Abstract
Commercially cultivated Limnospira (species formerly classified to genus Arthrospira) is a popular food/supplement consumed by millions of people worldwide for health benefits. The objective of the current research was to advance the standardization technology for Limnospira. Quantitative methods were established to detect fatty acids as potential chemical markers and immune-enhancing activity. Analysis of 20 different batches of biomass obtained from one commercial grower demonstrated that there was a statistically significant relationship between the sum of two fatty acids (linoleic and γ-linolenic) and Toll-like receptor (TLR)2/TLR1-dependent activation (R2 = 0.48, p = 0.0007). Investigation of 12 biomass samples sourced from growers in 10 different countries demonstrated that fatty acid content was again significantly correlated with biological activity (R2 = 0.72, p = 0.0005) and the content of fatty acids varied by twofold and activity by 12.5-fold. This large variation between different samples confirms the need to use the present standardization methods to ensure consistent and properly characterized biomass for consumers and for future scientific research.
Similar content being viewed by others
Introduction
Authentication and standardization have emerged as two critical tools for the rigorous scientific evaluation of botanical dietary supplements. Botanical authentication establishes the complete taxonomic identification and provides a foundation for future studies on the same test material. Lack of properly authenticated study material has resulted in published work that is misleading and potentially not reproducible1. Additionally, misidentification and adulteration with other species can impact the efficacy and safety of a botanical for consumers2. The objective of botanical standardization is to ensure that product material exhibits consistent and known levels of the biologically active compounds. Evaluation of material that is not standardized can be a major issue since botanicals often exhibit substantial variation in the level of their chemical constituents2, and this has contributed to a lack of consistent product efficacy in clinical trials3.
In the last 30 years several authors confirmed classification of Arthrospira and Spirulina into two independent genera, which was more or less widely accepted4,5. However, Arthrospira biomass is still widely referred to as “spirulina” by the general public, commercial marketing, and occasionally in the scientific literature. The use of incorrect taxonomic nomenclature can create confusion for data interpretation. In addition, cases of incorrect species designations within culture collections have also contributed to confusion regarding the correct identification of Arthrospira biomass material6. Recent taxonomic advances have resulted from investigating the Arthrospira type species based on a polyphasic approach using morphology, ecology, cell wall ultrastructure, and 16S rRNA sequence analysis. Based on this detailed characterization, Limnospira has been proposed as a new genus containing species formally part of Arthrospira and including the commercially grown taxa used for food and botanical dietary supplements6. Thus, in this paper, we refer to this organism by its taxonomically correct genus name—Limnospira.
Certain cyanobacterial representatives are used for human consumption7. Arthrospira/Limnospira is among the cyanobacterial genera that have been used as food for hundreds of years. Its species have been harvested in alkaline lakes, for example in Africa around Lake Chad and by the Aztecs in Central Mexico under the names dihé or tecuitlatl, respectively8. Since 1973, when a pilot plant in Mexico started annual production of 150 tons of dry biomass9, it has been cultivated on an industrial scale under control conditions in numerous places across the globe. The product is usually marketed under the name Spirulina. Although early interest in commercializing this food was focused mainly on its nutrient and protein content, it has emerged as a popular botanical dietary supplement exhibiting various human health benefits. A growing body of evidence indicates that this botanical’s consumption is a particularly useful natural product with immune-enhancing benefits that provides resilience against respiratory viral infection10,11,12.
Despite the popularity of Limnospira as a botanical dietary supplement, little work has been achieved on standardization methods. In the US Pharmacopeia method13, fatty acid profile, chlorophyll A and carotenoids are listed as chemical entities useful for identification purposes. For composition, the USP method includes the content of β-carotene, total carotenoids, c-phycocyanin, and protein. Since many of these constitutents are also present within other natural products, it remains to be determined if any of these can be used as meaningful authentication markers and whether any are correlated with Limnospira-dependent activation of immune cells, a major biological activity attributed to this botanical14.
Previous studies identified Braun-type lipoproteins as the predominant macrophage-activating principal within Limnospira14. Based on this research, a botanical extract was commercially developed that preferentially enriches the level of these active macromolecules (trade name Immulina). Currently, Immulina is standardized using only an in vitro bioassay that quantitates general monocyte-activation (without specificity for receptor mechanism of action). Chemical standardization of macromolecules, such as lipoproteins, can be challenging because the biological activity of these substances is often poorly correlated to their chemical composition. A similar problem existed in pharmaceutical biologics, a problem eventually solved by bioassay-based standardization to afford the consistent quality of products. Therefore, proper characterization of biologics requires information from both chemical and bioassay methods15.
The objective of the present research was to advance the standardization technology for Limnospira botanical dietary supplements and food products by employing a combination of chemical- and bioassay-based approaches. Selection and establishment of quantitative methods employed the use of biomass that was taxonomically identified using the most current classification system. Since the lipid moiety is the integral structural component on Braun-type lipoproteins, we hypothesized that the fatty acid profile could be a potential chemical standardization marker predictive of immune-enhancing activity. To test this hypothesis, we evaluated 20 different lots of biomass obtained from one commercial grower as well as material sourced from commercial growers in 10 different countries.
Results and discussion
Taxonomic identification using sequence analysis and morphological examination
According to its morphological features (Fig. 1) the reference study material (obtained from Dongtai Cibainian Biological Engineering) was determined using16 as L. fusiformis, but with opposite spirality than the one cited in16, i.e. sinistral instead of dextral. In terms of this character the species would be L. maxima, however the comparison of sequences of 16S rRNA gene + 16S-23S ITS region has shown accordance with genome sequence of reference strain of L. fusiformis, i.e. SAG 85.79. In any case this is a difference to the designation used by the manufacturer—A. platensis. Regardless of generic assignment which has undergone formal nomenclature change, the species are different based on their morphology. Unfortunately, it is not possible to apply molecular comparison because of lack of reference sequence of A. platensis in public sequence repositories. The sequences available bearing this name cannot be considered as reference, since the material was not isolated from a site similar to the locus classicus of A. platensis17 in terms of ecology and geography. In any case, usefulness of trichome spirality as one of the features for species determination needs to be reviewed based strictly on reference material. The same applies on taxonomic assignment of sequence data available in public repositories. However, for such studies availability of reference strain of at least A. platensis is a necessity.
The analysis of powdered biomass sold as food supplement (Table 1) revealed that the microscopy is rather difficult tool for species delimitation, since post-harvest processing of the material leads to biomass disintegration in some extent. These steps cause loss of some important morphological features like style of trichome coiling, or their well-developed terminal parts. The microscopic appearance of powdered biomass may be influenced also by length of its storage. Additionally, the DNA of the powdered biomass was extracted and subsequently the 16S rRNA genes of the sold food supplements were sequenced. The obtained sequences were compared to the sequences extracted from the genome of the reference strain of the type species of the Limnospira genus—L. fusiformis SAG 85.79, as well as to the outgroup taxon Limnoraphis hieronymusii CN4-3 AB045906 that was shown in previous phylogenetic studies to be closely related to the genus Limnospira6. Comparison of 16S rRNA gene identities among these sequences (Table 2) revealed that only the outgroup taxon considerably differed from the rest. On the other hand, all the taxa sequenced in this study shared more than 99% sequence identity with the reference 16S rRNA gene of the type species’ reference strain L. fusiformis SAG 85.79 and more than 98.8% identity among each other. For specimens of Limnospira 1, 3, 4, 5, 7, 8, 9, and 11 (numbers according to Table 1) sequences of the 16S rRNA gene were obtained from three clones. Our data suggest that the variability among different rRNA operons within the organisms is more noticeable than variability among the orgasnisms themselves; thus, the obtained data do not indicate that there is more than one species present in the food supplements investigated here. However, the high operon variability within the organisms makes the taxonomic situation of the genus Limnospira even more confusing. The taxonomic status and position of the species currently recognized in the genus Limnospira were not completely clarified yet and will need more thorough study using more molecular markers that is out of scope of the current paper.
Quantitative fatty acid profiling as a chemical standardization method for Limnospira biomass and immulina extract
Hyphenated analytical methods are instrumental and routinely implemented in the quality assurance of botanical extracts in various matrices. For example, detection of volatile organics in an extract with gas chromatography (GC) coupled with mass spectrometry (MS) and compared with reference standards enables the unambiguous identity of the major components, overall composition and can serve as a chemical fingerprint to assure the overall quality of raw materials used in various finished products. Moreover, such chemical fingerprints can be helpful to differentiate the anomalies due to possible misidentification of closely related species, the role of soil conditions, (un)intentional adulteration with other species, or contamination with extraneous ingredients.
To determine if the type of lipids present in Limnospira biomass and the Immulina extract could serve as potential chemical standardization markers, a GC–MS method was developed using 36 different fatty acid methyl esters as reference standards (Supplementary Fig. S1 online). As reported earlier18,19, the methyl esters of corresponding palmitic, linoleic, and γ-linolenic acids (γ-LA) were identified as dominant fatty acids, along with palmitoleic, stearic, and oleic acids as minor constituents in the dried biomass reference material (Supplementary Fig. S2 online). Single-step transesterification with 5% hydrogen chloride in methanol was identified as a suitable derivatization method with more reproducible results than the USP derivatization method (Table 3). In our laboratory, even though the latter method produced relative percentages of fatty acids within acceptable distribution ranges13, high inter-assay and intra-assay coefficients of variations (16.70% and 8.32%, respectively) were observed for the measurement of total fatty acid content. The obtained discordant results were partially attributed to the number of steps and the quality of the boron trifluoride-methanol complex utilized for the USP monogram’s derivatization method. Nevertheless, the abundance (Supplementary Fig. S2 online) of these six fatty acids was in agreement with the overall molecular composition of galactolipids mono- and digalactosyl diacylglycerol (MGDG, DGDG) and sulfolipid sulfoquinovosyl diacylglycerol (SQDG) from Spirulina platensis published by Xue et al.20 (in which palmitic, linoleic and γ-LA were identified as major fatty acids) as well as the lipid profile of Limnospira biomass reported in the literature19,21,22,23.
The capillary column, J&W HP-88 (60 m × 0.25 mm i.d.), with a film thickness of 0.2 µm used in the current study, significantly increased retention, improved resolution, and reduced tailing FAMES with the GC analysis as compared to the column used in the USP method (details provided in Table 3). In our quantitation experiments, the selective ion monitoring (SIM) mode (except for methyl stearate) was identified to be at least 26–50 times more sensitive than the conventional full-scan MS mode with limits of detection (LOD) and limits of quantitation (LOQ) in 2–17 and 6–50 ppb levels, respectively (Supplementary Fig. S3 online, Supplementary Table S1 online). The selective ions, m/z 69 for methyl oleate; m/z 74 for palmitate, palmitoleate, and stearate methyl esters; m/z 79 for α-linolenate and γ-linolenate methyl esters; and m/z 81 for linoleate methyl ester were identified as suitable ions in SIM mode for enhanced detection sensitivity and quantification purposes. Together with retention indices, the SIM mode also provided good selectivity allowing unambiguous identification of the six FAMES that were detected in the TIC mode.
Using the developed GC–MS quantification method (Table 3), the lipid profile of the Immulina extract was investigated and compared to the dried biomass to assess whether the fatty acids in Immulina were concentrated or depleted. Only one of the minor FAME, methyl stearate, was significantly higher in the Immulina extract compared with the biomass, 0.05 mg/g versus 0.02 mg/g, respectively (p = 0.04, n = 3, two-tailed paired t-test). No statistically significant differences in the absolute quantities (mg/g biomass dry wt) of the other five FAMES, as well as total FAME content (mg/g biomass dry wt), was observed in Immulina compared to the corresponding biomass sourced from Dongtai Cibainian Biological Engineering, China (Fig. 2). These results indicate that the FAME profile of the biomass is preserved in Immulina and hence has significant implications for authentication of extract material. Since morphological features of botanicals are destroyed and DNA typically lost in extract material, chemical markers have emerged as a valuable tool for botanical extract authentication. The fatty acid signature can therefore not only be used as one of the methods for identification of Limnospira in biomass samples but also in Immulina samples (and potentially other extracts of this botanical).
Comparative content of each FAME in dried Limnospira and commercial extract, Immulina, extracted from the same batch of dried biomass. Bars represent the average ± SD of three samples from the same batch of material. Each sample was analyzed in duplicate in the GC–MS. No statistically significant differences were observed between sample means, except for methyl stearate (compared by paired t-tests). *p = 0.04.
To evaluate the extent of FAME variation, different batches of biomass from a single commercial grower as well as from growers throughout the world were analyzed (Supplementary Fig. S4 online).
Assessing fatty acid profile variation in different batches of biomass from a single source
Twenty batches (acquired over the last 14 years, Supplementary Table S2 online) of dried biomass sourced from a single source were subjected to transesterification to assess the total fatty acid content as well as individual FAMES. In all these samples, the methyl ester of α-linolenic acid (α-LA) was not detected, confirming that these samples are probably free from chlorella, which usually contains α-LA at levels > 0.5% of total FAME content24. The total FAME content in the 20 samples ranged from 3.57 to 5.47% of biomass dry weight, with an average of 4.43%, in which polyunsaturated acids (linoleic and γ-LA) ranged from 1.32 to 3.24%. Total FAME content was significantly correlated with the date of biomass acquisition (R2 = 0.6508, p < 0.0001, Supplementary Fig. S5 online). Further regression analysis of the six individual fatty acids revealed that significant positive correlations were only observed for methyl linoleate and methyl γ-linolenate (R2 = 0.8036, p < 0.0001 and R2 = 0.5896, p < 0.0001, respectively, Supplementary Fig. S6 online). This data indicates that the observed decrease in FAME content with biomass age was predominantly due to the reduction of the polyunsaturated FAMES. Since polyunsaturated are more unstable than saturated fatty acids, the reduced FAME content with biomass age may be due to air oxidation of these lipids. However, controlled stability investigations are necessary before drawing major conclusions. It is possible that other factors could have contributed to the observed results. For example, the biomass was grown in outdoor ponds and as a result there could have been variation in ambient temperature during the cultivation of different batches. This may be a confounding factor since an increase in temperature19 has been observed to increase overall FAME content in many algal samples. Although strain differences can affect overall variations in FAME composition18, it is unlikely that this was a factor since all 20 samples originated from the same commercial grower.
Assessing fatty acid profile variation in biomass sourced from 10 different countries
Accessing FAME variation in biomass obtained from different sources was achieved by analyzing product material acquired from 11 commercial growers in 10 different countries (Table 1, Supplementary Fig. S4 online). The total FAME content (sum of the 6 quantifiable fatty acids) varied between 3.14 to 5.94% of biomass dry wt (1.9-fold difference), slightly more than the variation observed for the 20 samples from a single grower (1.5-fold). However, lower content of the polyunsaturated FAMES (1.05–2.30%) was observed in the globally sourced samples. None of the samples contained detectable levels of α-LA.
Re-evaluating the acceptance criteria for Limnospira fatty acid profile
The acceptance criteria range for the fatty acid profile in the USP method is broadly used for multiple species of Arthrospira (A. platensis, A. maxima, and A. fusiformis). Our sample collection included A. platensis and A. maxima (as designated by the commercial growers). Although all 32 analyzed samples are within the USP monogram’s accepted range for the percentage of palmitic acid (the predominant FAME) and linoleic acid, anomalies were observed for other FAMES (Supplementary Tables S2 and S3 online). The USP lower limit for the content of stearic and oleic acids is 1% of total FAMES. Methyl stearate was identified in negligible amounts (less than 1%) in 29 samples. Similarly, the content of methyl oleate was lower than 1% in 8 out of the 32 samples. In contrast, about 50% of samples were higher than the upper accepted criteria limit of 27% for γ-LA by 1 to 11 percentage points. Deviations from the USP accepted criteria range were also observed in our biomass reference material (0.06% of methyl stearate and 29.56% for γ-LA), highlighting the need for further revisions of this monograph.
Additionally, the acceptance criteria for the USP method only express the content of each FAME as a percentage of the total fatty acid content. A potential limitation of this approach is that two samples could have a similar profile in the percentage of each FAME but differ significantly in the absolute quantities of these compounds. For example, biomass from Chile contains the same percentage of methyl palmitate as material from Greece (49.99 and 49.21%, respectively, Supplementary Table S3 online). However, the two biomasses differed significantly in the absolute quantity of methyl palmitate (23.1 mg/g versus 15.4 mg/g, calculated from Supplementary Table S3 online). To overcome this limitation, we included total fatty acid content per gram of dry biomass as a parameter in the data analysis (values for each sample are listed in Table 1, Supplementary Tables S2 and S3 online).
Activation of the TLR2/TLR1 signaling pathway as an in vitro bioassay for quantitation of immune-enhancing activity
Hantke and Braun25 identified a lipoprotein within E. coli that contained a unique S-glycerylcysteine residue modified with three fatty acids (palmitate and cis-vaccenate as major constituents). This lipoprotein is commonly referred to as Braun’s lipoprotein. Research over the past few decades has established the presence of these molecules in both Gram-negative and Gram-positive bacteria and determined that they are potent activators of immune cells through a Toll-like receptor (TLR)2-dependent pathway26. Our previous research determined that this class of lipoproteins fully accounts for the in vitro monocyte/macrophage stimulatory activity exhibited by Limnospira14. Therefore, our efforts to design a biological standardization assay for Limnospira focused on capturing the activity exhibited by these lipoproteins.
Selecting an appropriate in vitro bioassay for biological standardization is driven based on the product’s mechanism of action27. The Limnospira-derived lipoproteins, similar to other Braun-type lipoproteins, are activators of TLR2 signaling. This was previously demonstrated using two different approaches28. First, in blocking antibody studies, NF-kappa B activation by Immulina in THP-1 cells was suppressed by antibodies to CD14 and TLR2, but not by antibodies to TLR4. Second, in expression vector experiments, Immulina increased NF-kappa B-directed luciferase expression in cells co-transfected with a vector expressing proteins required for TLR2 but not TLR4 signaling. In the current research, additional supporting evidence was obtained by evaluating the activity exhibited by Immulina in cell lines engineered to stably co-express a TLR gene and an NF-kappa B inducible secreted embryonic alkaline phosphatase (SEAP) gene. Immulina exhibited a robust dose-dependent activation in cells selectively expressing TLR2, and no significant activity was detected in cells selectively expressing TLR4 at the concentrations tested (Supplementary Fig. S7 online).
To facilitate ligand specificity and signaling, TLR2 forms heterodimers. The TLR2/TLR1 heterodimer detects triacylated lipopeptides, whereas TLR2/TLR6 heterodimer detects diacylated lipopeptides. Immulina (extracted from the biomass reference material) likely contains triacylated lipoproteins as evidenced by its ability to enhance activation in HEK-Blue hTLR2/TLR1 cells (Fig. 3a). As expected for this cell line, activation is also observed with the positive control Pam3CSK4 (a synthetic triacylated lipopeptide), but no activity was observed with the negative control Pam2CSK4 (synthetic diacylated lipopeptide). No sample induced SEAP levels above background values when evaluated in the control cells HEK-Blue hTLR2 KO-TLR1/6 cells (Fig. 3b). Cyanobacteria such as Limnospira have characteristics similar to gram-negative bacteria. Therefore, the presence of triacylated lipoproteins in Limnospira is consistent with the literature that this lipoprotein structure is also the most common type found in gram-negative bacteria29. Free fatty acids do not appear to contribute to the detected activity since no TLR2/TLR1-dependent activation was observed for any of the six individual fatty acids that occur within Limnospira (activity for individual compounds tested at 1 µg/ml ranged from 2.7 to 2.9% and were similar to untreated cells at 2.7%, values expressed as percent of activity exhibited by Pam3CSK4 tested at 100 ng/ml).
Immulina extract activates the TLR2/TLR1 signaling pathway. Activity evaluated in HEK-Blue hTLR2-TLR1 cells (a) and control HEK-Blue hTLR2 KO-TLR1/6 cells (b). Response ratio ± SD is defined as OD of sample/OD of untreated cells. Synthetic triacylated lipopeptide Pam3CSK4 was used as a positive control, and the synthetic diacylated lipopeptide Pam2CSK4 served as the negative control.
Assessing variation in immune-enhancing activity in different batches of biomass from a single source
Based on the receptor mechanism of action for the Limnospira lipoproteins, the HEK-Blue hTLR2/TLR1 cell line was selected as the biological assay to quantitate immune-enhancing activity. Analysis of the 20 biomass samples obtained from a single commercial grower (Dongtai Cibainian Biological Engineering) over 14 years displayed a 27-fold variation in TLR2/TRL1-dependent activity (Supplementary Table S2 online). Similar to the chemical analysis for content of total fatty acids (Supplementary Fig. S5 online) and polyunsaturated acids (Supplementary Fig. S6 online), activity also exhibited a statistically significant decrease with sample age (R2 = 0.7644, p < 0.0001, Supplementary Fig. S5online). Average EC50 values and average total fatty acid (TFA) content for samples in the different time periods demonstrated a clear change overtime: 2021–2019 (EC50 10 µg/mL, 50 µg/g TFA), 2018–2015 (EC50 32 µg/mL, 44 µg/g TFA) and 2008–2007 (EC50 135 µg/mL, 38 µg/g TFA). Between 2021–2019 and 2008–2007 the average EC50 value increased by 13.5 times, indicating that storage time was likely a major factor responsible for the 27-fold variation in activity observed in these samples.
Assessing variation in immune-enhancing activity in biomass sourced from 10 different countries
Extracts from the 12 biomass samples sourced from commercial growers in 10 different countries exhibited a 12.5-fold variation in TLR2/TLR1-dependent activity (Table 1). Sample 6A and 6B are from the same grower and differ in activity by about threefold. This data, combined with variation observed between the 20 batches from Dongtai Cibainian Biological Engineering (Supplementary Table S2 online), indicates that there can be substantial variation between lots obtained from the same commercial supplier. Therefore, it is not valid to use the data in Table 1 to rank the companies according to producers of the highest activity biomass. Many of these commercial suppliers have proprietary strains, and a detailed analysis of many different lots would be required to evaluate whether the biomass from a particular company exhibits consistently high or low activity. Hence it remains to be determined whether the 12.5-fold difference in activity between commercial biomass samples is due to strain differences and/or environmental factors such as temperature, length of sunlight exposure, nutrient composition of growth media, etc. It is unlikely that the differences are due to biomass age since all product material was acquired during a 3-month period (November 2020 through January 2021). Evidence supporting this interpretation can be derived from the analysis of the 20 Limnospira samples obtained over 14 years which suggest that years (not months) are required to significantly change biomass activity (Supplementary Table S2 online).
Content of FAMES is significantly correlated with TLR2/TLR1-dependent activity: basis for a chemico-biological approach for standardization of Limnospira products
The raw materials used to manufacture botanical dietary supplements exhibit inherent variability in chemical composition due to numerous factors (e.g., genetic/phenotypic variations, differences in agronomic conditions during growth, harvesting practices, etc.). This variation in raw material can influence the level of active compounds, resulting in a lack of consistent quality if the finished products are not properly standardized to the bioactives.
The Limnospira samples analyzed in the current study were raw materials sourced from many of the largest growers and represent biomass cultivated in diverse environments from 10 different countries. Similar to the variability observed for other botanicals30, our analysis of these commercial biomass samples demonstrated a twofold variation in total fatty acid content and a 12.5-fold variation in TLR2/TLR1-dependent activity (Table 1). In addition, our analysis indicated that long-term storage of biomass samples results in decreases in both activity and total fatty acid content (Supplementary Table S2 online). The public health implication of this data is that consumers are ingesting products that can vary in immune-enhancing activity by over 10 times. In addition, this level of variation poses a challenge to conducting reproducible scientific research on these products. Therefore, an effort is warranted to advance the standardization technology for Limnospira botanical dietary supplements and food products.
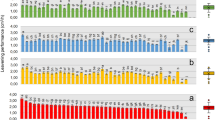
Regression analysis was performed to test our original hypothesis that the fatty acid profile could potentially serve as a relevant chemical standardization method to assess the relationship between total fatty acid content and immune-enhancing activity. The amount of total fatty acid (sum of the six quantifiable FAMES) was significantly correlated with TLR2/TLR1-dependent activity in the 20 biomass samples from Dongtai Cibainian Biological Engineering (R2 = 0.4735, p = 0.0008, Fig. 4a) as well as the 12 biomass samples from commercial growers in 10 different countries (R2 = 0.7002, p = 0.0007, Fig. 4c). Further regression analysis of the content of individual FAMES revealed that methyl linoleate and γ-LA were the only two compounds significantly correlated with activity in both sample sets (Table 4). The total content of the these two FAMES exhibited a similar correlation with TLR2/TLR1-dependent activity (R2 = 0.4777, p = 0.0007 (Fig. 4b) and R2 = 0.7192, p = 0.0005 (Fig. 4d), as compared to the correlation using the sum of all six FAMES. Overall, the data support our hypothesis and demonstrates that total FAME content (or just the content of methyl linoleate and γ-LA) can be used to predict the in vitro macrophage activation potential exhibited by Limnospira biomass samples.
Correlations between TLR2/TLR1 extract activity and content of fatty acids in biomass material obtained from Dongtai Cibainian Biological Engineering (20 batches, a and b) and commercial growers in 10 different countries (12 batches, c and d). The total content of 6 fatty acids represents the combined total of palmitate, palmitoleate, stearate, oleate, linoleate, and γ-linolenate in each sample (a, c). “LA” and “γ-LA” represent linoleate and gamma linolenate, respectively. EC50 values represent the concentration (µg/mL) of biomass material required to induce activation for the TLR2/TLR1 signaling pathway to levels 50% of those achieved by Pam3CSK4 (100 ng/mL).
Conclusion
In summary, we have identified an in vitro bioassay (detection of TLR2/TLR1 activity) and an analytical method (detection of FAME content) that can serve as a relevant approach for standardizing the immune-enhancing activity of Limnospira botanical dietary supplements and food products. Measurement of TLR2/TLR1-dependent activity detects Braun-type lipoprotein activity, the predominant compounds responsible for the in vitro macrophage activation potential of Limnospira. FAME content is correlated with activity and therefore expands the utility of this chemical marker to standardization efforts in addition to its original use in the USP method as a component of product identification. Utilizing the combination of an analytical method and bioassay provides an orthogonal approach to botanical standardization. The value of this combination approach has been highlighted for other botanicals such as hops (Humulus lupulus)31 and red clover (Trifolium pratense)32. Widespread adoption of chemico-biological standardization techniques to the botanical field would be expected to greatly enhance product quality and well-defined material for clinical studies on the efficacy of these products.
Methods
Biomass material
The biomass that was selected and taxonomically identified for reference material was obtained from Dongtai Cibainian Biological Engineering Company, LTD (established in 1994 and one of the largest producers of Limnospira in China). Limnospira dried powder (supplier lot # 20190501 and 20190820B) was used for sequence analysis. Live biomass was collected at the cultivation facility (located in Dongtai City, Jiangsu Province), and preserved in 2.5% formaldehyde for morphological analysis. The light microscopy analysis was performed on an Olympus BX53 microscope using bright field and differential interference contrast.
As part of our ongoing research on this botanical, twenty different batches of Limnospira dried samples were obtained from Dongtai Cibainian Biological Engineering over 14 years (2007–2021, Supplementary Table S2 online). Dried biomass samples were stored in the dark at room temperature in sealed containers at the National Center for Natural Products Research (NCNPR) and all samples were analyzed together in June and September, 2021 for fatty acid content and activation of the TLR2/TLR1 pathway, respectively.
From November 2020 through January 2021, twelve Limnospira dried samples were purchased from commercial growers worldwide (Table 1) that sell this biomass for food or use as a botanical dietary supplement. Biomass samples were stored in the dark at room temperature in sealed containers at the NCNPR, and all samples were analyzed together in March and May, 2021 for activation of the TLR2/TLR1 pathway and fatty acid content, respectively. Immulina extract was provided by Scandinavian Clinical Nutrition (Lot 2290006) and ChromaDex (Lots 2290020 and 2290021).
The twelve samples of powdered biomass listed in Table 1 were resuspended in distilled water and the reference sample were examined by a light microscopy for their morphological features, which were compared with known taxa listed in16. Subsequently their DNA was extracted using the UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA). The partial 16S rRNA gene together with the partial 16S-23S ITS region was amplified using the primers 16S27F/fD133,34 and 23S30R33. The PCR reaction was prepared as described in35. The presence of the PCR products was verified on 1.5% agarose gel and subsequently sequenced or stained with Sybr Green (Lonza, Rockland, ME, USA), separated on 1.5% low melting agarose gel, excised, and cloned with pGEM-T Easy vector system (Promega Corp., Madison, WI, USA). Plasmids were purified and commercially sequenced in both directions with the use of primers T7 (5′-TAA TAC GAC TCA CTA TAG GG-3′) and SP6 (5′-TAT TTA GGT GAC ACT ATA G-3′). In case the obtained reads were not long enough to assemble into a consensus sequence of the amplified region, the internal primer CYA781F(a)36 was used as well. Only for samples 2, 6a, and 10 sequence reads of sufficient length and quality were obtained by direct sequencing of PCR product purified by ExoSap (Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase; Thermo Fisher Scientific, Lithuania) with the use of the amplification primers 16S27F/fD133,34 and 23S30R33. For the rest of the samples cloning needed to be used, and thus 1 to 3 clones were obtained per sample. Sequences were then submitted to the NCBI GenBank database with the accession numbers listed in Table 1. The percent identities of the 16S rRNA gene sequences were calculated in Geneious Prime 2020.2.4 (http://www.geneious.com).
Chemistry
Derivatization of Limnospira biomass and Immulina
To characterize the overall lipid composition, dried biomass of Limnospira or its patented extract, Immulina, was subjected to saponification followed by esterification to convert free fatty acids into their corresponding methyl esters, hereafter referred to as fatty acid methyl esters (FAMES). All derivatization experiments (method A or method B, Table 3) were repeated at least twice, and each sample was injected twice into the GC–MS, resulting in at least quadruplicate results for every sample.
Saponification and esterification of biomass and Immulina (method A)
Following the USP method13, dried biomass from Limnospira and Immulina was subjected to two-step derivatization to corresponding FAMES. In brief, around 100 mg of sample was transferred to a 50 mL round-bottom flask, fitted with a reflux condenser and magnetic stir bar. To this flask, 100 mg of pyrogallic acid, 10 mL of 0.5 N methanolic sodium hydroxide solution were added and heated to reflux. After 15 min, 5.0 mL of 14% boron trifluoride—methanol was added through the reflux condenser into the round-bottom flask and continued to reflux for 2 more minutes. Around 5.0 mL of n-heptane was added through the condenser and refluxed for an additional 1.0 min. The flask was cooled to room temperature, removed the condenser, added 15 mL of saturated NaCl, closed with a stopper, shook vigorously, and the layers were allowed to separate. The supernatant n-heptane layer was transferred to a 2.0 mL vial, dried over anhydrous sodium sulfate, and subjected to GC–MS analysis.
Transesterification of biomass and Immulina (method B)
A transesterification method was developed based on earlier reports on total fatty acids in algae37,38. In a 20 mL scintillation vial containing 50.0 mg of biomass or Immulina, 1.5 mL of hydrogen chloride (5%) solution in methanol was added, stirred with a magnetic bar, and heated at 60 °C. After one hour, the resulting solution was cooled to room temperature, and 4.0 mL of n-heptane was added, stirred for 10 min, and allowed to settle as two separate layers. After 5 min, the supernatant n-heptane layer was carefully passed through anhydrous sodium sulfate. The resulting n-heptane (2.0 mL) was filtered through a 0.45 μm PVDF filter and collected in a clear glass vial for GC–MS analysis.
GC–MS analysis and quantitation of FAMES
Sample preparation
First, the samples prepared from derivatization experiments (Method A or B) were vortexed for 30 s to ensure sample homogeneity. Next, 480 µL of the sample was removed and transferred to a labeled GC vial. To the vial, 20 µL of the previously prepared internal standard (IS, methyl myristate) solution was added to the sample vial for a final concentration of 80 µg/mL. The vial was vortexed for 30 s and then analyzed by GC–MS.
GC/MS instrument conditions
All samples were analyzed using an Agilent 7890B gas chromatography (GC) instrument equipped with an Agilent 7693 autosampler and injector. A capillary column, J&W HP-88 (60 m × 0.25 mm i.d.) with a film thickness of 0.2 µm was connected to the GC and Agilent 5977A single quadrupole mass spectrometer (MS). Ultra-pure helium was used as the carrier gas at a flow rate of 1.2 mL/min. The samples were analyzed using the following oven program: starting from 60 °C held for 1 min, then programmed at 10 °C/min to 145 °C, then 1 °C/min to 190 °C, and finally at 5 °C/min to 240 °C. The inlet was operated in split mode at a temperature of 260 °C. The split ratio was 10:1 with 2 µL of each sample being injected. The transfer line from the GC to the MS was held at 260 °C, while the source and quadrupole were maintained at 230 °C and 150 °C, respectively, throughout the experiment.
Selective ion monitoring and confirmation
The 5977A mass spectrometer was operated with an electron energy of 70 eV, and the MS was operated in both scan and selective ion monitoring (SIM) modes. All scan mass spectra data were recorded at a rate of 5 Hz from m/z 40–400 after a 5-min solvent delay. The selective ions, m/z 69 for methyl oleate; m/z 74 for palmitate, palmitoleate, and stearate methyl esters; m/z 79 for α-linolenate and γ-linolenate methyl esters; and m/z 81 for linoleate methyl ester were identified as suitable ions in SIM mode with enhanced detection sensitivity and quantification purposes. Data were acquired utilizing Agilent MassHunter software (version B7.06.274), while quantification was achieved with Agilent MassHunter Quantitative Analysis (version 10.0.707.0). The NIST database (version 2.3) was utilized for tentative compound identification. The 6 quantifiable fatty acids were later confirmed with analytical reference standards.
Quantitative analyses
Methyl myristate in heptane was utilized as an IS, and the 36 reference standards (Supplementary Fig. S1 online) were obtained either from Sigma-Aldrich (St. Louis, MO USA), Thermo Fisher Scientific (Waltham, MA USA), or Cayman Chemicals (Ann Arbor, MI USA). The six quantifiable fatty acid methyl esters (FAMES) in Limnospira biomass, methyl palmitate, methyl palmitoleate, methyl stearate, methyl oleate, methyl linoleate, and methyl γ-linolenate were quantified using the GC–MS method. Methyl α-linolenate was also evaluated since this fatty acid was included in the USP method13,19 and it also served as a marker for detection of chlorella contamination24,39,40. A series of standard solutions were prepared for each FAME analyte to establish the calibration curves (Supplementary Fig. S3 online) using 0.1, 0.45, 0.75, 1, 4.5, 10, 25, 45, 75, 100, 200, 300, and 450 µg/mL solutions from a corresponding 5000 µL/mL solution in heptane and spiked with methyl myristate as IS with a final concentration of 80 µg/mL. Next, duplicate injections of each calibration curve solution were analyzed by GC–MS.
Biology
Sample preparation for evaluation of TLR2/TLR1-dependent activity
Crude extracts were prepared from the Limnospira dried biomass samples to enrich the levels of Braun-type lipoproteins. Raw material (1 g) was extracted with 7 mL of 50% ethanol at 80 °C for 45 min. Following centrifugation, the supernatant was collected, ethanol concentration adjusted to 72.5% by adding 1 volume of cold 95%, and the sample incubated at − 20 °C overnight. Precipitable material was collected by centrifugation and subsequently washed with cold 95% ethanol and the final extract dried. For TLR2/TLR1-dependent activity analysis, extracts were dissolved in Milli-Q water containing 2% sodium dodecyl sulfate at 25 mg/mL.
Receptor cell lines
Selective detection of pathogen recognition receptors (PRR) activation was accomplished using HEK-Blue hTLR2, HEK-Blue hTLR4, HEK-Blue hTLR2-TLR1, and HEK-Blue hTLR2 KO-TLR1/6 cells (InvivoGen, San Diego CA USA). These cell lines are engineered to stably co-express a PRR gene and an NF-kappa B inducible secreted embryonic alkaline phosphatase (SEAP) gene. Activation is detected by evaluating SEAP levels in the culture medium using the QUANTI-Blue reagent with optical density measurement at 635 nm. Experiments were performed according to manufacturer instructions. Positive controls were purchased from InvivoGen and included Pam3CSK4 (TLR2/TLR1 agonist) and Pam2CSK4 (TLR2/TLR6 agonist).
Data availability
Sequencing data is deposited in the NCBI GenBank database and available online using the accession numbers listed in Table 1. All other data and materials are available upon request by contacting either Dr. Nirmal Pugh (email: ndpugh@olemiss.edu, phone: + 1 662 915 1141) or Dr. Amar Chittiboyina (email: amar@olemiss.edu, phone + 1 662 915 1572).
References
Applequist, W. L. & Miller, J. S. Selection and authentication of botanical materials for the development of analytical methods. Anal. Bioanal. Chem. 405, 4419–4428. https://doi.org/10.1007/s00216-012-6595-1 (2013).
Shipkowski, K. A. et al. Naturally complex: Perspectives and challenges associated with botanical dietary supplement safety assessment. Food Chem. Toxicol. 118, 963–971. https://doi.org/10.1016/j.fct.2018.04.007 (2018).
Landis, S. C. et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 490, 187–191. https://doi.org/10.1038/nature11556 (2012).
Tomaselli, L., Palandri, R. M. & Tredici, M. R. On the correct use of the Spirulina designation. Algol. Stud. Archiv für Hydrobiol. 83, 539–548 (1996).
Komárek, J. & Lund, J. W. What is «Spirulina platensis» in fact?. Algol. Stud. Archiv für Hydrobiologie 85, 1–13 (1990).
Nowicka-Krawczyk, P., Muhlsteinova, R. & Hauer, T. Detailed characterization of the Arthrospira type species separating commercially grown taxa into the new genus Limnospira (Cyanobacteria). Sci. Rep. 9, 694. https://doi.org/10.1038/s41598-018-36831-0 (2019).
Chacón-Lee, T. & González-Mariño, G. Microalgae for “healthy” foods—Possibilities and challenges. Compr. Rev. Food Sci. Food Saf. 9, 655–675 (2010).
Ciferri, O. Spirulina, the edible microorganism. Microbiol. Rev. 47, 551–578. https://doi.org/10.1128/mr.47.4.551-578.1983 (1983).
Ahsan, M., Habib, B., Parvin, M., Huntington, T. C. & Hasan, M. R. A review on culture, production and use of spirulina as food for humans and feeds for domestic animals (2008).
Capelli, B. & Cysewski, G. R. Potential health benefits of spirulina microalgae. Nutrafoods 9, 19–26 (2010).
Ratha, S. K., Renuka, N., Rawat, I. & Bux, F. Prospective options of algae-derived nutraceuticals as supplements to combat COVID-19 and human coronavirus diseases. Nutrition 83, 111089. https://doi.org/10.1016/j.nut.2020.111089 (2021).
Pugh, N. D. et al. Oral administration of a Spirulina extract enriched for Braun-type lipoproteins protects mice against influenza A (H1N1) virus infection. Phytomedicine 22, 271–276. https://doi.org/10.1016/j.phymed.2014.12.006 (2015).
Food Chemicals Codex (FCC) monograph # 2181. Published by the United States Pharmacopeia (USP) and the National Formulary (NF). Accessed on June 23rd, 2020.
Nielsen, C. H. et al. Enhancement of natural killer cell activity in healthy subjects by Immulina(R), a Spirulina extract enriched for Braun-type lipoproteins. Planta Med. 76, 1802–1808. https://doi.org/10.1055/s-0030-1250043 (2010).
Meager, A. Measurement of cytokines by bioassays: Theory and application. Methods 38, 237–252. https://doi.org/10.1016/j.ymeth.2005.11.005 (2006).
Komárek, J. Cyanoprokaryota 2—Teil/2nd part: Oscillatoriales. Susswasserflora von Mitteleuropa 19, 1–759 (2005).
Gomont, M. A. Monographie des Oscillariées:(Nostacacées Homocystées). Vol. 15 (Masson, 1893).
Mühling, M., Belay, A. & Whitton, B. A. Variation in fatty acid composition of Arthrospira (Spirulina) strains. J. Appl. Phycol. 17, 137–146. https://doi.org/10.1007/s10811-005-7213-9 (2005).
Cohen, Z., Vonshak, A. & Richmond, A. Fatty acid composition of Spirulina strains grown under various environmental conditions. Phytochemistry 26, 2255–2258 (1987).
Xue, C. et al. Molecular species composition of glycolipids from Sprirulina platensis. Food Chem. 77, 9–13. https://doi.org/10.1016/S0308-8146(01)00315-6 (2002).
Ljubic, A., Safafar, H., Holdt, S. L. & Jacobsen, C. Biomass composition of Arthrospira platensis during cultivation on industrial process water and harvesting. J. Appl. Phycol. 30, 943–954 (2018).
Hultberg, M., Lind, O., Birgersson, G. & Asp, H. Use of the effluent from biogas production for cultivation of Spirulina. Bioprocess. Biosyst. Eng. 40, 625–631 (2017).
Hena, S., Znad, H., Heong, K. & Judd, S. Dairy farm wastewater treatment and lipid accumulation by Arthrospira platensis. Water Res. 128, 267–277 (2018).
Compositional guideline: Arthrospira platensis. https://www.tga.gov.au/compositional-guideline/arthrospira-platensis (2011).
Hantke, K. & Braun, V. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur. J. Biochem. 34, 284–296 (1973).
Nguyen, M. T. & Gotz, F. Lipoproteins of gram-positive bacteria: Key players in the immune response and virulence. Microbiol. Mol. Biol. Rev. 80, 891–903. https://doi.org/10.1128/MMBR.00028-16 (2016).
White, J. R. et al. Best practices in bioassay development to support registration of biopharmaceuticals. Biotechniques 67, 126–137. https://doi.org/10.2144/btn-2019-0031 (2019).
Balachandran, P., Pugh, N. D., Ma, G. & Pasco, D. S. Toll-like receptor 2-dependent activation of monocytes by Spirulina polysaccharide and its immune enhancing action in mice. Int. Immunopharmacol. 6, 1808–1814. https://doi.org/10.1016/j.intimp.2006.08.001 (2006).
Nakayama, H., Kurokawa, K. & Lee, B. L. Lipoproteins in bacteria: Structures and biosynthetic pathways. FEBS J. 279, 4247–4268. https://doi.org/10.1111/febs.12041 (2012).
Pferschy-Wenzig, E.-M. & Bauer, R. The relevance of pharmacognosy in pharmacological research on herbal medicinal products. Epilepsy. Behav. 52, 344–362 (2015).
Krause, E. et al. Biological and chemical standardization of a hop (Humulus lupulus) botanical dietary supplement. Biomed. Chromatogr. 28, 729–734. https://doi.org/10.1002/bmc.3177 (2014).
Piersen, C. E. et al. Chemical and biological characterization and clinical evaluation of botanical dietary supplements: A phase I red clover extract as a model. Curr. Med. Chem. 11, 1361–1374. https://doi.org/10.2174/0929867043365134 (2004).
Taton, A., Grubisic, S., Brambilla, E., De Wit, R. & Wilmotte, A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): A morphological and molecular approach. Appl. Environ. Microbiol. 69, 5157–5169. https://doi.org/10.1128/AEM.69.9.5157-5169.2003 (2003).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. https://doi.org/10.1128/jb.173.2.697-703.1991 (1991).
Muhlsteinova, R., Hauer, T., De Ley, P. & Pietrasiak, N. Seeking the true Oscillatoria: A quest for a reliable phylogenetic and taxonomic reference point. Preslia 90, 151–169 (2018).
Nübel, U., Garcia-Pichel, F. & Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63, 3327–3332. https://doi.org/10.1128/AEM.63.8.3327-3332.1997 (1997).
Cavonius, L. R., Carlsson, N.-G. & Undeland, I. Quantification of total fatty acids in microalgae: Comparison of extraction and transesterification methods. Anal. Bioanal. Chem. 406, 7313–7322 (2014).
Kim, B., Im, H. & Lee, J. W. In situ transesterification of highly wet microalgae using hydrochloric acid. Biores. Technol. 185, 421–425 (2015).
Otles, S. & Pire, R. Fatty acid composition of Chlorella and Spirulina microalgae species. J. AOAC Int. 84, 1708–1714 (2001).
Tudor, C., Gherasim, E. C., Dulf, F. V. & Pintea, A. In vitro bioaccessibility of macular xanthophylls from commercial microalgal powders of Arthrospira platensis and Chlorella pyrenoidosa. Food Sci. Nutr. 9, 1896–1906. https://doi.org/10.1002/fsn3.2150 (2021).
Acknowledgements
Authors would like to thank Botanical Therapeutics and Ningbo Green-Health Pharmaceuticals for coordinating the collection of live Limnospira biomass and its subsequent preservation from the commercial supplier Dongtai Cibainian Biological Engineering. The authors would also like to thank ChromaDex for providing the Immulina product. Research reported in this publication was supported by the Office of Dietary Supplements and National Center for Complementary and Integrative Health of the National Institutes of Health under Award Number U19AT010838. The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Institutes of Health. Additional funding was also provided by a grant from the USDA, Agricultural Research Service Specific Cooperative Agreement No. 58-6060-6-015.
Author information
Authors and Affiliations
Contributions
J.H. and S.H. conducted all derivatization experiments and prepared the samples for GC–MS analysis. J.L. and M.W. developed a suitable, well-resolved GC–MS method and conducted calibration experiments for quantification. I.A.K. and A.G.C. contributed to the design of chemical experiments, planning of research, data interpretation, and preparation and editing of the manuscript. J.Z. performed the cell culture experiments including sample preparation and analysis using the reporter cell line assays; N.D.P. contributed to the design/planning of the research, sample acquisition, statistical analysis and manuscript preparation. R.H. did all molecular analyses and contributed to preparation of manuscript; T.H. did all morpological analyses and contributed to preparation of manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
NDP and IAK acknowledge financial interest in Immulina. All other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huh, J., Zhang, J., Hauerová, R. et al. Utility of fatty acid profile and in vitro immune cell activation for chemical and biological standardization of Arthrospira/Limnospira. Sci Rep 12, 15657 (2022). https://doi.org/10.1038/s41598-022-19590-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19590-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.