Abstract
The muscle of aquatic crustaceans is perishable and susceptible to environmental contamination. Vibrio harveyi is a widely occurring pathogen in aquatic animals. Here, bath treatment with a virulent V. harveyi strain (which was added directly in the rearing water to imitate environmental contamination) isolated from the muscle of the whiteleg shrimp, Litopenaeus vannamei, caused the muscle of Li. vannamei to display a whitish-opaque appearance due to microscopic changes including muscle lysis, muscle fiber damage and microbial colonization. When administered orally by incorporating this isolate in feed (which is an imitation of infection via natural route), rather than direct invasion followed by colonization in the muscle, this isolate indirectly stimulated severe muscle necrosis in Li. vannamei via steering the enrichment of two important (human) pathogens, V. cholerae and V. vulnificus, and one environmental bacterium Pseudomonas oleovorans, based on the meta-taxonomic analyses. In addition to the scientifically proven viral diseases, our research proved that bacterial agents are also capable of causing muscle spoilage in crustaceans via changing the microbial composition, and that the crustaceans might be exploited as the wide-spectrum sensitive bio-detector to indicate the extent of microbial contamination.
Similar content being viewed by others
Introduction
The ubiquitous aquatic crustaceans are sensitive to versatile biological, physical and chemical pollutants; once affected, they display easy-to-be-recognized disease symptoms, such as a focal to extensive opaque-whitish coloration underneath the carapace. The microbial causative agents of this symptom are conventionally attributed to mainly viruses1,2. However, myonecrosis is not inevitably linked to viral infection3,4,5,6,7,8. Recently, a few studies have proven that bacteria belonging to the Vibrionaceae are also capable of causing muscle necrosis in crustaceans; these pathogens include Vibrio harveyi, V. parahaemolyticus, Photobacterium damselae subsp. damselae, etc.3,4,5,6,7,8. Meanwhile, with regard to food safety concerns, spoilage of the crustacean muscle is often related to the accumulation of and/or contamination by microbes, including (zoonotic) pathogens, such as V. cholerae, V. vulnificus and V. parahaemolyticus9.
Vibrio pathogens are ubiquitous in aquatic and terrestrial ecosystems, and able to exert their detrimental effects on a wide range of living organisms, including humans. This study aimed at testing the effects of a virulent V. harveyi isolate on the structure and microbiomes of the muscle of Li. vannamei, which may provide scientific support for the future construction and application of crustacean-based broad-spectrum bio-detectors to assess microbial contamination in aquatic ecosystems.
Results and discussion
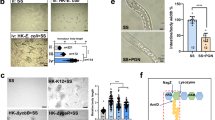
In order to select virulent Vibrio isolates, we previously isolated 18 Vibrio isolates from the rearing water and the organs (muscle, intestine, hepatopancreas or gill) of Li. vannamei10. Over 77% of these isolates were taxonomically clustered within the V. harveyi and V. parahaemolyticus clades (strain 1, 3, 4, 5, 6, 10, 11, 13, 17, 21, 22, 29, 43 and 53), and over 16% within the V. cholerae clade (strain 41, 45 and 51) (Fig. 1). To mimic an suboptimal aquatic environment, e.g. pollution by microorganisms and transport of shrimps at high density, a bath challenge test with Li. vannamei, focusing on 13 isolates that propagated well in LB broth, revealed that strain 1, which was originally isolated from the muscle of Li. vannamei10 and showed 99.92% (1288-bp query length of the 16S rRNA gene) homology to V. harveyi P411 in the BLASTn comparison on NCBI website and 99.6839% intra-species average nucleotide identity (ANI) to V. harveyi FDAARGOS_10712 according to the genome-based calculation by FastANI (version 1.32)13, exhibited the strongest virulence (Fig. 2a). This isolate was not re-isolated from the muscle of Li. vannamei in this test (data not shown), but induced severe whitish-opaque lesions in the muscle (Fig. 2b). Muscle deformation and muscle fiber fracture accompanied by the dispersion of (curved) rod-shaped and bacterial-sized cells were observed photomicrographically (Fig. 2b). Comparing with fishes, shrimps only possess primitive mucosal immunity in their gills14, which could not protect them from pathogens of high density15. In this test, Li. vannamei were immersed in the rearing water with high density of V. harveyi strain 1. This pathogen might invade the host via the gills, which were probably the major entrance for the bacteria, and directly go into the hemolymph and ultimately compromise the host.
Phylogenetic characterization of Vibrio isolates. The neighbor-joining28 consensus tree displays 16S rRNA gene sequences (≥ 1200 bp) of (i) 18 Vibrio isolates and (ii) all Vibrio type strains with good sequence quality (103 strains in total) downloaded from the Ribosomal Database Project27 (RDP, http://rdp.cme.msu.edu/). The phylogenetic analyses were performed in Mega 729 using the Kimura-2-parameter30 method with Gamma distribution (0.10) to calculate the evolutionary distances. The bootstrap values indicated at the nodes are based on 1 000 bootstrap replicates31. Branch values lower than 50% are hidden. The scale bar indicates an evolutionary distance of 0.005 nucleotide substitution per sequence position. Black, red, blue and green colors indicate the isolates of this study and the reference strains from human-related, aquatic and terrestrial/plant sources of isolation, respectively. Strains from (inter)tidal area, coastal sediment, mangrove soil and salt marsh mud are marked in green. The name of each reference strain is preceded by the accession number.
Pathogenicity of Vibrio isolates on Li. vannamei. (a) Mean mortality percentage of Li. vannamei after a 3-day challenge with 13 Vibrio isolates that proliferate fast in LB broth and (b) the consequential disease symptoms in the muscle of Li. vannamei cultivated in V. harveyi strain 1-treated water. (b) The muscle of the V. harveyi strain 1-treated Li. vannamei showed extensive whitish-opaque necrosis (upper right), which was not observed in that of the control treatment (upper left). Hematoxylin and eosin (H&E)-stained sections (bottom row) displayed the enlargement of the squared areas of the corresponding Li. vannamei. The necrotic muscle (lower right) arranged abnormally with ruptured muscle fibers, degraded muscle and (curved) rod-shaped and bacterial-sized cells (indicated by arrows). (c) Bacterial cell density in the muscle and hepatopancreas of Li. vannamei, which was determined on TCBS agar after 5 days of feeding with V. harveyi strain 1-treated feed in the first feeding test. *Statistically significant difference compared with the corresponding untreated control scored at the same time. Error bars represent S.E.M. (N = 3 (a) and 24 (c)). (d) Gross signs of Li. vannamei fed with V. harveyi strain 1-treated feed, which showed similar symptoms as described in (b).
Unlike most of the vertebrates, crustaceans have microbiota in their hemolymph and the microbiota-immune system balance is pivotal for maintaining their health16. In addition, unlike mammals, in which bacteria will be engulfed by macrophages and neutrophils, massive infection in crustaceans will provoke serious melanization; during this process, excessive reactive oxygen species will be secreted into the tissues, which would cause massive tissue damage17. According to the genomic analysis of the virulence factors, V. harveyi strain 1 harbors genes encoding toxins and toxin secretion, anti-immune strategies to overcome the host immune defenses, strategies to acquire nutritional factors from the host to propagate (which might destroy the microbiota-immunity balance), etc. (Table S1). Collectively, this genomic analysis and the aforementioned experimental results revealed that the pathogenic V. harveyi strain 1 might be capable of sabotaging the host immune system, breaking the microbiota-host immune system balance and forcing the host immune system secrete massive reactive oxygen species which cause tissue damage. Therefore, V. harveyi strain 1 was used as the infection inoculum in the following feeding tests.
In order to investigate the virulence of putative pathogens on aquatic animals, many studies used injection. However, this is not a natural route of infection and thus fails to provide realistic scenarios allowing the tested pathogen to exert authentic pathological activity. Naturally, the vast majority of microorganisms enter their host via feeding. Consequently, the in-taken pathogens can initiate an infection and/or necrosis directly or indirectly (e.g. via stimulating the enrichment or function of other microbes). Indeed, the gastrointestinal tract of Li. vannamei has its balanced microbial community composition. The destruction of this balance might negatively affect the immune system and unbalance the hemolymph microbial community in Li. vannamei, which could then compromise the shrimps. Therefore, in this research, we studied the potential pathogenicity of V. harveyi strain 1 on Li. vannamei via inclusion in the diet. Based on the colony counts on the Vibrio semi-selective medium TCBS agar (referred to as TCBS-bacteria from this point onwards), in the first feeding test, the cell density of TCBS-bacteria in the muscle of Li. vannamei fed with V. harveyi strain 1-treated feed was significantly higher compared to that in Li. vannamei fed with untreated feed (which contained no detectable TCBS-bacteria as shown in Fig. 3a); while the cell density in the hepatopancreas of the former treatment was higher than that of the later treatment, it showed no significant difference in both treatments (Fig. 2c). Correspondingly, the muscle of the treated Li. vannamei showed an extensive whitish-opaque necrotic appearance, while the change in the hepatopancreas was not seen with the unaided eye (Fig. 2d); these might be ascribed to the dissimilar anti-pathogen capability among different organs. Taxonomic analyses of the TCBS-bacteria growing out from these two organs of the treated Li. vannamei indicated that they were distantly related to V. harveyi strain 1 (Fig. S1). Moreover, a preliminary test revealed that the TCBS-bacteria were not present in the hemolymph collected from the Li. vannamei fed with V. harveyi strain 1-treated feed (data not shown). Therefore, rather than direct invasion and infection, the ingested V. harveyi strain 1 might cause disease in the host via an indirect path, even though this strain was originally isolated from the muscle of Li. vannamei.
Impact of V. harveyi strain 1 on feed pellets and on Li. vannamei. The feed pellets were treated with a suspension of V. harveyi strain 1 after pre-mixing with (suppressive feed) or without La. plantarum L75a-fermented broth. (a) Impact of V. harveyi strain 1 on the bacterial cell density in feed pellets, which was determined on TCBS agar every half hour in a duration of 2 h. *Statistically significant difference compared with the corresponding suppressive feed treatment scored at the same time. (b) Bacterial cell density in the feces and intestine of Li. vannamei after 3 h of feeding (left, the second feeding test) and in the intestine, muscle and hepatopancreas after 4 days of feeding (right, the third feeding test), which was determined on TCBS agar. The dotted line separated the results from the two independent tests. (a) and (b) share the same legends. (c) The health status of Li. vannamei fed with V. harveyi strain 1-treated feed for 4 days (the third feeding test): the diseased Li. vannamei (fed without La. plantarum supplementation, lower image) exhibited acute whitish-opaque muscle necrosis, while the shrimp fed with the suppressive feed (with La. plantarum supplementation, upper image) possessed clear and transparent muscle. These muscle samples were also determined for the percentage of abundance of bacterial species based on the 16S rRNA gene amplicon sequencing (d); the detected species without designated species names and/or with mean percentage lower than 3% were not shown; the first and second numbers in the legend represent the percentage of abundance of the corresponding species detected in the muscle of Li. vannamei fed with the suppressive feed and V. harveyi strain 1-treated diets, respectively. Error bars represent S.E.M. (N = 9 (a), 8 (b) and 3 (d)).
In further experiments, the whole of the fermented broth of Lactiplantibacillus plantarum L75a, which had been preliminarily proven to be antagonistic to several Vibrio isolates (including V. harveyi strain 1) and not pathogenic to Li. vannamei10, was introduced as the measure to inhibit Vibrio and to guarantee and stabilize the healthy status of Li. vannamei in the suppressive treatment (positive control, with V. harveyi strain 1 and La. plantarum L75a-fermented broth). Under in vitro condition during a 2-h treatment, V. harveyi strain 1 facilitated more TCBS-bacteria to grow in the unsuppressive feed pellets (with V. harveyi strain 1 and without La. plantarum L75a-fermented broth) compared to that in the suppressive feed pellets (Fig. 3a).
Albeit V. harveyi is a well-known pathogen associated with Li. vannamei and many other aquatic animals, and a few studies have demonstrated that it is capable of causing muscle necrosis in aquatic animals5,6,18, the impact of V. harveyi on muscle necrosis and the concomitant microbiome composition is hitherto unclear. Parallel to the above in vitro test, we also performed the second and third feeding tests. We found that the orally administered V. harveyi strain 1 led to unidentical but not significantly different cell density of TCBS-bacteria in the fecal matter, intestine, muscle and hepatopancreas of Li. vannamei fed with both diets (Fig. 3b). Remarkably, the presence of V. harveyi strain 1 consistently and conspicuously stimulated acute extensive muscle necrosis in Li. vannamei fed with the V. harveyi strain 1-treated feed (without La. plantarum L75a-fermented broth), which was not observed in Li. vannamei fed with the suppressive feed (Fig. 3c).
In addition to being the bio-indicator of microbial contamination, the coloration and appearance of the muscle of shrimps reflect the pathogen composition, which is primarily correlated to food safety19,20,21. Based on the 16S rRNA gene amplicon sequencing of the muscle of Li. vannamei from the third feeding test, the abundance of the 4 dominant species detected included V. cholerae, V. vulnificus, Pseudomonas oleovorans and Ph. damselae; the former three ones were more abundant in the muscle of the V. harveyi strain 1-unsuppressive treatment, while the later one was in reverse (Fig. 3d). With regard to the causative pathogens of zoonotic Vibriosis, V. cholerae and V. vulnificus are categorized in the ‘higher risk organisms’ whereas Ph. damselae and V. harveyi in the ‘lower risk organisms’22. Notably, V. cholerae is affiliated to foodborne diseases in a relatively higher frequency; while V. vulnificus and Ph. damselae have a higher correlation with contact zoonoses via open wounds, and both of them were sometimes found to be responsible for food spoilage23,24. Ps. oleovorans was rarely known as a pathogen, and only very few papers reported that it was the causal agent for human infections25,26.
In our study, although V. harveyi strain 1 was originally isolated from the muscle of Li. vannamei, when delivered orally to Li. vannamei in the in vivo feeding tests, it was not re-isolated from or detected in the muscle of this host (Figs. 3d, S1, S2). This may imply that, instead of direct invasion, V. harveyi strain 1 conferred harmful activity to Li. vannamei via facilitating/driving the foodborne pathogens, especially V. cholerae and V. vulnificus, to invade or proliferate, which compromised the health status of the host. Besides, in the treatment with suppressive feed, although a high density of Ph. damselae was detected in the muscle of Li. vannamei, it remained healthy. According to the previous studies, Ph. damselae subsp. damselae was capable of inducing myonecrosis in Li. vannamei3. Therefore, further studies need to be conducted to elucidate the mechanism of the tripartite interplay amongst the virulent microorganisms (e.g. Vibrio pathogens), the host microbiomes and the aquatic hosts, in order to properly apply crustaceans as the bio-detectors for microbial contamination. Importantly, since viruses are commonly asserted as the causative agents of muscle diseases in crustaceans1,2, and this study manifested that V. harveyi drove the bacterial pathogens in the shrimp muscle to induced easily-visualized disease symptom (myonecrosis), we thus suggest that the crustaceans might be (extensively) applied as straight-forward bio-detectors to indicate the state of (broad-spectrum) microbial contamination in the aquatic ecosystems, especially in the estuary regions that are prone to eutrophication.
In addition to the prospective for practical application, since the four independent in vivo tests (Figs. 2a–d, 3b–d) collectively asserted that the distinctly visible myonecrosis was not directly attributed to the presence of the originally applied pathogen V. harveyi strain 1 in the muscle of Li. vannamei, these suggested that the contribution could be the necessary cell density and other possible factors to improve or impair beneficial or prejudicial microbiota for causing myonecrosis. This milestone scientific discovery, which does not closely resemble the conventionally known and prevailingly accepted scientific theory, might help ameliorate the existing theory of etiology and pathology of bacterial infections and might be an important starting point of a new research theme in the scientific domain of etiology and pathology of bacterial infections.
Methods
Media preparation
The Vibrio semi-selective agar, Thiosulfate-Citrate-Bile salts-Sucrose agar (TCBS agar, Guangdong Huankai Microbial Sci. & Tech. Co., Ltd., Guangzhou, Guangdong, China)10, was prepared by filling in a stainless still beaker covered with cling wrap and boiling intermittently to avoid boiling over for 1–2 minutes10. The Luria–Bertani agar media (LB5 or LB10 agar, consisted of 0.5% or 1% NaCl, respectively; 1% tryptone (Oxoid Ltd., Hampshire, England); 0.5% yeast extract (Oxoid Ltd., Hants, UK); 1.5% agar (BioFroxx GmbH, Einhausen, Germany)) were applied to cultivate Vibrio isolates. The Luria–Bertani broth media (LB5 or LB10 broth) were not supplemented with agar.
Phylogeny of the Vibrio isolates
In order to select virulent Vibrio isolates, we previously isolated 18 Vibrio isolates from the organs (muscle, intestine, hepatopancreas or gill) of Li. vannamei and their rearing water10 with the salinity of 5 ppt. These Vibrio isolates are preserved in 20%-40% glycerol at − 80 °C. The GenBank accession numbers of 16S rRNA gene sequences of these isolates are MT974072-MT97408910. These sequences were subjected to phylogenetic analyses. The reference sequences of all 16S rRNA gene sequences of Vibrio type strains and of good quality were obtained from the Ribosomal Database Project27 (RDP, http://rdp.cme.msu.edu/index.jsp). A neighbor-joining28 consensus tree was performed in MEGA729 based on all the 16S rRNA gene sequences (≥ 1200 bp) of the isolates and reference strains. The evolutionary distances were modelled by the Kimura-2-parameter30, the non-uniformity of evolutionary rates amongst sites were determined using discrete Gamma distribution, and 1000 bootstrap replicates31 were exploited.
Pre-cultivation and transfer of Li. vannamei for in vivo tests10
The Li. vannamei used for the following in vivo tests were purchased from a commercial farm. Upon arrival in the aquaculture laboratory, the Li. vannamei were introduced in a recirculation aquaculture system (RAS) containing 0.5% seawater crystal (Yanzhibao™, Guangzhou, Guangdong, China) and cultivated for at least two weeks before use. The muscle of Li. vannamei was sampled and detected for absence of bacterial colony on TCBS agar before experiments. Prior to each in vivo test, the Li. vannamei were transferred within one hour in the RAS water in big buckets from the RAS to the testing system, which was located in a room with the ambient temperature of 28 ± 2 °C.
Pathogenicity of the Vibrio isolates on Li. vannamei via bath treatment
The pathogenicity of the Vibrio isolates was determined by a bath challenge test on Li. vannamei. Each testing unit, 500 ml glass bottle (with the diameter of 7.5 cm), contained 400 ml of the RAS water and was continuously aerated by an air stone connected to an air pump. Ten Li. vannamei of approximately 4 ± 1 cm in length were gently introduced into each testing unit and allowed to acclimatize overnight. To screen for the most virulent Vibrio isolate, all isolates that grew well in LB5 or LB10 broth (13 Vibrio isolates in total), were tested for their pathogenicity on Li. vannamei. Each isolate was pre-grown in LB5 at 28 °C at 200 r.p.m. for 1–2 days. The bacterial cells of each culture were washed three times by sterile RAS water and centrifugation at 5000×g for 5 min at room temperature to remove the culture supernatant. The washed cell pellets were resuspended in sterile RAS water. After determination of the cell density by a spectrophotometer at 600 nm, the cell suspension of each isolate was added into each testing unit to reach a final density of 105 cells ml−132. Each Li. vannamei was fed once daily with one pellet of shrimp feed (type: No. 0, Guangdong Yuehai feed group, Guangdong, China) with the diameter of approximately 2 mm. The dead Li. vannamei was removed immediately when observed. The survival of Li. vannamei was determined on day 1, 2 and 3. Each treatment was performed in triplicate. The control treatment was not supplemented with Vibrio isolate.
Histological analyses of the muscle of Li. vannamei
At the end of the aforementioned bath treatment, two Li. vannamei, untreated (control treatment) and bath-treated by the most virulent isolate, V. harveyi strain 1, were collected and surface-disinfected by 75% ethanol for 3 s. The muscle of each Li. vannamei was dissected and fixed in Bouin's solution for 5 days at room temperature. Then, each muscle sample was processed for examination of hematoxylin and eosin (H&E)-stained paraffin sections by light microscopy. Preparation and photomicrography of the sections were conducted by Wuhan Servicebio Technology Co., Ltd (Wuhan, Hubei, China) according to Servicebio’s in-house protocols.
The virulence factors of V. harveyi strain 1
The two contigs of the genome of V. harveyi strain 1 were respectively analyzed by blastn in the database ‘DNA sequences from VFDB full dataset’ on the virulence factor database (VFDB)33 website (http://www.mgc.ac.cn/VFs/). From the top 100 sequences producing significant alignments, sequences with identity lower than 80% or related to flagellar structure and function were excluded.
Pathogenicity of V. harveyi strain 1 on Li. vannamei via feeding (the first feeding test)
The Li. vannamei used for this test was not fed 12 h prior to the test. To circumvent light and perturbation without blocking aeration, each optimized testing unit (plastic box of 37 × 24 × 14 cm) was wrapped with black plastic and covered with a black lid. Each unit contained 8 l of chlorine dioxide-disinfected and dechlorinated RAS water, and was continuously aerated as mentioned above. Five Li. vannamei of 10 ± 2 cm were gently introduced into each testing unit and allowed to acclimatize for 4–5 h before the first feeding. The Li. vannamei was fed with V. harveyi strain 1-treated feed once or twice per day and the feed dry weight of each meal was 0.3% to 1% of their body weight.
To prepare V. harveyi strain 1-treated feed, the pregrown cells of V. harveyi strain 1 (in LB10 broth) were washed once and resuspended in sterile RAS water to a final density of 105 cells ml−1 based on the aforementioned methods. The dry feed of one meal of each testing unit was added in a Petri dish (with the diameter of 6 cm) containing 3–4 ml cell suspension of V. harveyi strain 1. After incubation for approximately one hour, the feed was then surface-dried by sterile paper and added into each testing unit. The Li. vannamei of the control treatment was fed with untreated feed. Each treatment was performed in quadruplicate (one testing unit represented one replicate). To circumvent water deterioration, dead shrimps, feces and uneaten feed were removed by nets.
After 5 days of cultivation, 8 Li. vannamei were collected from each treatment, photographed and surface-disinfected by 75% ethanol for 3 s. Under aseptic condition, the shell of each Li. vannamei was removed; and the muscle and hepatopancreas were dissected and put into a sterile 1.5-ml tube containing 200 and 400 µl sterile RAS water, respectively, and two sterile glass beads with the diameter of 3 mm. The muscle and hepatopancreas were weighed and homogenized by a Biologix® vortex mixer (Jinan, Shandong, China) at the maximal speed for 4 and 1 min, respectively. Next, to determine the cell density of bacteria in the two organs that could grow on TCBS agar (referred to as TCBS-bacteria from this point onwards), each homogenized sample was serially diluted by sterile RAS water and spot-inoculated (5 µl per droplet) on TCBS agar in triplicate. The TCBS-bacterial cell density was determined after the TCBS agar plates were incubated at 28 °C for 18 h. To investigate if V. harveyi strain 1 was present in the organs, single colonies (with a predilection of yellowish colonies whose morphology was identical to that of V. harveyi strain 1) of each treatment and each organ were randomly picked, grown in LB5 broth and sent for 16S rRNA gene sequencing using the forward primer to generate the synthesis of the sequences from the 3’ end. Good-quality sequences (≥ 850 bp) of these colonies were subjected to phylogenetic analyses based on the aforementioned methods.
Lactiplantibacillus plantarum L75a
According to the preliminary tests, the fermented broth of La. plantarum L75a, isolated from the intestine of wild Scylla serrata from the South China Sea, was inhibitory to the proliferation of several Vibrio isolates (including V. harveyi strain 1) and was not pathogenic to Li. vannamei10. Therefore, this fermented broth was exploited as the antagonistic biological agent (in the suppressive treatment) against Vibrios in this study.
Impact of V. harveyi strain 1 on TCBS-bacteria in feed pellets (in vitro test)
According to the aforementioned methods, the V. harveyi strain 1-treated feed (V. harveyi-unsuppressive feed) was prepared by placing 30 feed pellets in a well of a 6-well cell culture cluster (Costar®, Corning, NY, USA) containing 2 ml cell suspension of V. harveyi strain 1. To prepare the V. harveyi-suppressive feed, the feed pellets were thoroughly pre-mixed with the whole of the La. plantarum L75a-fermented broth and distilled water at the ratio of 100/1/33 (w/w/w) before placing into the cell suspension of V. harveyi strain 1. Each treatment was conducted in triplicate. Every 30 min in the duration of 2 h, 2 feed pellets of each treatment were collected by the tweezers, surface-dried by a sterile paper, ground and homogenized by a blunt dissecting needle inside a sterile tube containing 50 µl of sterile 1% saline. To determine the TCBS-bacterial cell density in the feed, each feed suspension was serially diluted by sterile 1% saline and spot-inoculated (5 µl per droplet) on TCBS agar in triplicate. These procedures were conducted in a laminar flow hood to maintain axenic manipulation. The cell density of TCBS-bacteria in the feed was determined after the TCBS plates were incubated at 28 °C for 18 h.
Impact of V. harveyi strain 1 on TCBS-bacteria in the feces, intestine, muscle and hepatopancreas of Li. vannamei (the second and third feeding tests)
The experimental setup of the two tests below was similar to the aforementioned feeding test with some adjustments. In both tests, the Li. vannamei were fed with V. harveyi strain 1-treated feed pre-mixed with (suppressive feed) or without (unsuppressive feed) La. plantarum L75a-fermented broth. The V. harveyi strain 1-treated feed was prepared based on the methods described in the first feeding test.
In the second feeding test, each testing unit (semi-transparent plastic box of 30 × 20 × 15 cm) contained 5 Li. vannamei of 11 ± 2 cm in 6 l of disinfected and dechlorinated RAS water. After acclimation for 4–5 h, the Li. vannamei were fed with the aforementioned V. harveyi strain 1-suppressive or unsuppressive feed whose dry weight was 1% of their body weight. Each treatment was performed in triplicate (one testing unit represented one replicate). After three hours, fecal strings and intestine of Li. vannamei from each testing unit were collected in triplicate. Each sample was surface-dried by sterile paper, weighed, homogenized, suspended and serially diluted in sterile RAS water, spot-inoculated on TCBS agar in duplicate or triplicate, and determined for the TCBS-bacterial cell density according to the aforementioned methods of the first feeding test.
The third feeding test was performed for a longer duration to investigate the impact of V. harveyi strain 1 on TCBS-bacteria in the intestine, muscle and hepatopancreas of Li. vannamei. Each optimized testing unit, which was used in the first feeding test, contained 5 Li. vannamei of 11 ± 2 cm in 8 l of disinfected and dechlorinated RAS water. The first feeding was administered after acclimation for 4–5 h. The Li. vannamei were fed twice with the aforementioned V. harveyi strain 1-suppressive or unsuppressive feed, and the feed dry weight of each meal was 1% of their body weight. Each treatment was performed in triplicate and each replicate encompassed two testing units. After 4 days of cultivation, one Li. vannamei was collected from each replicate, and the intestine, muscle and hepatopancreas were collected and determined for their TCBS-bacterial cell density according to the aforementioned methods of the first feeding test. To investigate if V. harveyi strain 1 was present in the muscle of Li. vannamei fed with the unsuppressive feed (without La. plantarum supplementation), single colonies (with a predilection of yellowish colonies whose morphology was identical to that of V. harveyi strain 1) grown from such muscle type were randomly picked, grown in LB10 broth and sent for 16S rRNA gene sequencing. The resultant good-quality sequences, together with the Vibrio sequences detected by the 16S rRNA gene amplicon sequencing, were analyzed for their phylogeny based on the aforementioned methods.
Detection of bacterial community in the muscle of Li. vannamei fed with V. harveyi strain 1-treated feed
The muscle samples collected in the third feeding test were also sent for 16S rRNA gene amplicon sequencing. DNA extraction, PCR amplification of 16S rRNA gene (with an inclusion of 0.2 µl of BSA within each 20-µl mixture for the PCR reaction), Illumina MiSeq sequencing and processing of sequencing data were performed by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China) according to the protocol described by Wang and colleagues34. Briefly, the primers 338F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT) were used. The UPARSE (version7.1, http://drive5.com/uparse/) was applied to cluster the operational taxonomic units (OTUs) with a 97% similarity cutoff34.
Statistics and reproducibility
For each test, the resultant data was analyzed for normal distribution and homogeneity of variances. The impacts of the 13 Vibrio isolates on Li. vannamei, of V. harveyi strain 1 on the feed and on the muscle, hepatopancreas, intestine and feces of Li. vannamei, were analyzed by comparing the mortality percentage of the treated Li. vannamei (Fig. 2a, P < 0.05), by comparing the cell density of TCBS-bacteria in the treated feed (Fig. 3a, P < 0.05) and in the treated organs and feces of Li. vannamei (Fig. 2c, P < 0.05; Fig. 3b, P > 0.05), and by comparing the percentage of abundance of detected bacterial species in the muscle of Li. vannamei (Fig. 3d, P > 0.05), versus their corresponding control treatments scored at the same time using Mann–Whitney U-test, so as to ease the comparability amongst different tests. All the statistical tests were two-tailed.
Data availability
The accession numbers of the genome of V. harveyi strain 1 are CP080097 and CP080098 (BioProject PRJNA749085, BioSample SAMN20353411) in GenBank (www.ncbi.nlm.nih.gov).
References
Arulmoorthy, M. P., Anandajothi, E., Vasudevan, S. & Suresh, E. Major viral diseases in culturable penaeid shrimps: A review. Aquacult. Int. https://doi.org/10.1007/s10499-020-00568-3 (2020).
Lightner, D. V. Virus diseases of farmed shrimp in the western hemisphere (the Americas): A review. J. Invertebr. Pathol. 106, 110–130. https://doi.org/10.1016/j.jip.2010.09.012 (2011).
Singaravel, V. et al. Photobacterium damselae subsp. Damselae associated with bacterial myonecrosis and hepatopancreatic necrosis in broodstock Pacific white leg shrimp, Litopenaeus vannamei (Boone, 931). Aquacult. Int. 28, 1593–1608. https://doi.org/10.1007/s10499-020-00545-w (2020).
Raja, R. A. et al. Pathogenicity profile of Vibrio parahaemolyticus in farmed Pacific white shrimp, Penaeus vannamei. Fish Shellfish Immunol. 67, 368–381. https://doi.org/10.1016/j.fsi.2017.06.020 (2017).
Zhou, J. et al. A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease” of Litopenaeus vannamei shrimp. PLoS ONE 7, e29961. https://doi.org/10.1371/journal.pone.0029961 (2012).
Soto-Rodriguez, S. A. et al. Virulence of Vibrio harveyi responsible for the “Bright-red” Syndrome in the Pacific white shrimp Litopenaeus vannamei. J. Invertebr. Pathol. 109, 307–317. https://doi.org/10.1016/j.jip.2012.01.006 (2012).
Kiran, G. S. et al. Dietary administration of gelatinised polyhydroxybutyrate to Penaeus vannamei improved growth performance and enhanced immune response against Vibrio parahaemolyticus. Aquaculture 517, 734777. https://doi.org/10.1016/j.aquaculture.2019.734773 (2020).
Xie, J. et al. First report of Photobacterium damselae subsp. damselae infection in the mud crab Scylla paramamosain cultured in China. Aquaculture 530, 735880. https://doi.org/10.1016/j.aquaculture.2020.735880 (2021).
Elbashir, S. et al. Seafood pathogens and information on antimicrobial resistance: A review. Food Microbiol. 70, 85–93. https://doi.org/10.1016/j.fm.2017.09.011 (2018).
Gan, L. et al. Probiotics: their action against pathogens can be turned around. Sci. Rep. 11, 13247. https://doi.org/10.1038/s41598-021-91542-3 (2021).
Hatje, E. et al. First description of ‘Chalky back’ phenomenon in banana prawns (Fenneropenaeus merguiensis) and its possible association with Vibrio and Photobacterium species. FEMS Microbiol. Lett. 363, 019. https://doi.org/10.1093/femsle/fnw019 (2016).
Sichtig, H. et al. FDA-ARGOS is a database with public quality-controlled reference genomes for diagnostic use and regulatory science. Nat. Commun. 10, 3313. https://doi.org/10.1038/s41467-019-11306-6 (2019).
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. https://doi.org/10.1038/s41467-018-07641-9 (2018).
Alenton, R. R. R. et al. A hint of primitive mucosal immunity in shrimp through Marsupenaeus japonicus Gill C-type lectin. J. Immunol. 203, 2310–2318. https://doi.org/10.4049/jimmunol.1900156 (2019).
Wang, Z. et al. A novel research on isolation and characterization of Photobacterium damselae subsp. damselae from Pacific white shrimp, Penaeus vannamei, displaying black gill disease cultured in China. J. Fish Dis. 43, 551–559. https://doi.org/10.1111/jfd.13153 (2020).
Wang, X.-W., Xu, J.-D., Zhao, X.-F., Vasta, G. R. & Wang, J.-X. A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J. Biol. Chem. 289, 11779–11790. https://doi.org/10.1074/jbc.M114.552307 (2014).
Cerenius, L., Lee, B. L. & Soderhall, K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. https://doi.org/10.1016/j.it.2008.02.009 (2008).
Zhu, Z. M., Dong, C. F., Weng, S. P. & He, J. G. The high prevalence of pathogenic Vibrio harveyi with multiple antibiotic resistance in scale drop and muscle necrosis disease of the hybrid grouper, Epinephelus fuscoguttatus (f) x E-lanceolatus (o), China. J. Fish Dis. 41, 589–601. https://doi.org/10.1111/jfd.12758 (2018).
Cao, H.-J. et al. Sodium erythorbate, stable chlorine dioxide, and gellan gum glazing for shelf life extension of commercial peeled shrimp (Litopenaeus vannamei) during frozen storage. J. Food Process. Preserv. 43, 10. https://doi.org/10.1111/jfpp.14108 (2019).
Zhang, B., Ma, L.-K., Deng, S.-G., Xie, C. & Qiu, X.-H. Shelf-life of pacific white shrimp (Litopenaeus vannamei) as affected by weakly acidic electrolyzed water ice-glazing and modified atmosphere packaging. Food Control 51, 114–121. https://doi.org/10.1016/j.foodcont.2014.11.016 (2015).
Du, S. et al. Acidic electrolyzed water as a novel transmitting medium for high hydrostatic pressure reduction of bacterial loads on shelled fresh shrimp. Front. Microbiol. 7, 305. https://doi.org/10.3389/fmicb.2016.00305 (2016).
Austin, B. Vibrios as causal agents of zoonoses. Vet. Microbiol. 140, 310–317. https://doi.org/10.1016/j.vetmic.2009.03.015 (2010).
Yazgan, H., Ozogul, Y. & Kuley, E. Antimicrobial influence of nanoemulsified lemon essential oil and pure lemon essential oil on food-borne pathogens and fish spoilage bacteria. Int. J. Food Microbiol. 306, 108266. https://doi.org/10.1016/j.ijfoodmicro.2019.108266 (2019).
Novotny, L., Dvorska, L., Lorencova, A., Beran, V. & Pavlik, I. Fish: a potential source of bacterial pathogens for human beings. Vet. Med. 49, 343–358. https://doi.org/10.17221/5715-vetmed (2004).
Gautam, L. et al. Pseudomonas oleovorans sepsis in a child: The first reported case in India. Jpn. J. Infect. Dis. 68, 254–255. https://doi.org/10.7883/yoken.JJID.2014.174 (2015).
Faccone, D. et al. Human infections due to Pseudomonas chlororaphis and Pseudomonas oleovorans harboring new bla(VIM-2)-borne integrons. Infect. Genet. Evol. 28, 276–277. https://doi.org/10.1016/j.meegid.2014.10.012 (2014).
Cole, J. R. et al. Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. https://doi.org/10.1093/nar/gkt1244 (2014).
Saitou, N. & Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454 (1987).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. https://doi.org/10.1007/bf01731581 (1980).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Organ. Evol. 39, 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x (1985).
Liu, Y. et al. PHB-degrading bacteria isolated from the gastrointestinal tract of aquatic animals as protective actors against luminescent vibriosis. FEMS Microbiol. Ecol. 74, 196–204. https://doi.org/10.1111/j.1574-6941.2010.00926.x (2010).
Liu, B., Zheng, D., Zhou, S., Chen, L. & Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. https://doi.org/10.1093/nar/gkab1107 (2022).
Wang, X. et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 29, 787–803. https://doi.org/10.1038/s41422-019-0216-x (2019).
Acknowledgements
This research was supported by the Guangdong Marine Economy Promotion Projects (MEPP) Fund [Grant Number GDOE[2019]A26]; by the Guangdong Provincial Special Fund For Modern Agriculture Industry Technology Innovation Teams, Department of Agriculture and Rural Affairs of Guangdong Province [Grant Number 2021KJ150]; by the China Agriculture Research System [Grant Number CARS-47-G16]; by the Zhuhai Industry-University-Research Cooperation Project of Zhuhai Science and Technology Innovation Bureau [Grant Number ZH22017001200001PWC]; and by the Scientific Research Start-up Fund of South China Agricultural University [Grant Numbers 8000-218043, 8000-219242, 8000-220141]. The funders had no role in study design; in the collection, analysis and interpretation of data; in the writing and preparation of this article; or in the decision to submit this article for publication. We thank the bachelor students Jinghong Yan, Junfeng Wu, Jingjie Chen, Kun Xia and Jiayi Chen (College of Marine Sciences, South China Agricultural University, Guangzhou, China) for their support on shrimp cultivation, material preparation and shrimp in vivo experiments. We thank Mengqiu Chen for her suggestions and support on the analyses of the genome and the 16S rRNA gene amplicon sequence data.
Author information
Authors and Affiliations
Contributions
Study conceptualization, supervision and project administration by Y.L., L.G., Q.Q., W.-H.X. Sample collection and bacterial isolation by Y.L., L.G., W.-H.X. 16S rRNA gene amplification, analyses and BLAST by J.Z., W.-H.X., Y.L., Z.L., Q.G. Bacterial phylogenetic analyses by Y.L. with support from Y.Z., J.Z., Z.L., Q.G., W.-H.X., Z.W. Testing virulence of 13 Vibrio isolates by W.-H.X., Y.L. Sample collection for H&E-stained microscopy and image interpretation by Y.L., W.-H.X., C.L. Feed in vitro and in vivo testing led by J.Z. with support from W.-H.X., Jia.L., Jin.L., Y.Z., Y.L., Y.J., S.C., Z.W. Virulence factors analyses by Y.L., W.-H.X., Z.W. Results photography by J.Z., Y.L. Preliminary shrimp cultivation led by L.G., Y.J. and Q.G. with support from J.Z., W.-H.X., Jin.L., Jia.L., Z.L., Z.W. Formal analyses and figure preparation by Y.L. with support from J.Z., W.-H.X., Jia.L., L.G. Findings interpretation by Y.L. and L.G. with support from W.-H.X., J.Z., Jia.L., Z.W. Manuscript written by Y.L. with substantive contribution from T.D., J.Z., W.-H.X., L.G., Q.Q. Funding acquisition by L.G., Q.Q., Y.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gan, L., Zheng, J., Xu, WH. et al. Deciphering the virulent Vibrio harveyi causing spoilage in muscle of aquatic crustacean Litopenaeus vannamei. Sci Rep 12, 16296 (2022). https://doi.org/10.1038/s41598-022-20565-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20565-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.