes may no longer offer fitness benefits28. Troglomorphic arthropods have been useful systems for studying the evolution and genetic underpinnings and regulation of eye size18,29,30; however, in arachnids, the origins, development, and evolution of eye size remains largely unexplored12,31. Alternatively, large PMEs could have evolved multiple times in parallel in deinopids. While energetically costly to maintain, large eye size has been associated with enhanced survival, fitness, and prey capture in vertebrates32,33,34,35 and invertebrates36,37,38.
Trait evolution must be considered across geologic time scales in order to better understand macroevolutionary patterns. Teasing apart the role of vicariance—the geographic separation of populations over time leading to divergent species—from long-distance, often overwater, dispersal (long-distance dispersal—LDD) (e.g.40,41,42,43,44) in generating species distributions is at the center of biogeographic studies. Rigorous methods and model comparison frameworks have allowed researchers to explicitly test dispersal hypotheses45. Disjunct distributions of Southern Hemisphere taxa have largely been attributed to vicariance driven by the breakup of Gondwana, the southern portion of Pangaea, in the Mesozoic46. Deinopids were present on Gondwana prior to the breakup of the supercontinent, with the separation of Africa and South America generating the Eastern and Western Hemisphere deinopid clades15. Both vicariance and LDD have been important in shaping the distribution of deinopids in the Western Hemisphere15. Vicariance and LDD have also shaped the distributions of deinopids across the Eastern Hemisphere; however, the timing of this dispersal has not yet been extensively tested. Eastern Hemisphere deinopids contain both large-eyed Deinopis and Asianopis and small-eyed Menneus, and they are distributed throughout South Africa, Madagascar, Southeastern Asia, Indomalaya, and Australia. Because the Chamberland et al.15 molecular phylogeny of Menneus only contained species from South Africa, we have yet to ascertain when and where Menneus hypothesized eye size reduction, or Deinopis and Asianopis eye size increase, evolved. Did this evolutionary shift in eye size occur on the same continent and then the taxa dispersed (Menneus is monophyletic; trait conservatism) or did these shifts occur multiple times on different continents (Menneus is polyphyletic; trait convergence/parallel evolution) (Fig. 1)? With new Menneus and Deinopis specimens from Australia, we were able to more thoroughly test the historical biogeography of Eastern Hemisphere deinopids, while exploring the evolution of PME size across geographic time scales.
Main phylogenetic hypotheses of Deinopidae assessing the monophyly of Menneus. We included all alternative tree topologies, which include non-monophyly of Deinopis and Asianopis in Supplemental Fig. S1. Phylogenies are based on (a) morphological data, (b) incomplete molecular data. The two primary alternative hypotheses we tested with the addition of Menneus from Australia: (c) PME size is reduced once and (d) PME size is reduced twice, independently in Australian and South African Deinopis. Map was created using the base map form Wikimedia Commons (https://commons.wikimedia.org/wiki/File:BlankMap-World.svg) and country colors were modified using Adobe Illustrator (https://www.adobe.com/) (Map: Lisa Chamberland).
In the study, we have integrated a taxon-rich COI dataset with a locus-rich UCE dataset to infer phylogenetic relationships within the family Deinopidae and explicitly test the monophyly of Menneus, Asianopis, and Deinopis. We expanded taxon sampling beyond Chamberland et al.15 to include Menneus specimens from South Africa and Australia and Deinopis from Australia, South Africa, Taiwan, Madagascar, and Mexico (Fig. 2). Specifically, we seek to infer the first genus-level phylogeny of the family and explicitly evaluate the evolution of eye reduction in Menneus. We tested the hypotheses that the ‘regular’ sized PMEs of Menneus either (1) represent an ancestral state, (2) evolved once from a common ancestor shared with Deinopis that then dispersed to other continents, or (3) arose independently from Deinopis in South Africa and Australia. Finally, we examine the ancient Gondwanan biogeographic history of Deinopidae within the context of the newly derived phylogenetic hypothesis.
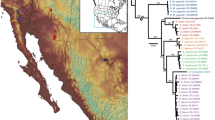
Sampling map of deinopids. Localities colored by current taxonomic genus. Map was created using the base map form Wikimedia Commons (https://commons.wikimedia.org/wiki/File:BlankMap-World.svg) Map: Lisa Chamberland).
Results
Phylogenetics
The taxon-rich cytochrome-oxidase I (COI) dataset comprised 258 deinopids and 4 Uloboridae outgroup individuals. Phylogenetic analysis of the COI dataset yielded weak nodal support for a number of important clades, including the placement of Menneus. To resolve the ambiguous phylogenetic relationships, we generated UCEs for 40 deinopid individuals across all major clades. We found strong support for Menneus monophyly and Deinopis paraphyly with the UCE and UCE + COI concatenated datasets (Figs. 3, 4; Table 1). Although the COI-only analyses did not support the monophyly of Menneus, nodal support was low (Supplementary Fig. S2). Furthermore, the tree topology tests on the UCE dataset supported the unconstrained tree, with Menneus monophyletic and Deinopis and Asianopis both paraphyletic, and rejected all four alternative tree topology hypotheses (see “Methods”; Table 1; Supplementary Table S1).
Maximum-likelihood phylogenetic tree of Deinopidae inferred using the COI + UCE (75% occupancy) matrix. Clades are colored by geographic location. The concatenated dataset resolves the uncertain placement of Australian Menneus and strongly supports the monophyly of Menneus sister to Eastern Hemisphere Deinopis. Numbers at nodes indicate bootstrap support values. Black stars indicate proposed genera (Deinopis, Menneus, and Asianopis).
We found strong support for Eastern and a Western Hemisphere deinopid clades across all phylogenetic analyses (Figs. 3, 4). The Western Hemisphere clade contained a Caribbean clade nested within a South and Central American grade, which was consistent with the phylogeny from Chamberland et al.15. There were nine nominal and nine putative species nested within the Eastern Hemisphere clade, including: three Menneus species from South Africa and three from Australia; two Deinopis species from Madagascar, three from South Africa, three from Australia, and at least one from Mexico; and five Asianopis species, including an undescribed Asianopis species from Taiwan (Fig. 3). The relationships of nominal genera differed between the COI only analyses and the analyses that contained UCE data; however, bootstrap support at these nodes were low and were most likely the result of using a single mitochondrial gene47,48,49,50,51,52. The addition of the UCE backbone to the COI dataset resulted in higher bootstrap support at the nodes. Thus, for all subsequent inferences and interpretations, we only considered the UCE only and UCE + COI phylogenies. The Eastern Hemisphere deinopid clade contained Menneus, Asianopis, and paraphyletic Eastern hemisphere Deinopis (Fig. 3). African, Australian, and Malagasy Deinopis were each monophyletic. We found D. cylindrica and an undescribed species, D. sp Hell’s Gates within an African clade. There were also two undescribed species within the Malagasy clade, D. sp Perinet and D. sp Andasibe-Mantadia. Finally, all phylogenetic analyses indicated three Deinopis clades in Australia, including D. subrufa in Eastern Australia, D. unicolor in Western Australia, and an undescribed species in Western Australia (Figs. 3, 4).
There is a well-supported relationship of a group of Mexican Deinopis sister to South African Deinopis (Fig. 3; Supplementary Figs. S3 and S4), thus supporting at least one Mexican group in the Eastern Hemisphere Deinopis + Asianopis clade. Individuals from this clade share morphological traits that characterize Eastern Hemisphere Deinopis and Asianopis, including bulky (as opposed elongated and narrow) abdomens. A second Mexican clade was sister to the entire Eastern Deinopis + Asianopis clade; however, this result was weakly supported and these individuals were represented by COI data only (Supplementary Fig. S5).
Within Asianopis, we recovered UCE data for A. wangi and A. wuchoai. A. wangi, A. wuchaou, A. zhuanghaoyuni, and A. sp from Taiwan all formed a clade sister to Australian Deinopis. Asianopis was paraphyletic in the UCE + COI phylogenetic inference with A. liukuensis sister to an Australian Deinopis + Asianopis clade. We were unable to recover UCEs for A. liukuensis; however, the COI only and COI + UCE concatenated phylogenetic reconstructions nested Australian Deinopis within the Asianopis clade. The A. liukuensis clade comprised specimens from China and India from Lin et al.16 and one specimen from Taiwan from the current dataset. Still, in all analyses, Asianopis (excluding A. liukuensis) was sister to Australian Deinopis.
Both the UCE only and COI + UCE analyses supported Menneus as monophyletic and sister to Eastern Hemisphere Deinopis + Asianopis clade. Both African and Australian Menneus were reciprocally monophyletic with three putative species in each geographic clade. We recovered UCEs for M. camelus and an undescribed species of Menneus from Africa, but only COI data for M. capensis.
Divergence time estimates and biogeography
Eastern and Western Hemisphere deinopids diverged in the lower Cretaceous and had an ancestral range in Africa + Australia + Asia (Fig. 4, Table 1). Following a vicariant divergence of Eastern and Western deinopids, Menneus diverged from the rest of the Eastern hemisphere deinopids generating a sympatric distribution of South African Menneus and Deinopis (Fig. 4). These vicariant and sympatric divergences generated three distinct clades: Western Hemisphere Deinopis, Eastern Hemisphere Deinopis + Asianopis, and Menneus.
Results from the biogeographic analyses supported LDD and vicariant divergences within Eastern Hemisphere Deinopis + Asianopis (Fig. 4). There are two Mexican deinopid clades that diverge from the South African Deinopis, one in the Upper Cretaceous and a second in the Eocene. In the absence of UCE data for the earlier diverging Mexican clade, the mechanism and direction of the dispersal was uncertain. Following the divergence of Mexican and South African Deinopis, there were two subsequent vicariant divergences: (1) African and Malagasy Deinopis diverged, (2) Malagasy Deinopis diverged from India + Asian Asianopis. Finally, there was a single LDD event from Asia to Australia resulting in the Australian Deinopis clade (Fig. 4). Both the UCE only (A. liukuensis absent) and UCE + COI concatenated biogeographic analyses indicated an ancestral range of Asianopis + Australian Deinopis in Asia. The inferred ancestral range of Australian Menneus was South Africa + Australia and diverged from South Africa around 56 Ma (24.22–94.35, Ma HPD 95%) (Fig. 4, Table 1). Since the date of divergence post-dates when Africa split from Australia53, Menneus likely dispersed via LDD from South Africa to Australia (Fig. 4). The 95% highest posterior density (HPD)54 varied widely across nodes, and is likely a consequence of having only one fossil to date the phylogeny (Table 1, Fig. S5).
Ancestral state reconstruction
There was a single shift in PME size, from large to small, in Deinopidae at the point of Menneus diversification (Fig. 5a–c, Supplementary Fig. S7). PME size was highly conserved and had a strong phylogenetic signal estimated D (− 2.11). The average PME diameter (scaled to carapace width at the PLEs) and total ocular distance of adult females were significantly larger for Deinopis and Asianopis compared to Menneus (Fig. 5; Supplementary Fig. S8). The tests of PME/carapace ratio and raw averages for the Mann–Whitney non-parametric test of mean difference between large and typical size PMEs were both significant p < 0.0001. In the phylogenetic principal component analysis (pPCA), the first principal component (PC) (PC1, 90.2% of the variance) was strongly affected by carapace measurements: length (− 0.983), width at widest point (− 0.963), width at PLE (− 0.934). The second PC (PC2, 4.3% of the variance), with a negative loading on PME diameter (− 0.469) and PME row width (− 0.410) and positive loadings on anterior median eye (AME row width (+ 0.214) and carapace length (+ 0.163) (Supplementary Table S2). The directionality of the vectors in the biplots are close together, indicating these traits are highly positively correlated (Fig. 5d). PC1 reflected body size whereas PC2 reflected PME and AME size and eye row width; however, while the three genera generally clustered in the pPCA, the combination of morphological characters used did not strongly differentiate the three genera. There was no significant difference between PME size and total ocular distance between Deinopis and Asianopis; however, Menneus PMEs and total ocular distances were significantly smaller (Fig. 5e,f). Average AME size was highest in Menneus, although this was not significant (Fig. 5g).
Summary of eye size data. (a) MCC phylogeny with branch width representing the log of PME to carapace ratio. The size of the dots represents raw eye diameter (mm) and are colored by eye type: PME (yellow), AME (blueish green), PLE (blue), ALE (yellowish green). (b) A. subrufa and (c) M. sp Gurragawee with eyes colored by type- also represented in dot phylogeny. (d) pPCA biplot of deinopid genus clusters (n = 29) based on carapace and eye dimensions. Clades, boxplots, and pPCA are colored by genus: Asianopis (magenta), Deinopis (orange); Menneus (purple). Boxplots represent (e) PME diameter scaled to carapace width, (f) total ocular distance, and (g) AME scaled to carapace width.
Discussion
Monophyly of Menneus and evidence for secondary loss of enlarged PME
Phylogenetic data (UCE and UCE + COI datasets) strongly support the monophyly of Menneus, whereas Deinopis and Asianopsis are rendered paraphyletic (Figs. 3, 4). These findings contradict the prevailing taxonomic hypothesis of the genus Deinopis (MacLeay 1839), as well as the more recently proposed new genus Asianopis16. Instead, all analyses support a deep divergence between Western Hemisphere Deinopis and Eastern Hemisphere deinopids (Deinopis + Asianopis), and the UCE and UCE + COI phylogenies support divergence between Menneus and Eastern Hemisphere Deinopis. Recent taxonomic studies have transferred a number of the Asian Deinopis species into Asianopis16,55,56; however, Australian and Malagasy Deinopis have yet to be moved into Asianopis. Based on the geographic and taxonomic coverage of the family in this study, including the deep divergences between the Eastern Hemisphere Deinopis + Asianopis clade, the Menneus clade, and the paraphyly of Deinopis with respect to Asianopis, we propose the transfer (see Taxonomic Section below) of the Eastern Hemisphere Deinopis into Asianopis Lin et al.56, thus retaining the three genera within Deinopidae: Deinopis in the Western Hemisphere, Asianopis in the Eastern Hemisphere, and Menneus (Simon 1876). Based on the phylogenetic hypothesis, we also propose the transfer of a number of Central American and Mexican Deinopis to Asianopis. Although these results highlight the need for further taxonomic treatment of the family and the formulation of a new diagnosis of Asianopis, we see no other working alternative that would adequately reconcile these phylogenetic results with the current taxonomy. That is, considering Asianopis and Menneus as junior synonyms of Deinopsis would fail to appropriately acknowledge the family’s phylogenetic structure, complex and worldwide biogeography, remarkable morphological divergence, and lineage divergence over major evolutionary time scales. The error bars around all major divergences are large, which is likely due to limited data and the use of a single fossil and broad priors to date the phylogeny. Still, major divergences are consistent with the previously published dated phylogeny of deinopids15.
We found support for a single reduction of PME size in deinopids (Fig. 5a, Supplementary Figs. S7, S9), indicating enlarged PMEs were an ancestral deinopid trait with subsequent size reduction in Menneus. Although it has been postulated that Deinopis species evolved large PMEs through adaptation to hunt exclusively at night, leaving Menneus to hunt at dusk and twilight, empirical testing of this hypothesis has been limited to two Australian species: D. subrufus and M. unifasciatus57. Extensive field observations indicate that Deinopis and Menneus hunt at the same time in low-light conditions (pers. obsv.). The observation that both genera are hunting at the same time and in the same locality, using the same hunting strategy, raises the question—what are the potential ecological shifts and evolutionary tradeoffs driving morphological divergence? The reduction in Menneus eyes size could be interpreted as regressive evolution. Regressive evolution, the loss or reduction of non-functional, formally adaptive characters, has been repeatedly observed in the eyes of subterranean23,58 and cave dwelling organisms59, animals of the deep ocean, and among species that make the switch from diurnal to nocturnal habitats60,61,62. Alternatively, although PME size are reduced in Menneus, they are not non-functional. Analyses of the anatomical structures have indicated Deinopis PMEs are more sensitive than Menneus17; however the extent of the visual sensitivity between the two genera remain largely unknown and understudied.
Among organisms where regressive evolution has been observed, there are three primary, not mutually exclusive, hypotheses that seek to explain character reduction: (1) relaxed selection or neutral mutations—the relaxation of selection leading to reduction (e.g. loss of eyes, reduction of eye size)63,64,65; (2) natural selection, in which reduced visual structures or the absence of eyes are advantageous66,67,68; (3) indirect selection, pleiotropy, or ‘genetic hitchhiking’, in which the gene related to trait reduction is linked to other adaptive trait(s) (e.g. olfaction or taste)29,69,70,71. D. spinosa has been shown to use auditory cues to capture their prey7. Future studies could, for example, explore whether Menneus has evolved morphological and/or behavioral characters that are advantageous for capturing aerial prey (e.g. additional sensitive vibratory sensors such as trichobothria). Furthermore, in D. spinosa, individuals use their large eyes to hunt cursorial prey, which ultimately increases their dietary breadth14; however, enlarged PMEs are metabolically costly. Stafstrom et al.72 found that there was a potential tradeoff between brain size and PME size. Deinopis PMEs are more complex13,73 and twice as sensitive compared to Menneus PMEs17. We postulate that relaxed selection in environments where such eyes do not offer major fitness benefits may explain this hypothesized secondary loss. Explicit tests of differential time and mode of hunting and evaluation of the underlying genetics of phenotypic differences may help tease apart potential evolutionary mechanisms. Finally, we propose an alternative hypothesis in which Menneus have far more sensitive PMEs than spiders with similarly sized eyes. If there was strong selection for increased eye size, neural connections, and light detection mechanisms in Deinopis, an energy cost could be spared by reducing eye size but retaining the high sensitivity. In that case there is a secondary reduction in eye size, but not, a reversal to the ancestral spider condition. Future detailed studies on receptor function, light sensitive cells, and visual/neural system communication could find that taking that advanced eye and making it smaller in Menneus would be advantageous even with certain anatomical simplifications from Deinopis. Deinopids have tremendous potential as a unique system for testing these alternative hypotheses and for studying visual evolution.
Early Gondwanan vicariance followed by long-distance dispersals
The disjunct geographic distributions of deinopids can be attributed to Upper Cretaceous vicariance during the breakup of West Gondwana, followed by subsequent transoceanic dispersal events. Initially formed around 500 Ma, Gondwana, the southern portion of the supercontinent Pangaea, began breaking away in the Middle Jurassic around 170 Ma74,75,76,77. However, the continents did not all break apart in succession. Instead, continents were simultaneously separating, with phases of these overlapping divergent events lasting tens of millions of years. After the initial separation, land bridges persisted between continental masses, providing passageways for biotic exchange78. Even amongst taxa with low vagility, there is mounting evidence for early Gondwanan vicariance followed by subsequent LDD events in taxa, including plants79,80,81,82, cave shrimp83, freshwater fish84 and arachnids85,86. Of course, in the epochs immediately following continental breakup, the distances between major landmasses were shorter and thus dispersal among these would be more likely than among the continents in their current positions. Following these early divergences driven by the breakup of West Gondwana, we concluded that subsequent deinopid diversification occurred via long-distance, trans-oceanic dispersal events.
Dispersal into Asia and Australia
With a largely tropical and sub-tropical range, deinopids do not follow the typical anti-equatorial distributions across the Southern Hemisphere found in many Gondwanan lineages87,88,89,90,91,92,93; nonetheless, vestiges of the ancient Gondwanan connections are still reflected in the biogeographic history of deinopids. Following the divergence from Menneus, Eastern Hemisphere Deinopis/Asianopis dispersed from South Africa to Madagascar where they subsequently spread into Asia. The Deinopis/Asianopis lineage dispersed and diversified throughout Asia, with later dispersal to Australia via Indonesia. The timing of the divergences and biogeographic analyses suggest that Deinopis may have arrived in Asia from India after India collided with Southeast Asia in the Eocene (Out of India hypothesis). The duration of the hypothesized IndoMadagascar landmass is debated. These may have separated as early as 130–125 Ma, but remained connected to India until around 84 Ma94,95 after which it is thought that fracture zones, plateaus, periodically emerged and facilitated stepping stone dispersal between Madagascar and India96. Such hypothesized stepping stone dispersal from Africa into Asia via India and Madagascar has been found in taxa with low vagility97,98,99. Alternatively, Deinopis could have dispersed through Northern Africa and Eurasia followed by subsequent extinction similar to sand scorpions100; however, our molecular phylogeny shows that the Madagascar clade is more closely related to Asia and India than Africa, consistent with the IndoMadagascar plus stepping stone hypothesis. Because this molecular dataset only included COI data for A. luikenisis from China and Taiwan, further sampling throughout India and Southeast Asia will be required to more rigorously test this dispersal hypothesis. Alternatively, these data support the hypothesis that Menneus dispersed to Australia via a single, trans-oceanic dispersal event, although divergence times of Australian and African Menneus post-date an Out of India hypothesis. With 37% of the total 46 described deinopid species in the Eastern Hemisphere (putative and genetic transfers), much of Africa, Asia, and Indomalaya remains under-sampled. More extensive sampling is required, particularly of Madagascar and Indomalaya, to more completely understand Menneus biogeographic history.
Crossing Wallace’s line
Wallace’s line is one of the most striking separations of biomes in the world. Wallace101 posited that the fauna of South America and Africa, despite being separated by the Atlantic Ocean, were more similar than the faunas of Asia and Australia. Based on our results, deinopids dispersed from Africa to Asia and then from Indomalaya to Australia 91–52 Ma (Fig. 4), which may reveal intriguing implications of dispersal across Wallace’s line. Furthermore, the Australian Deinopis diverged from its sister Indomalayan clade around 72 Ma (Fig. 4). This divergence pre-dates the Indo-Australian archipelago (IAA) and a deep sea and a large trench separated the two regions until around 40 Ma when islands began forming102. Recently the IAA has been discovered to be a more common route of dispersal than previously predicted in terrestrial animals102,103,104. We postulate that upon broader sampling in this region, we may find range expansions across the IAA in the Late Eocene to Early Miocene when landmasses were forming102.
South Africa, Madagascar, and the Neotropics
We found evidence for at least two trans-oceanic dispersals of South African Deinopis. First, Malagasy Deinopis diverged from South African Deinopis in the Paleocene, post-dating Lower Cretaceous continental drift and vicariance. Madagascar separated from Africa and moved southeastward in the Middle to Early Late Jurassic (160–155 Ma)105,106 and was one of the first of the Gondwanan landmasses to break away from the supercontinent. Madagascar was in its final position relative to Africa approximately 120 Ma (118–130 Ma)107,108. The relative contributions of Gondwanan vicariance and long-distance dispersal among Malagasy lineages remain a source of debate96,109,110,111. Madagascar’s biodiversity primarily comprises endemic flora and fauna that have either been present since Gondwana or were a product of long-distance dispersal events in the Cenozoic, mostly from Africa (400 km away), but including all major biogeographic regions96.
Second, we found evidence for at least one trans-oceanic long-distance dispersal between South Africa and Mexico in the Eocene. Discoveries of such extreme trans-oceanic dispersals are becoming increasingly common41,99,112 and can be attributed to salt-water tolerance113, long-distance flight114,115,116, and wind-dispersal in plants99 and ballooning spiders117,118. Exchanges between Africa, the Western Indian Ocean, and the Neotropics have been especially well documented in tropical flora119,120,121,122,123,124,125. Dispersal events from the Neotropics to Africa are facilitated by westerly winds125,126, whereas dispersals from Africa to the Neotropics, the rarer of the two dispersal scenarios125, typically follow the trade winds126. While there was at least one dispersal event between Africa to the Mexico, further sampling of Eastern Hemisphere deinopids in the Neotropics is necessary to rigorously test dispersal hypotheses.
Conclusions
Deinopids have a rich biogeographic history characterized by complex and ancient patterns of vicariance coupled with long distance dispersal since their emergence in the Cretaceous. While LDD has played an important role in shaping the distributions of deinopids, these events are not so prevalent as to completely obfuscate ancient Gondwanan signatures. The evolutionary history of Deinopidae provides fascinating examples of eye size reduction and unexpected trans-oceanic dispersals. Whereas most studies in eye size evolution are troglomorphic adaptations in the form of reductions/loss of eyes, here, we document an example of a single reversal to ‘normal’ sized eyes from the uniquely derived condition of grossly enlarged PMEs in ogre-faced spiders. The PME size reductions in Menneus provides a framework for considering and testing hypotheses of the potential selective forces that might be driving these divergent morphologies.
Taxonomic section
Based on phylogenetic structure and biogeographical distributions we propose the following new combinations (* indicates those taxa included in this study): Asianopis anchietae (Brito Capello127) NEW COMBINATION; Asianopis aspectans Pocock128 NEW COMBINATION; Asianopis aurita Pickard-Cambridge129 NEW COMBINATION; Asianopis camela Thorell130 NEW COMBINATION; Asianopis cornigera Gerstaecker131 NEW COMBINATION; Asianopis cylindrica* Pocock132 NEW COMBINATION; Asianopis fasciata Koch133 NEW COMBINATION; Asianopis fasciculigera Simon134 NEW COMBINATION; Asianopis giltayi Lessert135 NEW COMBINATION; Asianopis guineensis Berland & Millot136 NEW COMBINATION; Asianopis kollari Doleschall137 NEW COMBINATION; Asianopis labangan Barrion-Dupo & Barrion138 NEW COMBINATION; Asianopis longipalpula Strand139 NEW COMBINATION; Asianopis luzonensis Barrion-Dupo & Barrion138 NEW COMBINATION; Asianopis madagascariensis Lenz140 NEW COMBINATION; Asianopis mediocris Kulczyński141 NEW COMBINATION; Asianopis ornata Pocock142 NEW COMBINATION; Asianopis ravida Koch143 NEW COMBINATION; Asianopis reticulata Rainbow144 NEW COMBINATION; Asianopis schomburgki Karsch145 NEW COMBINATION; Asianopis schoutedeni Giltay146 NEW COMBINATION; Asianopis subrufa* Koch143 NEW COMBINATION; Asianopis tabidus Koch133 NEW COMBINATION; Asianopis unicolor* Koch143 NEW COMBINATION.
Methods
Taxon sampling
We collected a total of 42 deinopid individuals (29 Deinopis and 13 Menneus) from the east coast of Australia in January 2019 using standard aerial search and vegetation beating methods described in Coddington et al.147. Most of the specimens were collected at night as deinopids are highly cryptic and are most easily spotted when their nets reflect a blue hue under headlamp lights. The specimens were fixed in 95% ethanol in the field and kept at − 20 to − 80 °C in the lab. Collaborators also provided additional Deinopis specimens from Western Australia, Taiwan, Mexico, Madagascar, South Africa, and Colombia.
DNA extractions, sequence generation, and data processing
Detailed methods for molecular methods, sequence editing, and read processing are available in the Supplemental Methods S1. We extracted DNA from leg tissue for 65 individuals using the OMEGA BIO-TEK E.Z.N.A. DNA extraction kit. We amplified COI: cytochrome c oxidase subunit I, a mitochondrial locus. We obtained COI sequence data for seven species from GenBank for outgroups, following148. We included existing deinopid COI data available on DRYAD149 and Genbank from Lin et al.16 for additional global Deinopis samples and Asianopis samples, respectively. See Supplementary Table S3 for full taxon sample lists, GPS locality information, and Genbank accession numbers. Sequences were aligned in MAFFT150 and then edited by eye and checked for stop codons in Mesquite151.
For UCE data, we sampled at least two individuals from each major clade and their geographic representation, including from African Menneus, Australian Menneus, African Deinopis, Australian Deinopis, Malagasy Deinopis, Asianopis, North and Central American Deinopis, and Caribbean Deinopis. UCE libraries were hybridized to the Spider probeset148 150 bp paired-end reads were sequenced on the HiSeq4K at the University of California Davis DNA Technologies (See Supplemental Methods S1 for more detail). We obtained UCE data for four outgroup species from Kulkarni et al.148 (NCBI Sequence Read Archive, BioProject PRJNA575576). UCE and COI matrices were concatenated in AMAS152 to generate the UCE + COI combined dataset.
Phylogenetics
We inferred phylogenetic trees for the four molecular datasets: COI-only (258 deinopid individuals, four outgroup individuals; 1279 base pairs), UCE-only (40 deinopids, four outgroup individuals; 75% occupancy matrix; 1018 loci), COI + UCE concatenated, and COI with a UCE backbone phylogeny using Maximum Likelihood153 and coalescence-based methods154. In IQ-TREE 2153, we implemented the built-in model finder to consider invariant site and Gamma rate heterogeneity under the greedy strategy with the AICc criterion to determine molecular substitution models for each codon position. The COI dataset was partitioned by codon using the GTR + I + G and the UCE dataset was partitioned by locus using the GTR model. We ran four separate analyses on each dataset with 1000 pseudoreplicates of ultrafast bootstrapping155,156 on the codon-partitioned COI and COI-backbone datasets, and the loci-partitioned UCE, COI + UCE concatenated datasets. We generated trees from individual UCE loci using IQ-TREE. These trees were used to infer a coalescence-based species tree in IQ-TREE and ASTRAL-II v5.7.1154.
Tree topology tests
We implemented tree topology tests in IQ-TREE 2153 to evaluate the relationships of Asianopis, Deinopis, and Menneus, and to further assess the monophyly of these three groups. We tested the unconstrained tree and four alternative tree topologies (Supplementary Fig. S1) in IQ-TREE using the RELL approximation157, which uses bootstrap proportions, p-values with the Shimodaira-Hasegawa test158 and AU test159, and expected likelihood weights160. Using the UCE dataset, we inferred species trees in IQ-TREE for the datasets constrained to four alternative hypotheses (1) Menneus is paraphyletic (clades = New World Deinopis, Australian Deinopis + Menneus; African Deinopis + Menneus; Malagasy Deinopis; Asianopis); (2) Menneus is monophyletic and Deinopis and Asianopis form a clade (clades = Menneus, all Deinopis + Asianopis); (3) Menneus, New World Deinopis, Old World Deinopis, and Asianopis are each monophyletic; (4) Menneus, all Deinopis, and Asianopis are monophyletic (clades = Menneus, Deinopis, Asianopis) (Supplementary Fig. S1).
Molecular dating
We implemented MCMCTree in the PAML v4.8161 package on the UCE only dataset to estimate divergence times within Deinopidae. Prior to the analyses, we removed redundant subspecies so that only one individual per species (with the least amount of missing data) remained. To set up the model and model parameters, we assigned a birth–death model for the tree and an UCLN clock model (uncorrelated relaxed clock). A GTR substitution model was used with flat Dirichlet prior distribution. Previous phylogenetic dating analyses have used the fossil Palaeomicromenneus lebanensis to calibrate the stem of Deinopidae15,148,163. However, the taxonomy of this fossil is dubious164, and it has been transferred into the extinct family Salticoididae165; therefore we did not use this fossil as a calibration point in the dating inferences. Instead, we employed a molecular calibration point from recent phylogenetic reconstruction on Araneomorphs166 to conservatively use maximum age of 150 Ma as a soft constraint for the Deinopidae crown. We used a single fossil, Seppo koponeni to calibrate a soft constraint with the minimum root age of Deinopidae + outgroups of 132.9 and a maximum of 250 Ma. We ran two independent Markov chain Monte Carlo (MCMC) runs for 20,000 samples, resampling every 10,000 iterations for a total of 200,000,000 iterations with a burn-in set to discard the first 20,000,000 iterations. Outputs were assessed for stationarity in Tracer v1.7.1167 and visualized using FigTree v1.4.4168.
Ancestral range estimates
We estimated the ancestral ranges of deinopids and performed biogeographic analyses in BioGeoBEARS169 using the UCE-only MCC tree inferred in MCMCTree. We used eight isolated biogeographic areas including Africa (F), Asia (A), Australia (U), Neotropics (S), Nearctic (N), Madagascar (M), India (I), and the Caribbean (C), five of which were based on updated zoological regions of Holt et al.170 and on the most recent global biogeographic studies on spiders171. These regions were also selected due to their high endemism, geologic histories, and land availability. We included a time slice model based on the breakup of the continents following descriptions in Seton et al.’s172 of the historical geology and modeled after recent invertebrate dispersal models43,44 (Supplementary Table S4; See Supplemental Methods S1). We tested these models under the Dispersal‐Extinction‐Cladogenesis (DEC) and with the (+ j) parameter, which considers for jump dispersal or founder event speciation169,174. We compared Akaike information criterion (AIC) and relative likelihood scores across all models (a natural log of 2 was considered significant)175. We used the MCMCTree-generated UCE and UCE + COI dated phylogeny with outgroups pruned to only include Deinopidae specimens.
Ancestral state reconstruction and eye measurements
Measurements of adult female specimens were taken with a Leica M205 C scope using the micrometer scale tools in the Leica Application Suite (LAS Version 4.13.0) (Supplementary Table S5; See Supplemental Methods S1). We measured the total ocular distance and individual diameters of PME, anterior median eye (AME), anterior lateral eye (ALE), posterior lateral eye (PLE) diameter for all adult females. We also measured carapace length, width, and carapace width at the PLEs. A Mann–Whitney non-parametric test of mean difference was used to determine whether there was a statistically significant difference between large and small PME. We used a phylogenetic principal components analysis (pPCA) in R using the function phyl.pca in the R package phytools176 to determine if eye and carapace size, ten and three characters respectively, were partitioned among the three genera: Deinopis, Asianopis, and Menneus. We used a pruned MCC tree, which included 29 species throughout the tree that were absent of missing data. Data points included carapace length and width, eye diameter of all four types of eyes (PME, AME, PLE, and ALE) and the eye row width across each of these. We also calculated total ocular distance, the sum diameter of all eight eyes. We generated biplots in R of the scores of the 23 total traits obtained from the pPCA.
We tested the phylogenetic signal of the binary trait for eye size, “large” for Deinopis and “small/typical” for Menneus, and for the outgroups. We calculated D, which is a permutation-based model that measures the phylogenetic signal strength in a binary trait assuming the underlying continuous trait is evolving under Brownian Motion177 and was appropriate for eye size, which is a variable and potentially continuous trait within each genus.
We mapped the states for discrete characters, large and small PME, along the species level MCC phylogeny using the ace function in ape178 and in phytools176. To estimate ancestral states for discrete PME characters, we employed a continuous time Markov chain (Mk) model to give character probability distributions of ancestral states at internal nodes in the tree. We also used stochastic character mapping, an MCMC approach to sample character histories from their probability distributions.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. The UCE dataset generated and analyzed during the current study have been deposited at NCBI Short-Read Archive (https://www.ncbi.nlm.nih.gov/sra) as BioProject PRJNA802018. All COI sequences obtained in this study have been deposited at NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession numbers OM480748-OM480975.
References
Coddington, J. A., Kuntner, M. & Opell, B. D. Systematics of the spider family Deinopidae with a revision of the genus Menneus. (Smithsonian Contributions to Zoology, 2012).
Getty, R. M. & Coyle, F. A. Observations on prey capture and anti-predator behaviors of ogre-faced spiders (Deinopis) in southern Costa Rica (Araneae, Deinopidae). J. Arachnol. 24, 93–100 (1996).
Garrison, N. L. et al. Spider phylogenomics: Untangling the spider tree of life. PeerJ 4, e1719 (2016).
Coddington, J. A. Orb webs in “Non-orb weaving” ogre-faced spiders (Araneae: Dinopidae): A question of genealogy. Cladistics 2, 53–67 (1986).
Coddington, J. A. & Sobrevila, C. Web manipulation and two stereotyped attack behaviors in the ogre-faced spider Deinopis spinosus Marx (Araneae, Deinopidae). J. Arachnol. 15, (1987).
Blamires, S. J., Zhang, S. & Tso, I.-M. Webs: Diversity, Structure and Function. in Behaviour and Ecology of Spiders (eds. Viera, C. & Gonzaga, M. O.) 137–164 (Springer International Publishing, 2017). https://doi.org/10.1007/978-3-319-65717-2_6.
Stafstrom, J. A., Menda, G., Nitzany, E. I., Hebets, E. A. & Hoy, R. R. Ogre-faced, net-casting spiders use auditory cues to detect airborne prey. Curr. Biol. 30, 5033-5039.e3 (2020).
Spano, L., Long, S. M. & Jakob, E. M. Secondary eyes mediate the response to looming objects in jumping spiders (Phidippus audax, Salticidae). Biol. Lett. 8, 949–951 (2012).
Forster, L. M. Visual mechanisms of hunting behaviour in Trite planiceps, a jumping spider (Araneae: Salticidae). N. Z. J. Zool. 6, 79–93 (1979).
Land, M. F. The Morphology and Optics of Spider Eyes. in Neurobiology of Arachnids (ed. Barth, F. G.) 53–78 (Springer Berlin Heidelberg, 1985). https://doi.org/10.1007/978-3-642-70348-5_4.
Gainett, G. & Sharma, P. P. Genomic resources and toolkits for developmental study of whip spiders (Amblypygi) provide insights into arachnid genome evolution and antenniform leg patterning. EvoDevo 11, 18 (2020).
Foelix, R. F. Biology of spiders. (Oxford University Press, 2011).
Blest, A. D. & Land, M. F. The physiological optics of Dinopis subrufus L. Koch: A fish-lens in a spider. Proc. R. Soc. Lond. B Biol. Sci. 196, 197–222 (1977).
Stafstrom, J. A. & Hebets, E. A. Nocturnal foraging enhanced by enlarged secondary eyes in a net-casting spider. Biol. Lett. 12, 20160152 (2016).
Chamberland, L. et al. From Gondwana to GAARlandia: Evolutionary history and biogeography of ogre-faced spiders ( Deinopis ). J. Biogeogr. 45, 2442–2457 (2018).
Lin, Y. et al. Asianopis gen. nov., a new genus of the spider family Deinopidae from Asia. ZooKeys 911, 67–99 (2020).
Blest, A. D., Williams, D. S. & Kao, L. The posterior median eyes of the dinopid spider Menneus. Cell Tissue Res. 211, (1980).
Protas, M. E., Trontelj, P. & Patel, N. H. Genetic basis of eye and pigment loss in the cave crustacean Asellus aquaticus. Proc. Natl. Acad. Sci. 108, 5702–5707 (2011).
Klaus, S. et al. Rapid evolution of troglomorphic characters suggests selection rather than neutral mutation as a driver of eye reduction in cave crabs. Biol. Lett. 9, 20121098 (2013).
Langecker, T. G. & Longley, G. Morphological adaptations of the texas blind catfishes Trogloglanis pattersoni and Satan eurystomus (Siluriformes: Ictaluridae) to their underground environment. Copeia 1993, 976 (1993).
Wilkens, H. & Strecker, U. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): genetic evidence from reduced eye-size and pigmentation: Convergence of astyanax cavefish evolution. Biol. J. Linn. Soc. 80, 545–554 (2003).
David-Gray, Z. K., Janssen, J. W. H., DeGrip, W. J., Nevo, E. & Foster, R. G. Light detection in a ‘blind’ mammal. Nat. Neurosci. 1, 655–656 (1998).
Leys, R., Cooper, S. J. B., Strecker, U. & Wilkens, H. Regressive evolution of an eye pigment gene in independently evolved eyeless subterranean diving beetles. Biol. Lett. 1, 496–499 (2005).
Bosselaers, J. Nesticus henderickxi (Araneae, Nesticidae), a new blind troglobitic spider from Crete. Bull.-Br. Arachnol. Soc. 11, 9–14 (1998).
Bloom, T. et al. Discovery of two new species of eyeless spiders within a single Hispaniola cave. J. Arachnol. 42, 148–154 (2014).
Ribera, C., Elverici, M., Kunt, K. & Özkütük, R. Typhlonesticus gocmeni sp. n., a new cave-dwelling blind spider species from the Aegean region of Turkey (Araneae, Nesticidae). ZooKeys 419, 87–102 (2014).
Hedin, M. High-stakes species delimitation in eyeless cave spiders (Cicurina, Dictynidae, Araneae) from central Texas. Mol. Ecol. 24, 346–361 (2015).
Soares, D. & Niemiller, M. L. Extreme adaptation in caves. Anat. Rec. 303, 15–23 (2020).
Protas, M. et al. Multi-trait evolution in a cave fish, Astyanax mexicanus: QTL analysis of cave adaptation. Evol. Dev. 10, 196–209 (2008).
Re, C. et al. Common genetic basis of eye and pigment loss in two distinct cave populations of the isopod Crustacean Asellus aquaticus. Integr. Comp. Biol. 58, 421–430 (2018).
Morehouse, N. I., Buschbeck, E. K., Zurek, D. B., Steck, M. & Porter, M. L. Molecular evolution of spider vision: New opportunities familiar players. Biol. Bull. 233, 21–38 (2017).
Beston, S. M. & Walsh, M. R. Natural selection favours a larger eye in response to increased competition in natural populations of a vertebrate. Funct. Ecol. 33, 1321–1331 (2019).
Garamszegi, L. Z., Møller, A. P. & Erritzøe, J. Coevolving avian eye size and brain size in relation to prey capture and nocturnality. Proc. R. Soc. Lond. B Biol. Sci. 269, 961–967 (2002).
Ross, C. F. & Kirk, E. C. Evolution of eye size and shape in primates. J. Hum. Evol. 52, 294–313 (2007).
Hall, M. I. The anatomical relationships between the avian eye, orbit and sclerotic ring: Implications for inferring activity patterns in extinct birds. J. Anat. 212, 781–794 (2008).
Bauer, T., Desender, K., Morwinsky, T. & Betz, O. Eye morphology reflects habitat demands in three closely related ground beetle species (Coleoptera: Carabidae). J. Zool. 245, 467–472 (1998).
Moser, J. C. et al. Eye size and behaviour of day- and night-flying leafcutting ant alates. J. Zool. 264, 69–75 (2004).
Somanathan, H., Kelber, A., Borges, R. M., Wallén, R. & Warrant, E. J. Visual ecology of Indian carpenter bees II: Adaptations of eyes and ocelli to nocturnal and diurnal lifestyles. J. Comp. Physiol. A 195, 571–583 (2009).
Simpson, G. G. Tempo and mode in evolution. (Columbia University Press, 1984).
Rasmussen, C. & Cameron, S. A. Global stingless bee phylogeny supports ancient divergence, vicariance, and long distance dispersal: Stingless bee phylogeny. Biol. J. Linn. Soc. 99, 206–232 (2009).
Givnish, T. J. et al. Ancient Vicariance or Recent Long-Distance Dispersal? Inferences about Phylogeny and South American-African Disjunctions in Rapateaceae and Bromeliaceae Based on ndh F Sequence Data. Int. J. Plant Sci. 165, S35–S54 (2004).
Kropf, M., Comes, H. P. & Kadereit, J. W. Long-distance dispersal vs vicariance: The origin and genetic diversity of alpine plants in the Spanish Sierra Nevada. New Phytol. 172, 169–184 (2006).
Toussaint, E. F. A. & Short, A. E. Z. Biogeographic mirages? Molecular evidence for dispersal-driven evolution in Hydrobiusini water scavenger beetles: Biogeography of the beetle tribe Hydrobiusini. Syst. Entomol. 42, 692–702 (2017).
Toussaint, E. F. A., Bloom, D. & Short, A. E. Z. Cretaceous West Gondwana vicariance shaped giant water scavenger beetle biogeography. J. Biogeogr. 44, 1952–1965 (2017).
Gillespie, R. G. et al. Long-distance dispersal: A framework for hypothesis testing. Trends Ecol. Evol. 27, 47–56 (2012).
Raven, P. H. & Axelrod, D. I. Plate tectonics and the Astralasian paleobiogeography. Science 176, 1379–1386 (1972).
Capesius, I. & Bopp, M. New classification of liverworts based on molecular and morphological data. Plant Syst. Evol. 207, 87–97 (1997).
Gontcharov, A. A. Are Combined analyses better than single gene phylogenies? A case study using SSU rDNA and rbcL sequence comparisons in the Zygnematophyceae (Streptophyta). Mol. Biol. Evol. 21, 612–624 (2003).
Hoef-Emden, K., Marin, B. & Melkonian, M. Nuclear and nucleomorph SSU rDNA phylogeny in the cryptophyta and the evolution of cryptophyte diversity. J. Mol. Evol. 55, 161–179 (2002).
Nei, M., Kumar, S. & Takahashi, K. The optimization principle in phylogenetic analysis tends to give incorrect topologies when the number of nucleotides or amino acids used is small. Proc. Natl. Acad. Sci. 95, 12390–12397 (1998).
Nickrent, D. L., Parkinson, C. L., Palmer, J. D. & Duff, R. J. Multigene phylogeny of land plants with special reference to bryophytes and the earliest land plants. Mol. Biol. Evol. 17, 1885–1895 (2000).
Poe, S. & Swofford, D. L. Taxon sampling revisited. Nature 398, 299–300 (1999).
Scotese, C. R., Boucot, A. J. & McKerrow, W. S. Gondwanan palaeogeography and pal˦oclimatology. J. Afr. Earth Sci. 28, 99–114 (1999).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Basumatary, P., Caleb, J. T., Das, S. & Brahma, D. Redescription of the net-casting spider Asianopis goalparaensis (Tikader et Malhotra, 1978) comb.n. (Araneae: Deinopidae) from India. Arthropoda Sel. 29, 325–329 (2020).
Lin, Yejie., Jäger, P., Mikhailov, K. G. & Chen, Haifeng. Notes on two Asian species of the spider family Deinopidae (Aranei). Arthropoda Sel. 29, 467–469 (2020).
Austin, A. D. & Blest, A. D. The biology of two Australian species of dinopid spider. J. Zool. 189, 145–156 (1979).
Mohun, S. M. et al. Identification and characterization of visual pigments in caecilians (Amphibia: Gymnophiona), an order of limbless vertebrates with rudimentary eyes. J. Exp. Biol. 213, 3586–3592 (2010).
Wilkens, H. Regressive evolution: ontogeny and genetics of cavefish eye rudimentation: Genetics of cavefish eye rudimentation. Biol. J. Linn. Soc. 92, 287–296 (2007).
Martin, G. R. et al. Kiwi Forego vision in the guidance of their nocturnal activities. PLoS ONE 2, e198 (2007).
Jacobs, G. H. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision—A significant trend in the evolution of mammalian vision. Vis. Neurosci. 30, 39–53 (2013).
Tierney, S. M., Langille, B., Humphreys, W. F., Austin, A. D. & Cooper, S. J. B. Massive parallel regression: A précis of genetic mechanisms for vision loss in diving beetles. Integr. Comp. Biol. 58, 465–479 (2018).
Darwin, C. The Origin of Species by Means of Natural Selection. (Bantam Books, 1959).
Jeffery, W. R. Regressive evolution in Astyanax Cavefish. Annu. Rev. Genet. 43, 25–47 (2009).
Calderoni, L. et al. Relaxed selective constraints drove functional modifications in peripheral photoreception of the cavefish P. andruzzii and provide insight into the time of cave colonization. Heredity 117, 383–392 (2016).
Moran, D., Softley, R. & Warrant, E. J. The energetic cost of vision and the evolution of eyeless Mexican cavefish. Sci. Adv. 1, e1500363 (2015).
Cartwright, R. A., Schwartz, R. S., Merry, A. L. & Howell, M. M. The importance of selection in the evolution of blindness in cavefish. BMC Evol. Biol. 17, 45 (2017).
Herman, A. et al. The role of gene flow in rapid and repeated evolution of cave-related traits in Mexican tetra Astyanax mexicanus. Mol. Ecol. 27, 4397–4416 (2018).
Borowsky, R. & Wilkens, H. Mapping a cave fish genome: Polygenic systems and regressive evolution. J. Hered. 93, 19–21 (2002).
Jeffery, W. R. Adaptive evolution of eye degeneration in the Mexican Blind Cavefish. J. Hered. 96, 185–196 (2005).
Keene, A. C., Yoshizawa, M. & McGaugh, S. E. Biology and evolution of the Mexican cavefish. (Academic Press/Elsevier, 2015).
Stafstrom, J. A., Michalik, P. & Hebets, E. A. Sensory system plasticity in a visually specialized, nocturnal spider. Sci. Rep. 7, 46627 (2017).
Blest, A. D. The rapid synthesis and destruction of photoreceptor membrane by a dinopid spider: a daily cycle. Proc. R. Soc. Lond. B Biol. Sci. 200, 463–483 (1978).
Blakey, R. C. Gondwana paleogeography from assembly to breakup—A 500 m.y. odyssey. in Special Paper 441: Resolving the Late Paleozoic Ice Age in Time and Space vol. 441 1–28 (Geological Society of America, 2008).
Eagles, G. & König, M. A model of plate kinematics in Gondwana breakup. Geophys. J. Int. 173, 703–717 (2008).
Royer, J. Y. et al. Indian Ocean plate reconstructions since the Late Jurassic. Indian Ocean Plate Reconstr. Late Jurass. Synth. Results Sci. Drill. Indian Ocean Geophys Monogr Ser 70, 471–475 (1992).
Gibbons, A. D., Whittaker, J. M. & Müller, R. D. The breakup of East Gondwana: Assimilating constraints from Cretaceous ocean basins around India into a best-fit tectonic model: The Enderby basin/east Gondwana breakup. J. Geophys. Res. Solid Earth 118, 808–822 (2013).
Sereno, P. C., Wilson, J. A. & Conrad, J. L. New dinosaurs link southern landmasses in the Mid–Cretaceous. Proc. R. Soc. Lond. B Biol. Sci. 271, 1325–1330 (2004).
Barker, N. P., Weston, P. H., Rutschmann, F. & Sauquet, H. Molecular dating of the ‘Gondwanan’ plant family Proteaceae is only partially congruent with the timing of the break-up of Gondwana. J. Biogeogr. 34, 2012–2027 (2007).
Michalak, I., Zhang, L.-B. & Renner, S. S. Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae: Biogeography of Hernandiaceae. J. Biogeogr. 37, 1214–1226 (2010).
Korall, P. & Pryer, K. M. Global biogeography of scaly tree ferns (Cyatheaceae): evidence for Gondwanan vicariance and limited transoceanic dispersal. J. Biogeogr. 41, 402–413 (2014).
Luebert, F. et al. Historical biogeography of Boraginales: West Gondwanan vicariance followed by long-distance dispersal?. J. Biogeogr. 44, 158–169 (2017).
Jurado-Rivera, J. A. et al. Phylogenetic evidence that both ancient vicariance and dispersal have contributed to the biogeographic patterns of anchialine cave shrimps. Sci. Rep. 7, 2852 (2017).
Capobianco, A. & Friedman, M. Vicariance and dispersal in southern hemisphere freshwater fish clades: A palaeontological perspective: Vicariance and dispersal in freshwater fishes. Biol. Rev. 94, 662–699 (2019).
Derkarabetian, S., Baker, C. M. & Giribet, G. Complex patterns of Gondwanan biogeography revealed in a dispersal-limited arachnid. J. Biogeogr. 48, 1336–1352 (2021).
Chousou-Polydouri, N. et al. Giant Goblins above the waves at the southern end of the world: The biogeography of the spider family Orsolobidae (Araneae, Dysderoidea). J. Biogeogr. 46, 332–342 (2019).
Croizat, L., Nelson, G. & Rosen, D. E. Centers of origin and related concepts. Syst. Biol. 23, 265–287 (1974).
Rosen, D. E. Vicariant patterns and historical explanation in biogeography. Syst. Zool. 27, 159 (1978).
Brundin, L. A. Limnic Diptera in their bearings on the problem of transantarctic faunal connections. Pac. Basin Biogeogr. Symp. 425–434 (1963).
Cracraft, J. Avian evolution, Gondwana biogeography and the Cretaceous–Tertiary mass extinction event. Proc. R. Soc. Lond. B Biol. Sci. 268, 459–469 (2001).
McCulloch, G. A., Wallis, G. P. & Waters, J. M. A time-calibrated phylogeny of southern hemisphere stoneflies: Testing for Gondwanan origins. Mol. Phylogenet. Evol. 96, 150–160 (2016).
Baker, C. M., Boyer, S. L. & Giribet, G. A well-resolved transcriptomic phylogeny of the mite harvestman family Pettalidae (Arachnida, Opiliones, Cyphophthalmi) reveals signatures of Gondwanan vicariance. J. Biogeogr. 47, 1345–1361 (2020).
Brundin, L. A. Transantarctic relationship and their significance, as evidenced by chironomid midges. With a monograph of the subfamilies Podonominae and Aphroteniinae and the austral Heptagyiae. K Sven. Vetensk Akad Handl 11, 1–472 (1966).
Storey, M. et al. Timing of Hot Spot—Related Volcanism and the Breakup of Madagascar and India. Science 267, 852–855 (1995).
Torsvik, T. H. et al. Late Cretaceous India-Madagascar fit and timing of break-up related magmatism. Terra Nova 12, 220–224 (2000).
Agnarsson, I. & Kuntner, M. The Generation of a biodiversity hotspot: Biogeography and phylogeography of the Western Indian Ocean Islands. in Current topics in phylogenetics and phylogeography of terrestrial aquatic systems vol. 33 (2012).
Conti, E., Eriksson, T., Schönenberger, J., Sytsma, K. J. & Baum, D. A. Early tertiary out-of-India dispersal of Crypteroniaceae: Evidence from phylogeny and molecular dating. Evolution 56, 1931–1942 (2002).
Zhou, L., Su, Y. C. F., Thomas, D. C. & Saunders, R. M. K. ‘Out-of-Africa’ dispersal of tropical floras during the Miocene climatic optimum: evidence from Uvaria (Annonaceae): Out-of-Africa dispersal during Miocene climatic optimum. J. Biogeogr. 39, 322–335 (2012).
Renner, S. S. Multiple Miocene Melastomataceae dispersal between Madagascar, Africa and India. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 359, 1485–1494 (2004).
Cain, S., Loria, S. F., Ben-Shlomo, R., Prendini, L. & Gefen, E. Dated phylogeny and ancestral range estimation of sand scorpions (Buthidae: Buthacus) reveal Early Miocene divergence across land bridges connecting Africa and Asia. Mol. Phylogenet. Evol. 164, 107212 (2021).
Wallace, A. R. On the Zoological Geography of the Malay Archipelago. (Read Books Ltd., 1960).
Lohman, D. J. et al. Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 42, 205–226 (2011).
Stelbrink, B., Albrecht, C., Hall, R. & von Rintelen, T. The biogeography of Sulawesi revisited: Is there evidence for a Vicariant origin of taxa on Wallace’s “anomalous island”?: Biogeography of Sulawesi. Evolution 66, 2252–2271 (2012).
de Bruyn, M. et al. Borneo and Indochina are Major Evolutionary Hotspots for Southeast Asian Biodiversity. Syst. Biol. 63, 879–901 (2014).
Rabinowitz, P. D., Coffin, M. F. & Falvey, D. The separation of Madagascar and Africa. Science 220, 67–69 (1983).
Coffin, M. F. & Rabinowitz, P. D. Reconstruction of Madagascar and Africa: Evidence from the Davie Fracture Zone and Western Somali Basin. J. Geophys. Res. 92, 9385 (1987).
Barron, E. J. Cretaceous plate tectonic reconstructions. Palaeogeogr. Palaeoclimatol. Palaeoecol. 59, 3–29 (1987).
Lawver, L. A., Gahagan, L. M. & Coffin, M. F. The Development of Paleoseaways Around Antarctica. in Antarctic Research Series (eds. Kennett, J. P. & Warkne, D. A.) 7–30 (American Geophysical Union, 2013). https://doi.org/10.1029/AR056p0007.
Yoder, A. D. & Nowak, M. D. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu. Rev. Ecol. Evol. Syst. 37, 405–431 (2006).
Warren, B. H., Strasberg, D., Bruggemann, J. H., Prys-Jones, R. P. & Thébaud, C. Why does the biota of the Madagascar region have such a strong Asiatic flavour?. Cladistics 26, 526–538 (2010).
Janssens, S. B. et al. Dispersing towards Madagascar: Biogeography and evolution of the Madagascan endemics of the Spermacoceae tribe (Rubiaceae). Mol. Phylogenet. Evol. 95, 58–66 (2016).
Takayama, K., Tamura, M., Tateishi, Y., Webb, E. L. & Kajita, T. Strong genetic structure over the American continents and transoceanic dispersal in the mangrove genus Rhizophora (Rhizophoraceae) revealed by broad-scale nuclear and chloroplast DNA analysis. Am. J. Bot. 100, 1191–1201 (2013).
Hsu, M.-H., Lin, J.-W., Liao, C.-P., Hsu, J.-Y. & Huang, W.-S. Trans-marine dispersal inferred from the saltwater tolerance of lizards from Taiwan. PLoS ONE 16, e0247009 (2021).
Lim, B. K. Review of the origins and biogeography of bats in South America. Chiropt. Neotropical 15, (2009).
Hosner, P. A., Tobias, J. A., Braun, E. L. & Kimball, R. T. How do seemingly non-vagile clades accomplish trans-marine dispersal? Trait and dispersal evolution in the landfowl (Aves: Galliformes). Proc. R. Soc. B Biol. Sci. 284, 20170210 (2017).
Yi, X. & Latch, E. K. Systematics of the New World bats Eptesicus and Histiotus suggest trans-marine dispersal followed by Neotropical cryptic diversification. Mol. Phylogenet. Evol. 175, 107582 (2022).
Ceccarelli, F. S. et al. Around the world in eight million years: Historical biogeography and evolution of the spray zone spider Amaurobioides (Araneae: Anyphaenidae). PLoS ONE 11, e0163740 (2016).
Luo, Y. et al. Global diversification of anelosimus spiders driven by long-distance overwater dispersal and neogene climate oscillations. Syst. Biol. 69, 1122–1136 (2020).
Rouhan, G. et al. Molecular phylogeny of the fern genus Elaphoglossum (Elaphoglossaceae) based on chloroplast non-coding DNA sequences: Contributions of species from the Indian Ocean area. Mol. Phylogenet. Evol. 33, 745–763 (2004).
Vasco, A., Moran, R. C. & Rouhan, G. Circumscription and phylogeny of the Elaphoglossum ciliatum group (E. sect. Lepidoglossa , Dryopteridaceae) based on cpDNA sequences. TAXON 58, 825–834 (2009).
Rouhan, G., Hanks, J. G., McClelland, D. & Moran, R. C. Preliminary phylogenetic analysis of the fern genus Lomariopsis (Lomariopsidaceae). Brittonia 59, 115–128 (2007).
Labiak, P. H., Sundue, M. & Rouhan, G. Molecular phylogeny, character evolution, and biogeography of the grammitid fern genus Lellingeria (Polypodiaceae). Am. J. Bot. 97, 1354–1364 (2010).
Sundue, M. A. et al. Global phylogeny and biogeography of grammitid ferns (Polypodiaceae). Mol. Phylogenet. Evol. 81, 195–206 (2014).
Labiak, P. H. et al. Phylogeny and historical biogeography of the lastreopsid ferns (Dryopteridaceae). Am. J. Bot. 101, 1207–1228 (2014).
Bauret, L. et al. Madagascar sheds new light on the molecular systematics and biogeography of grammitid ferns: New unexpected lineages and numerous long-distance dispersal events. Mol. Phylogenet. Evol. 111, 1–17 (2017).
Moran, R. C. & Smith, A. R. Phytogeographic relationships between neotropical and African-Madagascan pteridophytes. Brittonia 53, 304–351 (2001).
Capello, B. Descripção de algumas especies novas ou pouco conhecidas de crustaceos e arachnidios de Portugal e possessões portuguezas do Ultramar. Mem. Acad. Real Sci. Lisb. 4, 1–18 (1867).
Pocock, R. I. On the scorpions, pedipaips, and spiders from tropical West Africa represented in the collection of the British Museum. Proc. Zool. Soc. Lond. 14, 833–885 (1899).
Pickard-Cabridge, F. O 313–424 (Arachnida - Araneida and Opiliones. in Biologia Centrali-Americana, 1902).
Thorell, T. Studi sui Ragni Malesi e Papuani. III. Ragni dell’Austro-Malesia e del Capo York, conservati nel Museo Civico di Storia Naturale di Geniva. Ann Mus Civ Stor Nat Genova 17, 1–720 (1881).
Gerstaecker, K. E. A., Decken, K. K. von der & Kersten, Otto. Die Gliedertheir-fauna des Sansibar-gebietes : nach dem von dr. O. Kersten während der v. d. Decken’schen Ost-Afrikanischen expedition im Jahre 1862 gesammelten material / bearb. von A. Gerstaecker. (C. F. Winter, 1873). https://doi.org/10.5962/bhl.title.10149.
Pocock, R. I. XXV. The Arachnida from the Province of Natal, South Africa, contained in the collection of the British Museum. Ann. Mag. Nat. Hist. 2, 197–226 (1898).
Koch, L., Keyserling, G. E. & Koch, Carl. Die Arachniden Australiens nach der Natur beschrieben und abgebildet. (Verlag von Bauer & Raspe, 1879). https://doi.org/10.5962/bhl.title.121660.
Simon, E. Etude sur les arachnides du Tonkin (1re partie). Bull. Sci. Fr. Belg. 42, 69–147 (1909).
Lessert, R. Araignées du Congo recueillies au cours de l’expédition organisée par l’American Museum (1909–1915). Quatrième et dernière partie. Rev. Suisse Zool. 37, 613–672 (1930).
Berland, L. & Millot, J. Les araignées de l’Afrique occidental français. II, Cribellata. Ann. Société Entomol. Fr. 108, 149–160 (1940).
Doleschall, L. Tweede Bijdrage tot de kennis der Arachniden van den Indischen Archipel. Acta Soc. Sci. Indica-Neerlandica 5, 1–6 (1859).
Barrion-Dupo, A. L., A. & Barrion, A. T. Discovery of the family Deinopidae from the Philippines, with descriptions of three new species of Deinopis Macleay, 1839. Philipp. Entomol. 31, 1–23 (2018).
Strand, E. Arachnida. in Wissenschaftliche ergebnisse der Deutschen Zentral-Africa-Expedition, 1907–1908 : unter Führung Adolf Friedrichs, herzogs zu Mecklenburg. vol. 4 325–474 (1913).
Lenz, H. Beiträge zur Kenntniss der Spinnenfauna Madagascars. Zool. Jahrb. Z. Für Syst. Geogr. Biol. Thiere 1, 379–408 (1886).
Kulczyński, W. Araneae musei nationalis Hungarici in regionibus Indica et Australia a Ludovico Biro collectae. Ann. Musei Natl. Hung. 6, 426–494 (1908).
Pocock, R. I. Some new African spiders. Ann. Mag. Nat. Hist. 7, 315–530 (1902).
Koch, L., Keyserling, G. E. & Koch, Carl. Die Arachniden Australiens nach der Natur beschrieben und abgebildet /. (Verlag von Bauer & Raspe, 1878). https://doi.org/10.5962/bhl.title.121660.
Rainbow, W. J. Contribution to a knowledge of Papuan Arachnida. Rec. Aust. Mus. 3, 108–118 (1899).
Karsch, F. Exotisch-araneologisches. Z. Für Gesammten. Naturwissenschaften 51(322–333), 771–826 (1878).
Giltay, L. Notes arachnologiques africaines. I. Une espèce nouvelle de Dinopis (Dinopis Schoutedeni nov. sp.) du Congo belge. II. Myrmarachne foenisex Sim., araignée myrmécophile oecophylliforme, au Congo belge. Rev. Zool. Bot. Afr. 18, 23–27 (1929).
Coddington, J. A., Griswold, C., Silva, D., Penaranda, E. & Larcher, S. F. Designing and testing sample protocols to estimate biodiversity in tropical ecosystems. Unity Evol. Biol. Proc. Fourth Int. Congr. Syst. Evol. Biol. 2, (1991).
Kulkarni, S., Wood, H., Lloyd, M. & Hormiga, G. Spider-specific probe set for ultraconserved elements offers new perspectives on the evolutionary history of spiders (Arachnida, Araneae). Mol. Ecol. Resour. 20, 185–203 (2020).
Chamberland, L. et al. From Gondwana to GAARlandia: Evolutionary history and biogeography of ogre‐faced spiders (Deinopis). 2170412 bytes (2020) 10.5061/DRYAD.GXD2547GZ.
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Maddison, W. P. & Maddison, D. R. Mesquite: A modular system for evolutionary analysis. (2021).
Borowiec, M. L. AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4, e1660 (2016).
Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Mirarab, S. & Warnow, T. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31, i44–i52 (2015).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Kishino, H., Takashi, M. & Hasegawa, M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol. 31, 151–160 (1990).
Shimodaira, H. & Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116.
Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002).
Strimmer, K. & Rambaut, A. Inferring confidence sets of possibly misspecified gene trees. Proc. R. Soc. Lond. B Biol. Sci. 269, 137–142 (2002).
Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Penny, D. A new deinopoid spider from Cretaceous Lebanese amber. Acta Palaeontol. Pol. 48, (2003).
Scharff, N. et al. Phylogeny of the orb-weaving spider family Araneidae (Araneae: Araneoidea). Cladistics 36, 1–21 (2020).
Magalhaes, I. L. F., Azevedo, G. H. F., Michalik, P. & Ramírez, M. J. The fossil record of spiders revisited: implications for calibrating trees and evidence for a major faunal turnover since the Mesozoic. Biol. Rev. 95, 184–217 (2020).
Wunderlich, J. On the evolution and the classification of spiders, the Mesozoic spider faunas, and descriptions of new Cretaceous taxa mainly in amber from Myanmar (Burma)(Arachnida: Araneae). Beitr. Zur Araneol. 9, e408 (2015).
Fernández, R. et al. Phylogenomics, diversification dynamics, and comparative transcriptomics across the spider tree of life. Curr. Biol. 28, 1489-1497.e5 (2018).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Rambaut, A. FigTree. (2018).
Matzke, N. J. BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. (2013).
Holt, B. G. et al. An update of Wallace’s zoogeographic regions of the world. Science 339, 74–78 (2013).
Turk, E., Čandek, K., Kralj-Fišer, S. & Kuntner, M. Biogeographical history of golden orbweavers: Chronology of a global conquest. J. Biogeogr. 47, 1333–1344 (2020).
Seton, M. et al. Global continental and ocean basin reconstructions since 200Ma. Earth-Sci. Rev. 113, 212–270 (2012).
Matzke, N. Statistical comparison of DEC and DEC+J is identical to comparison of two ClaSSE submodels, and is therefore valid. https://osf.io/vqm7r (2021) 10.31219/osf.io/vqm7r.
Matzke, N. J. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst. Biol. 63, 951–970 (2014).
Ree, R. H. & Sanmartín, I. Conceptual and statistical problems with the DEC +J model of founder-event speciation and its comparison with DEC via model selection. J. Biogeogr. 45, 741–749 (2018).
Revell, L. J. phytools: An R package for phylogenetic comparative biology (and other things): phytools: R package. Methods Ecol. Evol. 3, 217–223 (2012).
Fritz, S. A. & Purvis, A. Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits: Selectivity in extinction risk. Conserv. Biol. 24, 1042–1051 (2010).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Huelsenbeck, J. P., Nielsen, R. & Bollback, J. P. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158 (2003).
Acknowledgements
We would like to acknowledge the many people who have contributed ideas. We thank Jay Stafstrom for sharing his expertise on spider vision and for thoughtful discussions on deinopid ecology. We thank Lauren Esposito for assisting in securing permits and collecting specimens as part of the Caribbean Island Biogeography (CARBIO) project. We thank Lacie Newton for her help with the molecular work. We are especially grateful to the volunteers who helped collect Australian deinopids, including Michael Doe and Adam Fletcher of Project Maratus, Jeremy Jones, Nick Lambert, Matt Elmer, Matthew Hourston, and Robert Whyte, and for sharing their knowledge on Australian spiders. We thank the Australian landowners who gave us permission to collect spiders on their property and for hosting us. We thank Mariella Herberstein, George Binns, and Jonas Wolff for assisting with the Australian collections. We thank Dave Barrington and Greta Binford for their discussions on biogeography and evolution. Finally, we appreciate the feedback from two anonymous reviewers whose comments greatly improved the manuscript.
Funding
This article was funded by The John Wheeler Graduate Student Research and Development Award and Evert and Marion Schlinger Foundation.
Author information
Authors and Affiliations
Contributions
L.C. designed the study, acquired, analyzed, and interpreted the data, wrote the original draft, prepared the figures. L.C. and J.S. performed UCE acquisition. J.S. contributed to the supplemental methods S1 and manuscript revisions. I.A. and J.E.B revised the manuscript and contributed to project design and execution. J.E.B. and I.A. funded the study. I.L.Q. and T.R. performed data collection, photographed specimens, and provided corrections to the manuscript. All authors contributed critically to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chamberland, L., Agnarsson, I., Quayle, I.L. et al. Biogeography and eye size evolution of the ogre-faced spiders. Sci Rep 12, 17769 (2022). https://doi.org/10.1038/s41598-022-22157-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22157-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.