Abstract
Alcohol consumption is associated with a high increased lipid profile and this association may depend on genetic risk factors. In this study, we aimed to assess the effects of genetic variation associated with alcohol consumption on lipid profiles using data from two Korean population studies. We performed a genotype association study using the HEXA (n = 51,349) and KNHANES (n = 9158) data. Genotype analyses of the two sets of Korean population data showed associations of increased total cholesterol and high-density lipoprotein (HDL)-cholesterol with CETP rs708272. The HEXA and KNHANES populations revealed differences in HDL cholesterol according to the presence of CETP rs708272, independent of ALDH2 rs671 and alcohol consumption. In contrast, total cholesterol levels were associated with alcohol consumption and ALDH2 rs671 in men with CETP rs708272 (CT and TT genotypes). Furthermore, in drinkers with ALDH2 rs671 (GA and AA genotypes), higher total cholesterol was associated with the CETP rs708272 TT minor homozygous genotype based on both HEXA and KNHANES data. Our findings demonstrated that alcohol consumption and genetic variation in either CETP or ALDH2 may be associated with cholesterol levels. We hope these findings will provide a better understanding of the relationship between alcohol consumption and cholesterol according to each individual’s genetic background.
Similar content being viewed by others
Introduction
Alcohol consumption is a major environmental factor that is associated with health problems. Greater alcohol consumption exacerbates the lipid profile1, which has been linked to various diseases, including dyslipidemia and cardiovascular diseases2,3. Previous studies have reported that alcohol consumption is associated with improved high-density lipoprotein cholesterol (HDL-C) levels, but also with increased triglyceride (TGs)4 and low-density lipoprotein (LDL)5. The association between alcohol consumption and total cholesterol remains controversial, with several studies either showing no effect6 or increased cholesterol levels. Thus, it is necessary to clarify the association between alcohol consumption and lipid profiles and the related mechanisms in large groups representing various races/ethnicities.
Cholesterol levels are associated with both genetic and environmental factors. Among them, cholesteryl ester transfer protein (CETP; also called plasma lipid transfer protein), which circulates in plasma, is mainly bound to HDL-C and facilitates the transfer of cholesteryl ester particles to apolipoprotein B (including LDL and very low-density lipoprotein (V-LDL)) in exchange for TGs7. In general, changes in the activity and concentration of CETP decrease the plasma HDL-C levels and increase the plasma LDL-cholesterol (LDL-C) levels. Additionally, genetic variations are related to its regulation8. CETP rs708272 (also called Taq1B) is a C-to-T substitution at the 279th nucleotide of intron 1 and is the most thoroughly studied variant of this gene. A previous study identified an association between increased HDL-C levels or decreased CETP activity and allele type (B1, B2)9,10. Allele B1 affects the size and function of the CETP protein and HDL-C level. In contrast, allele B2 is associated with lower-molecular-weight CETP and increased HDL-C. Moreover, increased HDL-C levels were associated with reduced CETP activity due to alcohol consumption11, and high alcohol consumption increased HDL-C levels in B2 carriers12.
Facial flushing in response to alcohol consumption, known as Asian flushing or the Asian glow, is a reaction specific to East Asian populations13. This response can estimate the activities of ALDH2 and ADH1B, which are the most efficient enzymes associated with aldehydes. Higher activity of ADH (conferred by Arg48) or lower activity of ALDH2 (conferred by Lys487) leads to accumulation of acetaldehyde following alcohol consumption. In particular, the ALDH2 rs671 variant slows the degradation of acetaldehyde, and the accumulated acetaldehyde can cause serious inconvenience. In addition, ALDH2 rs671 had different effects on HDL-C and total cholesterol according to genotype; lower HDL-C and higher total cholesterol levels were observed with the ALDH2 rs671 genotype when compared to those observed in its absence14. Furthermore, carriers with the ALDH2 rs671 GG genotype had substantially higher mean alcohol consumption and HDL-C levels than those with the other genotypes15. The GG genotype carriers in the Shandong province of China had a higher mean lipid level and total cholesterol disorder rate than those of the other genotype carriers16.
In this regard, ALDH2 variation should be considered as the main genetic proxy for the differences in cholesterol according to alcohol consumption. Moreover, it is important in alcohol research because of its direct relation to alcohol metabolism. However, few studies have addressed the influence of alcohol consumption on cholesterol according to CETP and ALDH2 variant type status. Accordingly, we aimed to investigate the association between alcohol consumption and cholesterol according to CETP and ALDH2 variants in the Korean population.
Results
Data were obtained from the Health Examinees Study (HEXA) and Korean National Health and Nutrition Examination Survey (KNHANES). The characteristics of the study participants, according to sex, are presented in Table 1. Age, systolic/diastolic blood pressure, fasting glucose level, total cholesterol level, HDL-C level, TG level, aspartate transaminase (AST) level, alanine transaminase (ALT) level, and alcohol consumption rate differed by sex in both the HEXA and KNHANES study populations (p < 0.05). The frequency of ALDH2 rs671 differed significantly by sex in the HEXA population, but not in the KNHANES population. The general characteristics of age, systolic/diastolic blood pressure, HDL-C level, TG level, and AST and ALT levels differed according to alcohol consumption in both study populations (Online Supplementary Table S1).
The participants’ clinical characteristics are summarized according to CETP rs708272 [CC, CT, or TT] status in Table 2. The total cholesterol and HDL-C levels of CETP rs708272 risk T allele carriers were significantly higher than others in the HEXA and KNHANES populations (p < 0.05). In addition, the risk allele T carriers had a higher LDL-C levels in the KNHANES study (p < 0.05). When categorized by sex, LDL-C levels were significantly different between men in both the HEXA and KNHANES studies. (Online Supplementary Tables S2 and S3). Men carrying CETP rs708272 TT allele had higher LDL-C levels than those carrying the CT of the TT genotype. On the other hand, almost all general characteristics according to the ALDH2 rs671 genotype, except for sex and total cholesterol, showed differences in both study populations (Online Supplementary Table S4).
Table 3 shows the total cholesterol and HDL-C levels according to alcohol consumption in carriers of CETP rs708272. HDL-C levels differed significantly between CETP rs708272 by alcohol consumption and sex in the HEXA and KNHANES study populations. In the HEXA population, CETP rs708272 was associated with a higher HDL-C level in drinkers than in non-drinkers of both sexes. However, HDL-C level did not differ with the association between the CETP rs708272 TT minor homozygous genotype and alcohol consumption in both sexes in the KNHANES population. In the HEXA population, total cholesterol varied significantly with alcohol consumption in men with CETP rs708272, whereas in women, there was no association between drinking and CETP rs708272. In addition, in the KNHANES population, total cholesterol was not associated with CETP rs708272 among non-drinkers and drinkers. In the HEXA population, total cholesterol differed significantly among men with CETP rs708272 depending on alcohol consumption. In contrast, the KNHANES population, there was no association between drinking status and CETP rs708272. In addition, CETP rs708272 was associated with higher total cholesterol levels in male drinkers than in non-drinkers in the HEXA population, whereas no association was found in the KNHANES population. In women, total cholesterol differed significantly among CETP rs708272 in non-drinkers in both the HEXA and KNHANES populations. Total cholesterol was inversely associated with alcohol consumption according to CETP rs708272 in the HEXA population. However, the only risk TT minor homozygous genotype of CETP rs708272 was inversely associated with higher total cholesterol in drinkers in the KNHANES population. Conversely, the association of cholesterol with alcohol consumption in the CETP rs708272 major homozygous genotype group disappeared.
In the case of ALDH2 rs671, HDL-C showed a significant difference according to the ALDH2 rs671 genotype in non-drinkers and drinkers in the HEXA population. Nevertheless, only drinkers showed differences in the KNHANES population. Moreover, ALDH2 rs671 [GA + AA] genotype was associated with lower HDL-C levels in drinkers than those in non-drinkers in the HEXA population. Total cholesterol in the HEXA and KNHANES populations differed according to the ADLH2 rs671 genotype in drinkers (Online Supplementary Table S5).
The effects of CETP rs708272 and alcohol consumption on HDL-C according to ALDH2 rs671 are shown in Table 4 and Online Supplementary Fig. S1. HDL-C differed significantly according to CETP rs708272 independent of ALDH2 rs671 and alcohol consumption, but CETP rs708272 and HDL-C were slightly associated with the ALDH2 rs671 [GA + AA] genotypes. In addition, those with the ALDH2 rs671 [GA + AA] genotype exhibited trends toward lower HDL-C compared with those with the ALDH2 rs671 GG major homozygous genotype. Still, overall, the HDL-C level was higher in men drinkers than in non-drinkers, regardless of the ALDH2 rs671 genotype.
Table 5 shows the association of total cholesterol with alcohol consumption and ALDH2 rs671 according to CETP rs708272. In men, total cholesterol was associated with alcohol consumption and ADLH2 rs671 when CETP rs708272 CT heterozygous and TT minor homozygous genotypes were present in both HEXA and KNHANES populations. Additionally, drinkers with ALDH2 rs671 exhibited the highest total cholesterol level with the CETP rs708272 TT risk minor homozygous genotype in both the HEXA and KNHANES populations. On the other hand, as a result of analyzing the association between the CETP rs708272 genotype and alcohol consumption according to ALDH2 variation in women, the total cholesterol level was high in the presence of the CETP rs708272 risk T allele, but there was no significant difference, unlike in men. In particular, male drinkers with the CETP rs708272 TT risk minor homozygous genotype had the highest total cholesterol level when they had the ALDH2 rs671 [GA + AA] genotype.
Discussion
Several studies have reported associations between CETP rs708272, alcohol consumption, and cholesterol levels. In addition, ALDH2 rs671, which is closely related to alcohol consumption in East Asians, is well known to be associated with HDL-c according to alcohol consumption17, but studies involving CETP variants have not been reported. We investigated the association between alcohol consumption and cholesterol levels according to CETP and ALDH2 variants and their combinations in two large-scale Korean populations.
Previous studies have shown that CETP rs708272 increases HDL-C levels. In a meta-analysis of 98 published studies18, CETP rs708272 was associated with higher mean HDL-C levels. In that study, HDL-C level was higher in the CETP rs708272 risk allele (TT and CT genotype) carriers than in the CC major homozygous genotype carriers. In 2018, Cai et. al. reported that the HDL level of participants with CETP rs708272 TT genotype was higher than those of CC genotype, and the HDL level of male participants with only T allele was significantly higher19. Also, some studies have shown a gene × alcohol consumption interaction for CETP. Based on the limited evidence, alcohol consumption appears to be associated with lower CETP activity20. In the association between alcohol consumption and HDL-C, changes in other metabolic pathways including an increased transport rate of apolipoproteins, reduced hepatic lipase activity, and so on may be likely21,22,23,24,25. However, our study found no interaction between CETP rs708272 and alcohol consumption in determining HDL-C. This is in line with previous studies carried out in a Mediterranean population26, healthy men27, and insulin-dependent men28. The metabolic mechanisms underlying an increase in HDL-C following the interaction between CETP and alcohol consumption are poorly understood, and more research is needed.
In this study, we observed that increased HDL-C was associated with alcohol consumption independent of ALDH2 rs671; this result is similar to that of a previous meta-analysis, which revealed an association between alcohol consumption and HDL-C. However, Wakabayashi et al. reported that the effect of alcohol consumption on HDL-C was different in drinkers with an alcohol flushing response29. The beneficial effects of an association between alcohol consumption and HDL-C according to ALDH2 rs671 remains controversial because several other studies have shown that HDL-C is influenced by alcohol consumption differently in drinkers with an alcohol flushing response. According to some data, the ALDH2 rs671 A allele carriers, GA, and AA genotypes have a 2.6-fold higher risk of hypo-HDL-cholesterolemia than the GG homozygous genotype30. These conflicting findings may be due to the small size of the study population. Further studies using larger populations are needed to clarify whether ALDH2 affects the relationship between alcohol consumption and HDL-C.
The effects of alcohol consumption on total cholesterol may differ according to the CETP or ALDH2 status. Previous studies have shown that alcohol consumption in subjects with the ALDH2 rs671 risk A allele was associated with increased total cholesterol compared to alcohol consumption with the ALDH2 rs671 GG homozygous genotype31. Similar to previous studies, our study found that drinkers with the ALDH2 [GA + AA] genotype had higher total cholesterol levels than non-drinkers with the ALDH2 GG homozygous genotype. CETP variants also showed an association with total cholesterol increase32, but no significant difference was found in total cholesterol levels according to alcohol consumption in CETP rs70827233. The total cholesterol levels in our study were the highest in both men and women with CETP rs708272 risk TT genotype. Still, there was no difference according to ALDH2 rs671 genotype, while the presence of ALDH2 rs671 risk alleles in the drinking men group with CETP rs708272 risk TT genotype was associated with changes in total cholesterol levels.
This was an observational study of the association between alcohol consumption and cholesterol according to CETP and ALDH2 variants. Residual confounding is a critical issue that should be considered in observational studies. We replicated two Korean population cohorts (HEXA and KNHANES) to address residual confounding. In the present study, we revealed differences in HDL-cholesterol according to the presence of CETP rs708272, independent of ALDH2 rs671 and alcohol consumption, and found that the presence of CETP rs708272 risk allele in men was associated with an increase in HDL cholesterol following alcohol intake. Furthermore, total cholesterol levels were associated with alcohol consumption and ALDH2 rs671 in men with CETP rs708272 CT and TT genotypes with strong genetic effects. This study provides information about genetic variants of CETP and ALDH2 in the Korean population. It clarifies the probable mechanism underlying the differential association between alcohol consumption and cholesterol (total cholesterol and HDL-C) according to CETP and ALDH2 variants.
Overall, our work would be helpful in clarifying the clinical implications of improving the translational value for preventive management of public health policies, and further realizing personalized or precision medicines that can prevent individual diseases or realize early detection, treatment decisions, and prognosis. It can also be used as a predictor of population assessment and vulnerable groups, while providing evidence for health policy development and health insurance applications.
Methods
Study population
Data were obtained from two studies conducted by the Korea Disease Control and Prevention Agency (KDCA) as part of the Korean Genome and Epidemiology Study (KoGES) and the KNHANES. Written informed consent was obtained from all participants, and the research project was approved by KDCA. The study protocol was approved by the Institutional Review Board of the Korean National Institute of Health (2019-03-01-PE-A). HEXA and KNHANES have been previously described in detail34. All study protocols were carried out by the approved guidelines.
The HEXA cohort subjects (n = 53,754) were invited to health examination centers in eight regions (metropolitan areas or major cities) and enrolled in 38 health examination centers and hospitals in Korea between 2004 and 2013. Individuals with missing data for the CETP rs708272 genotype (n = 149) or alcohol consumption (n = 2256) were excluded. A total of 51,349 participants were included in this cross-sectional study.
KHNANES is a nationally representative survey of the health and nutritional status of the Korean population. We used data from a population of 15,000 people aged 20–60 years collected during the 2009–2015 period. Individuals with missing data for alcohol consumption (n = 401), cholesterol (HDL and total cholesterol, n = 153), and those under 40 years of age (n = 5288) were excluded. In total, 9158 participants were included in this cross-sectional study.
Anthropometric and biochemical analyses
In HEXA, age and smoking status information was obtained using interview-based questionnaires. Body mass index (BMI) was calculated as weight divided by height squared. The lipid profile (total cholesterol, TGs, and HDL-C) and fasting glucose, ALT, and AST levels were measured using a Hitachi 747 chemistry analyzer (Hitachi, Ltd., Tokyo, Japan). Systolic/diastolic blood pressure was measured twice using a standard mercury sphygmomanometer, and the results were averaged.
In the KNHANES, data were collected by various means, including interview-based and self-reported questionnaires, physical examinations, and assessments of nutritional status. The calculation of BMI and blood pressure (systolic and diastolic) was the same as that for HEXA. The lipid profile (total cholesterol, TGs, and HDL-C) and plasma glucose, ALT, and AST levels were measured using a Hitachi Automatic Analyzer 7600 (Hitachi, Tokyo, Japan).
Alcohol consumption data were collected using an interview-based questionnaire in HEXA and KNHNES. Subjects were asked whether they had ever consumed at least one alcoholic drink every month, and if they had, whether they were former-drinker or current drinkers35.
Genotyping
HEXA cohort subjects were genotyped using a Korean chip designed by the Center for Genome Science at the Korean National Institute of Health. The chip was provided and its use was approved by the National Biobank of Korea, the Centers for Disease Control and Prevention, Republic of Korea (No. 2019-022). The genotyping protocol and quality control process has been described in detail previously36.
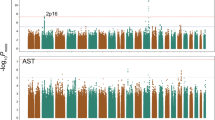
We preliminarily analyzed the association between cholesterol and genes using 128 SNPs related to alcohol metabolism which were selected from the KNHANES by TaqMan genotyping assay as previously described37. Only CETP rs708272 and ALDH2 rs671 were related to cholesterol in two sets of Korean population data, which led to the use of this study.
Statistical analysis
Statistical analyses were performed using SAS software package (ver. 9.4; SAS Institute Inc., Cary, NC, USA). Data are presented as the mean ± standard deviation or number (%). Logarithmic transformation was applied to variables with a non-Gaussian distribution. Student’s t-tests were conducted to compare clinical characteristics according to sex, and chi-square tests were used for categorical variables (ALDH2, CETP, and alcohol consumption). Analysis of variance was conducted to compare alcohol consumption according to CETP and ALDH2 genotypes. Multivariable linear regression models were used to assess differences in variables between CETP and ALDH2 genotypes and the combined effects of CETP and ALDH2 on the association between alcohol consumption and cholesterol, adjusted for age, BMI, and smoking. Statistical tests were two sided, and values of p < 0.05 were considered indicative of statistical significance.
References
O’Keefe, E. L., DiNicolantonio, J. J., O’Keefe, J. H. & Lavie, C. J. Alcohol and CV health: Jekyll and hyde J-curves. Prog. Cardiovasc. Dis. 61, 68–75. https://doi.org/10.1016/j.pcad.2018.02.001 (2018).
El-Lebedy, D. et al. Anti-apolipoprotein A-1 autoantibodies as risk biomarker for cardiovascular diseases in type 2 diabetes mellitus. J. Diabetes Complicat. 30, 580–585. https://doi.org/10.1016/j.jdiacomp.2016.02.014 (2016).
Menotti, A. et al. The role of HDL cholesterol in metabolic syndrome predicting cardiovascular events. The Gubbio population study. Nutr. Metab. Cardiovas. 21, 315–322. https://doi.org/10.1016/j.numecd.2009.11.001 (2011).
Yoo, M. G., Kim, H. J., Jang, H. B., Lee, H. J. & Park, S. I. The association between alcohol consumption and beta-cell function and insulin sensitivity in Korean population. Int. J. Env. Res. Pub. He. 13, 1. https://doi.org/10.3390/ijerph13111133 (2016).
Perissinotto, E. et al. Alcohol consumption and cardiovascular risk factors in older lifelong wine drinkers: The Italian longitudinal study on aging. Nutr. Metab. Cardiovas. 20, 647–655. https://doi.org/10.1016/j.numecd.2009.05.014 (2010).
Brien, S. E., Ronksley, P. E., Turner, B. J., Mukamal, K. J. & Ghali, W. A. Effect of alcohol consumption on biological markers associated with risk of coronary heart disease: Systematic review and meta-analysis of interventional studies. BMJ 342. https://doi.org/10.1136/bmj.d636 (2011).
Barter, P. J. et al. Cholesteryl ester transfer protein: A novel target for raising HDL and inhibiting atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23, 160–167. https://doi.org/10.1161/01.atv.0000054658.91146.64 (2003).
Kuivenhoven, J. A. et al. Heterogeneity at the CETP gene locus. Influence on plasma CETP concentrations and HDL cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 17, 560–568. https://doi.org/10.1161/01.atv.17.3.560 (1997).
Guo, S. X. et al. Associations of cholesteryl ester transfer protein TaqIB polymorphism with the composite ischemic cardiovascular disease risk and HDL-C concentrations: A meta-analysis. Int. J. Environ. Res. Public Health 13. https://doi.org/10.3390/ijerph13090882 (2016).
Nagano, M. et al. Molecular mechanisms of cholesteryl ester transfer protein deficiency in Japanese. J. Atheroscler. Thromb. 11, 110–121. https://doi.org/10.5551/jat.11.110 (2004).
Hannuksela, M., Marcel, Y. L., Kesaniemi, Y. A. & Savolainen, M. J. Reduction in the concentration and activity of plasma cholesteryl ester transfer protein by alcohol. J. Lipid Res. 33, 737–744. https://doi.org/10.1016/S0022-2275(20)41437-3 (1992).
Fumeron, F. et al. Alcohol intake modulates the effect of a polymorphism of the cholesteryl ester transfer protein gene on plasma high density lipoprotein and the risk of myocardial infarction. J. Clin. Invest. 96, 1664–1671. https://doi.org/10.1172/JCI118207 (1995).
Eng, M. Y., Luczak, S. E. & Wall, T. L. ALDH2, ADH1B, and ADH1C genotypes in Asians: A literature review. Alcohol Res. Health 30, 22–27 (2007).
Nakamura, Y. et al. Genetic variation in aldehyde dehydrogenase 2 and the effect of alcohol consumption on cholesterol levels. Atherosclerosis 164, 171–177. https://doi.org/10.1016/s0021-9150(02)00059-x (2002).
Millwood, I. Y. et al. Conventional and genetic evidence on alcohol and vascular disease aetiology: A prospective study of 500 000 men and women in China. Lancet 393, 1831–1842. https://doi.org/10.1016/S0140-6736(18)31772-0 (2019).
Han, S. et al. Acetaldehyde dehydrogenase 2 rs671 polymorphism affects hypertension susceptibility and lipid profiles in a Chinese population. DNA Cell Biol. 38, 962–968. https://doi.org/10.1089/dna.2019.4647 (2019).
Au Yeung SL. et al. Is aldehyde dehydrogenase 2 a credible genetic instrument for alcohol use in Mendelian randomization analysis in Southern Chinese men? Int. J. Epidemiol. 42, 318–328. https://doi.org/10.1093/ije/dys221 (2013).
Thompson, A. et al. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA 299, 2777–2788. https://doi.org/10.1001/jama.299.23.2777 (2008).
Cai, G., Shi, G. & Huang, Z. Gender specific effect of CETP rs708272 polymorphism on lipid and atherogenic index of plasma levels but not on the risk of coronary artery disease: A case-control study. Medicine 97, e13514. https://doi.org/10.1097/MD.0000000000013514 (2018).
Boekholdt, S. M. et al. Cholesteryl ester transfer protein TaqIB variant, high-density lipoprotein cholesterol levels, cardiovascular risk, and efficacy of pravastatin treatment: individual patient meta-analysis of 13,677 subjects. Circulation 111, 278–287. https://doi.org/10.1161/01.CIR.0000153341.46271.40 (2005).
De Oliveira, E. S. E. R. et al. Alcohol consumption raises HDL cholesterol levels by increasing the transport rate of apolipoproteins A-I and A-II. Circulation 102, 2347–2352. https://doi.org/10.1161/01.cir.102.19.2347 (2000).
Gottrand, F. et al. Moderate red wine consumption in healthy volunteers reduced plasma clearance of apolipoprotein AII. Eur. J. Clin. Invest. 29, 387–394. https://doi.org/10.1046/j.1365-2362.1999.00484.x (1999).
Malmendier, C. L. & Delcroix, C. Effect of alcohol intake on high and low density lipoprotein metabolism in healthy volunteers. Clin. Chim. Acta 152, 281–288. https://doi.org/10.1016/0009-8981(85)90103-2 (1985).
Perret, B. et al. Alcohol consumption is associated with enrichment of high-density lipoprotein particles in polyunsaturated lipids and increased cholesterol esterification rate. Alcohol Clin. Exp. Res. 26, 1134–1140. https://doi.org/10.1097/01.ALC.0000026101.76701.12 (2002).
Riemens, S. C. et al. Higher high density lipoprotein cholesterol associated with moderate alcohol consumption is not related to altered plasma lecithin: Cholesterol acyltransferase and lipid transfer protein activity levels. Clin. Chim. Acta 258, 105–115. https://doi.org/10.1016/s0009-8981(96)06451-0 (1997).
Corella, D. et al. Association of TaqIB polymorphism in the cholesteryl ester transfer protein gene with plasma lipid levels in a healthy Spanish population. Atherosclerosis 152, 367–376. https://doi.org/10.1016/s0021-9150(99)00477-3 (2000).
Talmud, P. J. et al. Genetic and environmental determinants of plasma high density lipoprotein cholesterol and apolipoprotein AI concentrations in healthy middle-aged men. Ann. Hum. Genet. 66, 111–124. https://doi.org/10.1017/S0003480002001057 (2002).
Dullaart, R. P. et al. High-density lipoprotein cholesterol is related to the TaqIB cholesteryl ester transfer protein gene polymorphism and smoking, but not to moderate alcohol consumption in insulin-dependent diabetic men. Scand. J. Clin. Lab. Invest. 58, 251–258. https://doi.org/10.1080/00365519850186643 (1998).
Wakabayashi, I. & Masuda, H. Influence of drinking alcohol on atherosclerotic risk in alcohol flushers and non-flushers of Oriental patients with type 2 diabetes mellitus. Alcohol 41, 672–677. https://doi.org/10.1093/alcalc/agl067 (2006).
Imatoh, T. et al. ALDH2 polymorphism rs671, but not ADH1B polymorphism rs1229984, increases risk for hypo-HDL-cholesterolemia in A/A carriers compared to the G/G carriers. Lipids 53, 797–807. https://doi.org/10.1002/lipd.12087 (2018).
Yoo, M. G. et al. Association between the incidence of hypertension and alcohol consumption pattern and the alcohol flushing response: A 12-year follow-up study. Alcohol 89, 43–48. https://doi.org/10.1016/j.alcohol.2020.07.001 (2020).
Heidari-Beni, M., Kelishadi, R., Mansourian, M. & Askari, G. Interaction of cholesterol ester transfer protein polymorphisms, body mass index, and birth weight with the risk of dyslipidemia in children and adolescents: the CASPIAN-III study. Iran J. Basic. Med. Sci. 18, 1079–1085. https://doi.org/10.22038/IJBMS.2015.6045 (2015).
Zhou, Y. J., Yin, R. X., Deng, Y. J., Li, Y. Y. & Wu, J. Z. Interactions between alcohol intake and the polymorphism of rs708272 on serum high-density lipoprotein cholesterol levels in the Guangxi Hei Yi Zhuang population. Alcohol 42, 583–591. https://doi.org/10.1016/j.alcohol.2008.08.004 (2008).
Kweon, S. et al. Data resource profile: The Korea national health and nutrition examination survey (KNHANES). Int. J. Epidemiol. 43, 69–77. https://doi.org/10.1093/ije/dyt228 (2014).
Yun, J. H., Yoo, M. G., Park, J. Y., Lee, H. J. & Park, S. I. Association between KCNJ11 rs5219 variant and alcohol consumption on the effect of insulin secretion in a community-based Korean cohort: A 12-year follow-up study. Sci. Rep. 11, 4729. https://doi.org/10.1038/s41598-021-84179-9 (2021).
Moon, S. et al. The Korea biobank array: Design and identification of coding variants associated with blood biochemical traits. Sci. Rep. 9, 1382. https://doi.org/10.1038/s41598-018-37832-9 (2019).
Hatta, F. H. M. & Aklillu, E. P450 (cytochrome) oxidoreductase gene (POR) common variant (POR*28) significantly alters CYP2C9 activity in Swedish, but not in Korean healthy subjects. OMICS 19, 777–781. https://doi.org/10.1089/omi.2015.0159 (2015).
Acknowledgements
This study was supported by the Korea National Institute of Health (2019-NI-088–01). Data used in the study were obtained from the Korean Genome Analysis Project (4845–301), the Korean Genome and Epidemiology Study (4851–302), and the Korean Biobank Project (4851–307, KBP-2019–022) supported by the Korean Center for Disease Control and Prevention. We would like to thank Editage (www.editage.co.kr) for English language editing.
Author information
Authors and Affiliations
Contributions
All authors had full access to the data. H.-J.L. were responsible for the integrity of the data and the accuracy of the data analyses. H.-J.L. conceived and designed the study. M.-G.Y. performed the statistical analyses, and M.-G.Y., J.H.Y, S.K.K. and H.-J.L interpreted the data. J.H.Y and M.-G.Y. wrote the initial draft of the manuscript, and J.H.Y., and H.-J.L. reviewed/edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoo, MG., Yun, J.H., Koo, S.K. et al. The effect of the association between CETP variant type and alcohol consumption on cholesterol level differs according to the ALDH2 variant type. Sci Rep 12, 15129 (2022). https://doi.org/10.1038/s41598-022-19171-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19171-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.