Abstract
The southern green stink bug (SGSB) Nezara viridula L. is one of the most common stink bug species in the United States and can cause significant yield loss in a variety of crops. A suitable marker for the assessment of gene-editing tools in SGSB has yet to be characterized. The white gene, first documented in Drosophila, has been a useful target to assess the efficiency of introduced mutations in many species as it controls pigmentation processes and mutants display readily identifiable phenotypes. In this study we used the RNAi technique to investigate functions and phenotypes associated with the white ortholog in the SGSB and to validate white as a marker for genetic transformation in this species. This study revealed that white may be a suitable marker for germline transformation in the SGSB as white transcript knockdown was not lethal, did not impair embryo development and provided a distinguishable phenotype. Our results demonstrated that the white ortholog in SGSB is involved in the pathway for ommochrome synthesis and suggested additional functions of this gene such as in the integument composition, management of hemolymph compounds and riboflavin mobilization.
Similar content being viewed by others
Introduction
The white gene was the first X-linked gene characterized in Drosophila that helped to identify the role chromosomes play in heredity1,2,3 and has been used for more than a century as a pleiotropic model to study a number of regulatory mechanisms in eukaryotes4,5,6,7,8,9,10,11,12,13,14.
In Drosophila, the protein subunit encoded by white is needed to form functional ATP-binding cassette (ABC) transporters most commonly known for moving ommochrome and pteridine pigment precursors across membranes8,15,16,17,18,19,20 and for controlling the appearance of pigments in compound eyes, ocelli, testes and Malpighian tubules4,5,15,21. The lack of pigmentation particularly in the compound eyes of Drosophila, has been a readily identifiable phenotype of white mutants that drives a general interest in characterizing orthologous sequences of this gene as a marker for genetic transformation across different insect taxa22,23,24,25,26,27,28,29,30,31,32,33. Stink bugs (Hemiptera:Pentatomidae) represent an increasingly important pest complex in which a marker for genetic transformation has yet to be characterized.
Most stink bugs are polyphagous34,35 and can cause significant yield loss in a variety of crops36,37,38,39,40,41,42,43,44,45,46,47,48. The introduction of transgenic crops targeting primary pests has been increasing the incidence of several stink bug species across the southern United States where the southern green stink bug (SGSB) Nezara viridula L. is considered one of the most common and primary stink bug pest35,49,50,51. Documentation of gene-editing systems in SGSB would open multiple avenues for continued research related to stink bug management52 and the white ortholog in this species may be a useful target to assess the efficiency of introduced mutations. Nevertheless, the function and phenotypes associated with the white ortholog in SGSB have never been documented and some previous investigations targeting white orthologs of hemipteran species suggest a diverse and contrasting capacity of using this gene as a marker for genetic transformation.
RNA interference (RNAi) experiments performed with the brown planthopper, Nilaparvata lugens (Stål), revealed that knockdown of white ortholog transcripts caused a significant reduction of ommochrome and pteridine pigments in their compound eyes and ocelli30,53 and was a reliable marker to confirm germline transformation28. By comparison, knockdown of white transcripts in the tarnished plant bug Lygus hesperus Knight, caused a whole body color depletion and some unexpected negative effects, such as lethargy, reduced body size, and softer and stickier cuticles indicating that white would be a less suitable marker for genetic transformation in this species31. A gene-editing approach targeting the white ortholog in the silverleaf whitefly Bemisia tabaci (Gennadius) showed that this marker allowed easy screening of white mutant nymphs and adults as they developed distinct eye colors relative to wild type, and allowed an investigation of the heritability of mutations generated33. More recently, RNAi and gene editing investigations performed with the milkweed bug, Oncopeltus fasciatus (Dallas), revealed that homozygous white mutations caused translucent patches in the integument, loss of pigments throughout the body, but were lethal during embryo development32. Embryonic lethality caused by the disruption of the white orthologues is not common among insect species but has been suggested in Lepidoptera27 and Hemiptera54, which is a major limitation of its use to document germline transformation.
Validation of gene editing markers is a necessary initial step to rule out lethality and unwanted deleterious phenotypes. RNAi is a gene silencing mechanism that is commonly used to investigate gene function and has been successfully triggered in stink bug species through microinjections of gene-specific double-stranded RNA (dsRNA)55,56,57 including in the SGSB58,59. Therefore, in our study we performed dsRNA microinjections targeting the white ortholog in SGSB aiming (1) to provide an initial evaluation of the phenotypes associated with the knockdown of this gene, and (2) to validate white as a marker for genetic transformation in the SGSB.
Methods
Insects
A reference SGSB population was established in the Insect Resistance lab, Entomology and Nematology Department, University of Florida, Gainesville, FL from a lab strain initiated from field collections that had been under laboratory rearing for many generations60. Insects were maintained on snap peas, sweet corn, peanuts, and provided with water through moisturized cotton in environmental chambers at 25 ± 1 °C, 65% RH and 14 light: 10 dark h photoperiod. Food and water were replaced twice a week.
RT-qPCR primer efficiency test
SGSB transcriptome sequences of three target genes (white, vermilion, and cinnabar) known to be involved in eye pigmentation of D. melanogaster15 and of four candidate SGSB reference genes (β-Actin, α-Tubulin, EF1-α, and GAPDH ) were generously provided by Syngenta Ltd., Jealott’s Hill International Research Centre, Bracknell, Berkshire, UK. Identities of sequences were first verified by similarity search in BLASTx against the brown marmorated stink bug Halyomorpha halys (Stål) database (NCBI taxid:286706) (Supplementary Table S1) and later confirmed by the sequencing of SGSB-derived PCR products. Additionally, a phylogenetic tree was constructed in Geneious prime 2022.1.1. for white, scarlet and brown amino acid sequences that have been previously annotated in genomic studies and/or were found in BLAST results for the SGSB sequence targeted in this study, which confirmed the white identity (Supplementary Figure S1). Primers for target and reference genes (Supplementary Table S2) were designed using PrimerQuest® Tool61 and obtained from Integrated DNA Technologies, IDT (Coralville, IA). Total RNA was extracted from three SGSB pooled samples (12 eggs, two second instar nymphs and legs of two adults) using a Qiagen RNeasy mini kit (Germantown, MD, Cat. No. 74104) following manufacturer instructions. RNA quality and quantity were verified by spectrophotometry using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific™, Waltham, MA). cDNAs were synthesized from 500 ng of total RNA using the Cloned AMV First Strand Synthesis kit (Invitrogen™ Life Technologies™, Carlsbad, CA, Cat. No. 12328-032). Samples were diluted tenfold and evenly mixed to obtain a single representative SGSB cDNA sample. The efficiencies of primers designed for target and reference genes of the SGSB were estimated from a fourfold serial dilution series of the pooled cDNA sample. Quantitative Real Time PCR (RT-qPCR) reactions were performed in a 20 μL final reaction volume containing 10 μL of SsoAdvanced™ universal SYBR® Green supermix (Bio-Rad, Hercules, CA, Cat No.172-5271), 1 μL cDNA, 0.3 μM of each primer and complemented with nuclease-free water. Reactions were carried out in triplicates in a CFX96™ Real-Time System (Bio-Rad Laboratories, Singapore) using hard-shell 96 well plates (Bio-Rad Laboratories, Hercules, CA, Cat. No. HSP9601). Temperature profiles included an initial heating step at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing at 60 °C for 30 s, one additional denaturation step at 95 °C for 5 s, and a final melting cycle from 65 to 95 °C (0.5 °C increments) for 5 s. A standard curve was generated for each primer pair and the regression correlation coefficient (R2) and PCR efficiency (E) for each standard curve were calculated62.
Double stranded RNA (dsRNA) preparation
SGSB genomic DNA was extracted from one SGSB adult leg using a Qiagen DNeasy kit (Germantown, MD, Cat No. 69504) according to the manufacturer’s instructions. Polymerase chain reactions (PCR) were performed in a 50 μL final reaction volume containing 10 ng of template DNA, 1 × PCR Buffer (Tris pH 9.2, 16 mM ammonium sulfate, 1.75 mM MgCl2), 0.35 mM dNTP, 1 unit of Taq Polymerase (Thermo Fisher Scientific™, Vilnius, LT, Cat. No. EP0402), 0.2 μM of the forward (5′-AATGATAATGTCTTACCTCTAGACC-3′) and reverse (5′- TTTAGAATGTGCTTCCTACCG-3′) primers to amplify a 191 bp SGSB white exonal fragment. The amplified PCR product was analyzed by 1% agarose gel electrophoresis, purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific™, Vilnius, LT, Cat. No. K0691) and confirmed by sequencing (Genewiz, South Plainfield, NJ). A T7 promoter was linked to the purified white DNA fragment by PCR using the gene-specific primers with the T7 sequence (5′-TAATACGACTCACTATAGGGAG-3′) appended to the 5′ end of each primer. Similarly, a non-specific green fluorescence protein (GFP) gene fragment of 503 bp was amplified from pGLO™ plasmid (Bio-Rad, Hercules, CA, Cat. No. 1660405EDU) by PCR using the forward (5′-T7- AGGTGATGCTACATACGGAAAG-3′) and reverse (5′-T7-ACAGGTAATGGTTGTCTGGTAAA-3′) primers. The PCR reactions were carried out using a C1000 Touch™ Thermal Cycler (Bio-Rad Laboratories, Singapore) and the temperature profiles included an initial heating step at 94 °C for 2 min, followed by 39 cycles of denaturation at 94 °C for 15 s, annealing at 60 °C for 30 s, and extension at 68 °C for 30 s. PCR products generated were then used for dsRNA synthesis using MEGAscript™ RNAi Kit (Invitrogen™ by Thermo Fischer Scientific™, Vilnius, LT, Cat. No. AM1626) according to manufacturer instructions (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_072987.pdf). The quality and quantity of purified dsRNAs were verified by NanoDrop.
RNAi experiments
Selection of reference genes
A preliminary test was performed to verify the stability of candidate reference genes during RNAi-based microinjection experiments in SGSB. Two doses of white and GFP dsRNAs (150 and 750 ng) were injected in the abdomen of newly emerged adult SGSB females in a final volume of 3 µL using glass microneedles attached to a pneumatic microinjector (Narishige model IM-11-2, Tokyo, JP). Solutions were prepared in the MEGAscript™ elution buffer. Females injected with elution buffer-only were included as negative controls. A 1% v/v green food dye (McCormick & Co, Hunt Valley, MD) was added to all solutions to facilitate visualization of injections. Insects were immobilized on ice for about 5 min before injections. Injected females were maintained separated by treatment under SGSB rearing conditions in plastic cups (9.5 D × 7.5 H cm) covered with tulle fabric. Total RNA was extracted from one middle leg of each treated female 4 days after treatment (4 DAT) using the Qiagen RNeasy Mini Kit and quantified using a NanoDrop spectrophotometer. cDNAs were synthesized using the High-Capacity cDNA reverse transcription kit (Applied Biosystems by Thermo Fisher Scientific™, Vilnius, LT, Cat. No. 4368813) from 70 ng of RNA, following the manufacturer’s instructions. cDNA of six individual females from each treatment were used as biological replicates to verify expression levels of white and reference genes by RT-qPCR. The four candidate reference genes (β-Actin, α-Tubulin, EF1-α, and GAPDH) were tested during RT-qPCR analysis. RT-qPCR reactions were carried out as previously described (subsection “RT-qPCR primer efficiency test”). The stability of reference genes was examined by BestKeeper63, NormFinder64, GeNorm65, and the comparative ΔCt method66 on a web-based platform RefFinder (https://www.heartcure.com.au/reffinder/?type=reference). The white relative gene expression was calculated following Pfaffl67 using a geometric normalization factor estimated for multiple reference genes65. The statistical analysis was performed using a generalized mixed model in SAS 9.4 software68 with a completely randomized experimental design. Multiple comparisons of treatment means were performed using Fisher’s least significant difference procedure at significance level α = 0.05.
Knockdown of white transcript
RNAi experiments were conducted with SGSB to verify white knockdown efficacy overtime and corresponding phenotypic responses. Abdominal microinjections of white and GFP dsRNA (150 ng) were performed in each sex of newly emerged adults (< 24 h-old) and fifth instar nymphs. Nymphs were injected either less than 3 days prior to adult emergence (< 3 DPE) or more than 4 days prior to emergence (4 < DPE). Insects treated with elution buffer-only were used as negative controls. Newly emerged adults received 3 μL solution (50 ng dsRNA/μL) while nymphs received 2 μL (75 ng dsRNA/μL). Treated insects were maintained in plastic cups using standard rearing conditions. Food and water were replaced twice a week. Phenotypic changes were monitored and photographed using either a Canon EOS 7D digital camera and MP-E 65 mm lens or a Canon EF 100 mm f/2.8 Macro USM Lens at the Insect ID Lab at the University of Florida. Total RNA was extracted from one leg of each treated adult at 4, 10 and 30 days after treatment (DAT). For treated nymphs, total RNA was extracted after molting into adults at 2, 10 and 30 days after emergence (DAE). RNA extractions, cDNA syntheses, RT-qPCR reactions and white relative gene expression analyses were performed as previously described in the methods section. cDNA of four individual insects from each sex, treatment and time-point combination were used as biological replicates. Two reference genes (β-Actin and α-Tubulin) were used for the data normalization. The statistical analyses of white relative expression were performed using a generalized mixed model in SAS 9.4 software with a completely randomized experimental design. Sex, treatment, and time-points were adopted as fixed factors in the statistical model (2 × 3 × 3 and 2 × 4 × 3 factorial treatment designs for adults injected at < 24 h-old or during the nymphal stage, respectively). Multiple comparisons of treatment means were performed using Fisher’s least significant difference procedure at significance level α = 0.05.
Adult mortality and parental RNAi effect
Newly emerged adults of each sex of SGSB were treated following the methods described above (subsection “Knockdown of white transcript”). Nine to ten replicates of each treatment (150 ng white or GFP dsRNA, and elution buffer control) were performed. Treatment replicates were evenly represented by groups of six to ten insects. Treated insects were kept individually in small polystyrene boxes (5.9 L × 5.9 W × 7.8 H cm, Althor Products, Windsor Locks, CT, Cat. No. SM-1207) provided with food and water and covered with tulle fabric. Approximately 10 days after treatment, three untreated insects of the opposite sex were transferred into each box to allow mating. Adult mortality, egg laying, and phenotypic changes in progeny were monitored daily. To estimate the lethality of white knockdown in adult SGSB, the proportion mortality obtained from each treatment replicate at two different time-points (10 and 23DAT) were analyzed with a Beta-binomial distribution69,70 using a logit link function with a generalized mixed model in SAS 9.4 software. Multiple comparisons of treatment means were performed using Fisher’s least significant difference procedure at significance level α = 0.05. A completely randomized experimental design was adopted with a 2 × 3 factorial treatment design. Sex and treatments were adopted as fixed factors within each time point in the statistical model.
Collected eggs were transferred to Petri dishes (35 D × 10 H mm, Thermo Fisher Scientific, Roskilde, DK, Cat. No. 153066) and maintained in a growth chamber at 25 ± 1 °C, 65% RH and 14 light: 10 dark h photoperiod. Phenotypic changes of eggs and enclosed nymphs were monitored and recorded using a Canon EOS 7D digital camera and MP-E 65 mm lens at the Insect ID Lab at the University of Florida. Three eggs were detached from each egg cluster at the time of collection and stored at − 80 °C for subsequent RNA extraction to quantify white relative expression. Similarly, < 12 h-old first instar nymphs were either used for RNA extraction or maintained under rearing conditions. Total RNA was extracted from pooled samples of three < 24 h-laid eggs using the RNAqueous™-Micro Kit (Invitrogen™ by Thermo Fischer Scientific™, Vilnius, LT, Cat. No. AM1931) and from pooled samples of two first instar nymphs using the Qiagen RNeasy Mini Kit following manufacturer instructions. cDNA syntheses, RT-qPCR reactions and white relative gene expression analyses were performed as described in subsection 2.4.1. Three cDNA egg samples and four nymph cDNA samples originating from adult treatments were used as biological replicates of RT-qPCR reactions. Two reference genes (α-Tubulin and EF1-α) were used for the geometric normalization of white relative expression.
Expression of ommochrome pathway genes
cDNA of SGSB adults at 30 days after emergence (30 DAE) that had been treated with white dsRNA either as newly emerged adults or prior to adult emergence (subsection “Knockdown of white transcript”) and pooled cDNA samples from first instar nymphs (< 12 h-old) from white dsRNA-treated mothers (subsection “Adult mortality and parental RNAi effect”) were used to measure the relative expression of two genes (vermilion and cinnabar) known to be involved in the metabolic pathway for the conversion of tryptophan to ommochromes in Drosophila15. These genes were assessed for potential differential regulation as an additional mechanism to prevent ommochrome precursor accumulation in the SGSB. Four adults from each sex and treatment combination and three pooled cDNA samples from nymphs were used as biological replicates. cDNA from buffer- and GFP dsRNA-injected treatments were used as wild-type controls. RT-qPCR reactions and relative expression analyses were performed as described in subsection “Selection of reference genes”. using two reference genes (β-Actin and α-Tubulin) for data normalization. The statistical analyses of gene relative expression were performed using a generalized mixed model in SAS 9.4 software with a completely randomized experimental design. Sex and treatments were adopted as fixed factors in the statistical model of the adult test (2 × 4 factorial treatment design), while treatment was the only fixed factor in the model testing first instar nymphs (unsexed). Multiple comparisons of treatment means were performed using Fisher’s least significant difference procedure at significance level α = 0.05.
Metabolomic analysis of ommochrome biosynthesis
Ommochrome precursors (tryptophan, kynurenine and 3-hydroxy-DL-kynurenine) and riboflavin were evaluated in SGSB samples collected from RNAi experiments. The metabolites were measured in adults at 30 days after emergence (30 DAE) and in first instar nymphs at < 12 h after hatching. Adults that had been treated with white dsRNA either as newly emerged adults or prior to adult emergence (subsection “Knockdown of white transcript”) were tested as separated groups. First instar nymphs tested were the progeny of SGSB females treated with white dsRNA at < 24 h after emergence. Wild-type insects were used as controls. Pooled samples of six adults (1:1 sex ratio) or 60 nymphs (unsexed) were used as biological replicates. Six biological replicates of 25 mg ground tissue were prepared for each treatment. Adult samples were prepared using a porcelain mortar and pestle while nymph samples were prepared with polypropylene disposable pestles in 1.5 mL centrifuge tubes. All samples were processed in liquid nitrogen and stored at -80 °C until analysis. Extractions were performed as previously described71. Ultra-high-performance liquid chromatography-high-resolution mass spectrometry (UHPLC-HRMS) was carried out on a Q Exactive mass spectrometer coupled to a Vanquish LC System (Thermo Fisher Scientific, Waltham, MA, USA) by reverse phase gradient elution using an ACE Excel 2 C18-PFP column (2.1 mm X 100 mm, 2 μm; part # EXL-1010-1002U) in full scan positive (injection volume 2 μL) and negative (injection volume 4 μL) ion modes. Identification was assigned to features by m/z (≤ 5 ppm) and retention time (< 0.2 min), using a method-specific metabolite library produced from pure standards previously analyzed using the above-mentioned chromatographic gradient. Analytical standards of L-Tryptophan (CAS: 73-22-3), L-kynurenine (CAS: 2922-83-0), 3-hydroxy-DL-kynurenine (CAS: 484-78-6) and riboflavin (CAS: 83-88-5) were purchased from Sigma-Aldrich Corp., St. Louis, MO (Cat No. T0254, K8625, H1771, PHR1054, respectively). The data were acquired, processed, normalized, filtered and metabolites identified using MZmine 2 software72 as previously described71.
Results
RT-qPCR primer efficiency and selection of reference genes
The PCR efficiency of all primers designed for target and reference genes of the SGSB is provided in Supplementary Table S2, and all exhibited appropriate efficiency. The estimated E values were all within a 92–100% range. Also, a consistently high linear correlation (> 99%) between the log of cDNA dilution factors and mean RT-qPCR Ct values was found for all primers tested. From the reference genes tested, α-Tubulin, β-Actin, and EF1-α transcripts were the most stably expressed across different dsRNA treatments (Supplementary Table S3). Consequently, these three transcripts were chosen as suitable reference options to be used for geometric mean normalization of white relative gene expression in subsequent RT-qPCR analyses.
Significant knockdown of the white transcript (\({F}_{\left(\mathrm{4,25}\right)}=35.37;p<0.0001)\) was observed in SGSB adult females injected with 150 and 750 ng white dsRNA relative to GFP dsRNA- and elution buffer-treated females (Supplementary Figure S2). There was no significant difference in the levels of transcript knockdown between the two white dsRNA doses tested (\({t}_{25}=0.02;p=0.9826)\) with both providing > 92% knockdown at 4 DAT. Therefore, further RNAi-based experiments were all conducted using 150 ng dsRNA.
RNAi experiments
Knockdown of white transcript
For knockdown experiments with newly emerged SGSB adults, there were no significant three-way interaction between sex, treatments and time points (\({F}_{\left(\mathrm{4,54}\right)}=0.09;p=0.9843)\) or two-way interactions between sex and time (\({F}_{\left(\mathrm{2,58}\right)}=0.65;p=0.5277\)), and sex and treatment (\({F}_{\left(\mathrm{2,58}\right)}=0.91;p=0.4077\)) on the efficiency of white transcript knockdown. As there was no significant main effect of sex (\({F}_{\left(\mathrm{1,62}\right)}=3.9;p=0.0529\)), this factor was removed from the statistical model used. Similarly, time was removed from the model because there was no significant interaction effect between time and treatments (\({F}_{(\mathrm{4,63})}=0.4;p=0.8067)\) or a main effect of time \(({F}_{(\mathrm{2,67})}=1.11;p=0.3353)\) on the level of white transcript knockdown. Overall, a significant level of knockdown (~ 87%) \(({F}_{\left(\mathrm{2,69}\right)}=63.64;p<0.0001)\) was observed at four, ten and 30 DAT in both sexes of SGSB adults treated with white dsRNA relative to GFP dsRNA- and buffer-treated controls (Fig. 1A).
Transcript knockdown efficacy overtime of RNAi-based microinjection targeting the white gene of the SGSB. (A) SGSB adults had been treated at < 24 h after emergence. Significant white knockdown \(({F}_{\left(\mathrm{2,69}\right)}=63.64;p<0.0001)\) was observed in adults treated with white dsRNA relative to GFP dsRNA- and buffer-treated controls at 4, 10 and 30 days after treatment (DAT). (B) SGSB adults had been treated during fifth nymphal stage at either less than three or more than 4 days prior to adult emergence (DPE). Significant white knockdown \(({F}_{\left(\mathrm{3,92}\right)}=17.61;p<0.0001)\) was observed at 2, 10 and 30 days after adult emergence (DAE) in SGSB adults treated with white dsRNA either at < 3 DPE or > 4 DPE relative to GFP dsRNA- and buffer-treated controls. In each graph, bars discriminate white normalized relative expression means ± SE per time point and above bars show overall means ± SE per treatment across time. Treatment means followed by the same letter were not statistically different (Fisher’s LSD test, α = 0.05).
For SGSB adults that had been treated during the fifth nymphal stage, there was no significant three-way interaction effect between sex, treatments and time (\({F}_{\left(\mathrm{6,72}\right)}=0.24;p=0.9612)\) or two-way interaction effects between sex and time (\({F}_{\left(\mathrm{2,78}\right)}=1.12;p=0.3315\)), and sex and treatment (\({F}_{\left(\mathrm{3,78}\right)}=0.74;p=0.5341\)) on the efficacy of white transcript knockdown. The main effect of sex was also not significant (\({F}_{\left(\mathrm{1,83}\right)}=0.01;p=0.9394\)), so this factor was removed from the statistical model used. As there was no significant interaction effect between time and treatments (\({F}_{(\mathrm{6,84})}=0.08;p=0.9977)\) and no significant main effect of time (\({F}_{(\mathrm{2,90})}=2.34;p=0.1021)\) on the level of white transcript knockdown, this factor was also removed from the statistical model. Overall, a significant level of white transcript knockdown (~ 76%) \(({F}_{\left(\mathrm{3,92}\right)}=17.61;p<0.0001)\) was observed at two, ten and 30 DAE in both sexes of SGSB adults treated during the fifth nymphal stage with white dsRNA either at < 3 DPE or > 4 DPE. This contrasts to no change in transcription for either the GFP dsRNA- or buffer-treated controls in all concurrent time points tested (Fig. 1B).
Adult phenotype
An observable change from wild-type green color to blueish green, as well as a more translucent integument was observed in 100% of both female (n = 150) and male (n = 150) SGSB adults treated with white dsRNA at < 24 h after emergence, which was not apparent in control treatments (Supplementary Figure S3). Changes in color patterns were apparent 10 DAT and were sustained throughout adult life. Fifth instar nymphs treated with white dsRNA (n = 300) molted into SGSB adults with distinct body colors as well that were sustained throughout adult life (Supplementary Figure S4).
Male and female nymphs injected with white dsRNA at < 3 DPE (n = 150) developed a translucent integument as adults and a blue-green body color, which was paler than the blueish green phenotype observed in SGSB treated at the early adult stage. Nymphs that were treated with white dsRNA > 4 DPE (n = 150) developed a yellow color along with a more translucent integument as adults. The phenotype observed in white dsRNA-treated nymphs was distinct from the wild-type control treatments from the day of adult emergence until the end of adult life. Although a body color variegation was the most obvious SGSB adult phenotype generated by the white transcript knockdown, minor color differences in their compound eyes and ocelli were also observed (Fig. 2A). The predominant brown pigmentation of SGSB compound eyes and ocelli was reduced in all white dsRNA treated insects, exposing underlying red pigments. The red pigmentation was more obvious in SGSB adults treated at the nymphal stage > 4 DPE relative to the other treatments.
Dissections of mature SGSB adults (~ 30 days old) revealed that white transcript knockdown generated clear phenotypic differences of internal organs in this species (Fig. 2). Malpighian tubules that are normally transparent-white in wild-type SGSB adults were orange-yellow in all white dsRNA treated insects (Fig. 2C). Conversely, the orange-yellow color of gonads and metathoracic scent reservoir appeared discolored in SGSB adults that had been treated with white dsRNA (Fig. 2D–F).
Adult mortality
The mean proportion mortality observed in white dsRNA-treated adults was not significantly different from the mortality observed in GFP dsRNA- and elution buffer-treated control groups either by 10 DAT \(({F}_{\left(\mathrm{2,53}\right)}=0.17;p=0.8421)\) or by 23 DAT \(({F}_{\left(\mathrm{2,54}\right)}=1.47;p=0.2394)\) (Supplementary Figure S5A-B, respectively). Furthermore, there was no significant interaction effect between sex and treatments on the mortality of SGSB following RNAi-based microinjections at either 10 DAT \(({F}_{\left(\mathrm{2,51}\right)}=0.57;p=0.5667)\) or 23 DAT \(({F}_{\left(\mathrm{2,51}\right)}=1.89;p=0.161)\). However, there was a significant main effect of sex on SGSB adult mortality at 10 DAT, when males showed higher mortality across all treatments \(({F}_{\left(\mathrm{1,53}\right)}=16.44;p=0.0002)\). No significant main effect of sex on SGSB adult mortality was observed at 23 DAT \(({F}_{\left(\mathrm{1,53}\right)}=3.61;p=0.0629)\).
Parental RNAi
Newly laid eggs (< 24 h) from adult females treated with white dsRNA that were mated with non-treated males showed a significant knockdown of white transcripts (> 99%) \(({F}_{\left(\mathrm{2,6}\right)}=9.1;p=0.0152)\) relative to eggs laid by GFP dsRNA- and buffer-treated female mating pairs (Supplementary Figure S6-B). However, there was no significant difference \(({F}_{\left(\mathrm{2,6}\right)}=0.05;p=0.9558)\) of white relative expression in eggs laid by non-treated females that were mated with white dsRNA-treated males relative to control groups (Supplementary Figure S6-A). Phenotypic differences were detected in eggs from white dsRNA-treated females before hatching when embryos were more developed. Near hatching, 100% of eggs (> 500 eggs) collected from adult females treated with elution buffer or GFP dsRNA showed a typical wild-type phenotype, where a transparent chorion revealed an enclosed orange embryo with red compound eyes and a red crescent moon-shape forehead mark (Fig. 3). In contrast, 100% of eggs (> 500 eggs) collected from white dsRNA-treated females had very little color prior to hatching with only the red eyes and a paler red forehead mark of the embryo visually detectable through the egg chorion (Fig. 3). The phenotype of eggs produced by untreated females that were mated with white dsRNA-treated males were similar to wild-type eggs, suggesting that the response was maternally inherited.
All first instar nymphs (> 500 nymphs) generated from white dsRNA treated SGSB adult females also showed a distinct phenotype compared to the controls (Fig. 3). Upon hatching, nymphs from control groups displayed a wild-type bright orange integument and red compound eyes that darkened to brown by 48 h after hatching. Conversely, all first instar nymphs from white dsRNA treated mothers showed considerably reduced integumental color and predominance of red eye pigments-only up to 48 h after hatching (Fig. 3). Although there were clear phenotypic differences between first instar nymphs of white dsRNA-treated SGSB adult females and those of GFP dsRNA- and buffer-treated female controls, the white relative expression levels at < 12 h after hatching were not significantly different \(({F}_{\left(\mathrm{2,9}\right)}=0.54;p=0.5999)\) amongst the different treatment groups (Supplementary Figure S6-C).
Expression of ommochrome pathway genes
Significant downregulation of vermilion \(({F}_{\left(\mathrm{3,28}\right)}=10.08;p<0.0001)\) was observed in SGSB adults that had been treated with white dsRNA relative to the wild type control, while the relative expression of cinnabar was not significantly different \(({F}_{\left(\mathrm{3,28}\right)}=1.67;p=0.1959)\) among the same treatment groups (Supplementary Figure S7-A). There was no significant two-way interaction effect between sex and treatments on the relative expression of either vermilion (\({F}_{\left(\mathrm{3,24}\right)}=0.24;p=0.8698\)), or cinnabar (\({F}_{\left(\mathrm{3,24}\right)}=1.03;p=0.3987\)). The main effect of sex was also not significant for vermilion (\({F}_{\left(\mathrm{1,27}\right)}=0.08;p=0.7750\)), or cinnabar (\({F}_{\left(\mathrm{1,27}\right)}=0.49;p=0.4884\)) in SGSB adults, so this factor had been removed from the statistical model. First instar nymphs from white dsRNA-treated mothers did not show significant differences in the expression of either vermilion (\({t}_{\left(4\right)}=0.3;p=0.7813\)) or cinnabar (\({t}_{\left(4\right)}=0.23;p=0.8283\)) relative to wild-type controls (Supplementary Figure S7-B).
Metabolomic analysis of ommochrome biosynthesis
Relative to wild type controls, SGSB adults that had been treated with white dsRNA generated significantly lower peak intensities of 3-hydroxy-DL-kynurenine \(({F}_{\left(\mathrm{3,20}\right)}=51.25;p<0.0001)\) and significantly higher peak intensities of riboflavin \(({F}_{\left(\mathrm{3,20}\right)}=23.08;p<0.0001)\), while tryptophan \(({F}_{\left(\mathrm{3,20}\right)}=1.23;p=0.3244)\) and kynurenine \(({F}_{\left(\mathrm{3,20}\right)}=0.52;p=0.6754)\) measurements were not significantly different (Fig. 4-A). Furthermore, riboflavin intensity was significantly higher in adults treated with white dsRNA during the fifth nymphal stage at < 3 DPE \(({t}_{\left(20\right)}=3.43;p=0.0026)\) and > 4DPE \(({t}_{\left(20\right)}=5.07;p<0.0001)\) than in adults treated at < 24 h after emergence (Fig. 4-A). First instar nymphs from SGSB females that had been treated with white dsRNA at < 24 h after emergence produced significantly higher peak intensities of tryptophan \(({t}_{\left(10\right)}=3.21;p=0.0093)\), kynurenine \(({t}_{\left(10\right)}=3.51;p=0.0056)\) and riboflavin \(\left({t}_{\left(10\right)}=6.46;p<0.0001\right)\) and significantly lower peak intensity of 3-hydroxy-DL-kynurenine \(({t}_{\left(10\right)}=9.25;p<0.0001)\) relative to the wild-type control (Fig. 4-B).
Metabolomic analysis of riboflavin and ommochrome biosynthesis in SGSB comparing white and wild type phenotypes observed during RNAi-based experiments. (A) adults at 30 days after emergence (DAE) that were treated with white dsRNA as < 24 h newly emerged adults or as nymphs either at < 3 days prior emergence (DPE) or > 4 DPE. Metabolites measured in adults were: tryptophan \(({F}_{\left(\mathrm{3,20}\right)}=1.23;p=0.3244)\) ; kynurenine \(({F}_{\left(\mathrm{3,20}\right)}=0.52;p=0.6754)\); 3-hydroxy-DL-kynurenine \(({F}_{\left(\mathrm{3,20}\right)}=51.25;p<0.0001)\); and riboflavin \(({F}_{\left(\mathrm{3,20}\right)}=23.08;p<0.0001)\). (B) first instar nymphs (< 12 h-old) originated from mothers treated with white dsRNA at < 24 h adult stage. Metabolites measured in first instar nymphs were: tryptophan \(({t}_{\left(10\right)}=3.21;p=0.0093)\); kynurenine \(({t}_{\left(10\right)}=3.51;p=0.0056)\); 3-hydroxy-DL-kynurenine \(({t}_{\left(10\right)}=9.25;p<0.0001)\); and riboflavin \(({t}_{\left(10\right)}=6.46;p<0.0001)\). Treatment means followed by the same letter were not statistically different (Fisher’s LSD test, α = 0.05).
Discussion
Microinjections of white dsRNAs into fifth instar nymphs and newly emerged SGSB adults effectively reduced white transcriptional levels throughout the SGSB adult life without causing lethality. Furthermore, a parental white knockdown effect was confirmed in SGSB by our RNAi experiments. Parental or transgenerational RNAi is the process of interfering with gene transcription in the progeny by treating parents with gene-specific dsRNAs. We found more than 99% knockdown of white transcripts in fertilized eggs of SGSB females treated with white dsRNAs, which significantly impacted the associated metabolism and phenotype of eclosed nymphs but did not prevent embryo development. Previous studies have demonstrated parental RNAi in Pentatomidae bugs such as the brown stink bug Euschistus heros (F.)55 and the brown marmorated stink bug H. halys56. Parental RNAi has also been investigated in a number of other heteropteran species such as kissing bugs73,74, water striders75,76 and milkweed bugs32,77 by treating females with dsRNAs. It should be noted that most parental RNAi cases documented for insects refers to a maternal effect where mothers were treated with dsRNAs. Little is known about paternal RNAi where fathers mediate the transgenerational response, and in some cases the words parental and paternal are incorrectly exchanged. Our results suggest that parental RNAi targeting the white transcription of SGSB was a maternal effect and may be achieved by delivering white dsRNAs into females only.
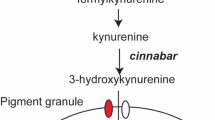
Gene expression knockdown allows the investigation of the targeted gene function and in our study, it revealed the physiological basis of the SGSB white in pigmentation of adults, embryos and first instar nymphs. At the aforementioned developmental stages, the wild-type colors of SGSB compound eyes, ocelli and integument were clearly disrupted after white transcript knockdown, and a subsequent metabolomics investigation confirmed an impact on the pathway for the conversion of tryptophan to ommochromes in this species. The white gene first discovered in Drosophila encodes a subunit of an ABC transporter recognized for loading pigment granules with pigment precursors such as 3-hydroxykynurenine1,6,78,79. The injection of white dsRNA into SGSB individuals significantly reduced the levels of 3-hydroxykynurenine supporting results reported for D. melanogaster Meigen79,80 and Ephestia kühniella (Zeller)81 where white null mutants exhibit reduced levels of this metabolite. Moreover, the progeny of SGSB females treated with white dsRNAs also showed reduced levels of 3-hydroxykynurenine confirming a persistent parental RNAi effect for the white gene.
The fact that 3-hydroxykynurenine is not properly transported across membranes of pigment granules in the absence of the white protein subunit20,79 could hypothetically cause a lethal accumulation of this compound in the insect hemolymph and/or cytosol82,83,84. It has been reported that insects carrying white mutations exhibit significantly reduced accumulation of 3-hydroxykynurenine in the body by an enhanced rate of excretion22,79,83. The current study examined genes involved in the upstream pathway for ommochrome synthesis during the conversion of tryptophan into N-formyl kynurenine (vermilion) and/or during the conversion of kynurenine into 3-hydroxykynurenine (cinnabar) and assessed potential differential regulation as an additional mechanism to prevent ommochrome precursor accumulation in the SGSB. Treatment with white dsRNAs resulted in significant downregulation of vermilion in SGSB adults while cinnabar expression was similar to untreated controls. However, in first instar progeny of females treated with white dsRNA where white transcript knockdown was achieved during embryo development but not after egg hatching, neither vermilion nor cinnabar were differentially expressed relative to controls. The levels of tryptophan and kynurenine in white dsRNA-treated adults did not differ from untreated controls while the levels of both metabolites were higher in first instar nymphs of white dsRNA treated females than in wild type nymphs. Previously, Drosophila mutants showed that the activity of vermilion encoded enzyme (tryptophan oxygenase) could be controlled by the concentration of kynurenine and that this enzyme is rate limiting in the catabolism of tryptophan16. Although the precise regulatory mechanisms remain uncertain, our results suggest that differential regulation of vermillion may be a SGSB response to white transcript knockdown that prevented tryptophan and kynurenine accumulation in adults. Furthermore, SGSB nymphs carrying the parental white RNAi effect could still maintain significantly low levels of 3-hydroxykynurenine regardless of the accumulation of its precursors tryptophan and kynurenine in their system, which suggests that regulation of other genes and/or an enhanced rate of excretion may prevent the 3-hydroxykynurenine accumulation in SGSB.
In Drosophila, the protein subunit encoded by white heterodimerizes with another subunit encoded by either scarlet or brown to form a functional ABC transporter for either ommochrome or pteridine pigment precursors, respectively15,18,19,20. Ommochromes are derived from tryptophan and are mostly brown, while pteridine pigments are derived from guanosine triphosphate and are characterized by yellow to red color variations85,86,87. Although we did not investigate pteridine related precursors and pigments in the SGSB, we found that white transcript knockdown in this species not only significantly reduced brown pigments in the eyes of adults and nymphs, but whole-body depletion of red-related pigments was apparent. Some wild populations of SGSB have been reported to carry genetic mutations conferring body color polymorphisms that range from the most frequent green morph to yellow and orange88,89,90. The color of SGSB adults is also known to change from green to russet during winter in high latitudes91,92. Furthermore, the pteridine erythropterin was detected in the integument of a closely related pentatomid stink bug species93. Thus, it is possible that pteridine pigments are present in the SGSB and that white may be involved in both ommochrome and pteridine pigment pathways in this species. Parental RNAi experiments performed on water striders where white transcription knockdown depleted all colors during embryo development confirmed that the white ortholog in these heteropterans is involved in both ommochrome and pteridine pathways75. It was further demonstrated in the same study that eye pigments of water striders are a combination of ommochromes and pteridines while body pigmentation was only associated with pteridines. Conversely, investigations on the blood-sucking heteropterans Triatoma infestans Klug and Rhodnius prolixus Stål suggest that only ommochrome pigments are present in the eyes of both wild-type and red-eyed mutants94,95. An investigation of pteridine precursors and derived pigments will be required to confirm the presence of this pigment pathway in SGSB and to characterize its spatial–temporal distribution across different tissues and life stages.
Some studies suggest that white may be involved not only in the transport of ommochrome and pteridine precursors but of other pigmented compounds, such as riboflavin, an orange-yellow B vitamin stored in the testes and Malpighian tubules of some insects85,96,97,98,99,100. Riboflavin has been associated with eye pigment pathways4,21,85,99,101. For example, the level of riboflavin was strongly reduced in testes of a white-eyed mutant strain of E. kühniella99 and Anopheles gambiae Giles23, and the testes and Malpighian tubules of white-eyed mutants of Drosophila4,6,21,80. More recently, investigations on mutant strains of the silkworm Bombyx mori L. showed that some mutations of the white and brown orthologs prevented an accumulation of riboflavin in Malpighian tubules102,103. The reproductive organs of more mature wild-type SGSB adults, such as testes and vas deferens of males and ovarioles of females are normally yellow or orange-yellow104. The metathoracic scent reservoir is another orange-yellow organ found in SGSB adults105. However, we observed that in adults that had been treated with white dsRNAs, especially earlier in the fifth nymphal stage, these organs were significantly discolored suggesting that the white encoded protein subunit may be involved in the transport of orange-yellow pigmented compounds into gonads and metathoracic scent reservoir of the SGSB. Dissections also revealed that SGSB adults that had been treated with white dsRNA developed orange-yellow Malpighian tubules instead of white-transparent ones normally found in wild-type adults.
Although we did not measure the pigmentation profile of SGSB organs individually to confirm that their differential pigmentation is due to riboflavin mobilization, we observed an overall accumulation of riboflavin in SGSB that had been treated with white dsRNAs and in their progeny suggesting that this compound was not being properly stored and/or catabolized relative to wild type controls. Bacterial symbionts are abundant in the hemolymph of hemipterans where they deliver essential amino acids and riboflavin to the hosts106,107,108,109,110. Thus, an impact on the transport of riboflavin could potentially result in the accumulation of this yellow compound in the hemolymph if not properly stored, and/or cause longer retention of this compound already deposited in specific organs, for example. It is possible that riboflavin is stored in the Malpighian tubules of SGSB during immature stages and then gradually transported to reproductive organs as they mature. Previous studies documented that in Lepidopteran species riboflavin content in Malpighian tubules and gonads are inversely correlated suggesting mobilization of this compound from one organ to the other99,111.
The relationship between white and riboflavin mobilization in the SGSB has yet to be clarified. Nevertheless, white has been implicated in the transport of cyclic GMP9, which is a signaling molecule involved in Malpighian tubule secretion in Drosophila112,113 and R. prolixus114. A microarray-based atlas of gene expression in multiple tissues in Drosophila named FlyAtlas (http://flyatlas.org)115 showed that white is most highly expressed in Malpighian tubules indicating a major local function of this gene. Interestingly, Malpighian tubules are responsible for regulating uric acid levels in different insect species116 and null white mutations were associated with transport defects of uric acid and its purine precursors in Diptera4,117 and Lepidoptera118. Reduced accumulation of uric acid in the epidermal cells of Bombyx and Anopheles larvae produced translucent skin and increased sensitivity to UV damage117,118,119,120. We found translucent skin in SGSB adults that had been treated with white dsRNAs, which could be an impairment of uric acid accumulation in their integument and will require further investigation.
The color of the adult hemolymph released during dissections was considerably different among the white SGSB phenotypes induced by RNAi treatments. SGSB adults that had been treated with white dsRNAs during the last nymphal stage developed a yellow integument and green hemolymph, while those treated near or after the adult molting period developed a blue-green integument, and discolored hemolymph similar to wild type SGSB adults. The green hemolymph of SGSB nymphs is made of a mixture of yellow and blue compounds121 and our results indicate that the white encoded protein subunit may be involved in their transport. Furthermore, some major physiological processes happening around 3 days prior to adult molting shifted the effect of white transcript knockdown on the integument greening of SGSB adults, turning it more blue than yellow. Perhaps an interaction between hormones released prior molting and pigmented compounds present in the hemolymph of SGSB nymphs that may be retained in the adult integument could have been impacted by white transcript knockdown. For example, copper-containing hemocyanins/hexamerins are blue proteins found in the hemolymph of many arthropods122,123,124 that are known to participate in the molting process125,126,127,128 and riboflavin transport111. Hexamerins with a blue cromophore (biliverdin) cause stage-dependent changes in the blue coloration of the hemolymph and fat body of the bean bug Riptorus clavatus (Thunberg) under the control of juvenile hormone129,130. Also, a hormone known to regulate red-related pigment transport in the integument and compound eyes of crustaceans has been previously detected in the corpora cardiaca of the SGSB131,132. Additional research is needed to characterize the origin and transport of integument pigments during SGSB adult molting and to clarify the involvement of white in the process.
This study revealed that the white ortholog in the SGSB is involved in the ommochrome pigment pathway and suggests additional functions of this gene such as riboflavin mobilization, management of hemolymph compounds and integument composition. Further investigations are needed for a complete characterization of this gene in the SGSB including verification of fitness cost and/or advantage associated with white transcript knockdown in this species. The fact that white transcript knockdown in the SGSB was not lethal or impaired embryo development while providing a readily identifiable phenotype may support the use of this gene as a marker for genetic transformation in this species, which would also allow additional investigations on the function and heritability of this gene.
Data availability
The datasets generated and /or analyzed during the current study are available from the corresponding author on reasonable request.
References
Morgan, T. H. Sex limited inheritance in Drosophila. Science 32, 120–122 (1910).
Morgan, T. H. Chromosomes and heredity. Am. Nat. 44, 449–496 (1910).
Morgan, T. H. Random segregation versus coupling in mendelian inheritance. Science 34, 384–384 (1911).
Sullivan, D. T., Bell, L. A., Paton, D. R. & Sullivan, M. C. Purine transport by malpighian tubules of pteridine-deficient eye color mutants of Drosophila melanogaster. Biochem. Genet. 17, 565–573 (1979).
Pirrotta, V., Steller, H. & Bozzetti, M. P. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 4, 3501–3508 (1985).
Hazelrigg, T. The Drosophila white gene: A molecular update. Trends Genet. 3, 43–47 (1987).
van Breugel, F. M. A. Differential riboflavin deposition in white and variegated white mutants of Drosophila hydei. Dev. Genet. 8, 45–58 (1987).
Mackenzie, S. M. et al. Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim. Biophys. Acta 1419, 173–185 (1999).
Evans, J. M., Day, J. P., Cabrero, P., Dow, J. A. T. & Davies, S.-A. A new role for a classical gene: White transports cyclic GMP. J. Exp. Biol. 211, 890–899 (2008).
Campesan, S. et al. The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington’s disease. Curr. Biol. 21, 961–966 (2011).
Green, E. W. et al. Drosophila eye color mutants as therapeutic tools for Huntington disease. Fly (Austin) 6, 117–120 (2012).
Bellen, H. J. & Yamamoto, S. Morgan’s legacy: Fruit flies and the functional annotation of conserved genes. Cell 163, 12–14 (2015).
Xiao, C. & Robertson, R. M. Timing of locomotor recovery from anoxia modulated by the white gene in Drosophila. Genetics 203, 787–797 (2016).
Hersh, B. M. More than meets the eye: A primer for “timing of locomotor recovery from anoxia modulated by the white gene in Drosophila melanogaster”. Genetics 204, 1369–1375 (2016).
Sullivan, D. T. & Sullivan, M. C. Transport defects as the physiological basis for eye color mutants of Drosophila melanogaster. Biochem. Genet. 13, 603–613 (1975).
Sullivan, D. T. & Kitos, R. J. Developmental regulation of tryptophan catabolism in Drosophila. Insect Biochem. 6, 649–655 (1976).
Sullivan, D. T., Bell, L. A., Paton, D. R. & Sullivan, M. C. Genetic and functional analysis of tryptophan transport in Malpighian tubules of Drosophila. Biochem. Genet. 18, 1109–1130 (1980).
Dreesen, T. D., Johnson, D. H. & Henikoff, S. The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol. Cell. Biol. 8, 5206–5215 (1988).
Ewart, G. D. & Howells, A. J. [15] ABC transporters involved in transport of eye pigment precursors in Drosophila melanogaster. Methods Enzymol. 292, 213–224 (1998).
Mackenzie, S. M., Howells, A. J., Cox, G. B. & Ewart, G. D. Sub-cellular localisation of the white/scarlet ABC transporter to pigment granule membranes within the compound eye of Drosophila melanogaster. Genetica 108, 239–252 (2000).
Nickla, H. Interaction between pteridine synthesis and riboflavin accumulation in Drosophila melanogaster. Can. J. Genet. Cytol. J. Can. Genet. Cytol. 14, 105–111 (1972).
Summers, K. M. & Howells, A. J. Functions of the white and topaz loci of Lucilia cuprina in the production of the eye pigment xanthommatin. Biochem. Genet. 18, 643–653 (1980).
Benedict, M. Q., Besansky, N. J., Chang, H., Mukabayire, O. & Collins, F. H. Mutations in the Anopheles gambiae pink-eye and white genes define distinct, tightly linked eye-color loci. J. Hered. 87, 48–53 (1996).
Kômoto, N., Quan, G.-X., Sezutsu, H. & Tamura, T. A single-base deletion in an ABC transporter gene causes white eyes, white eggs, and translucent larval skin in the silkworm w-3oe mutant. Insect Biochem. Mol. Biol. 39, 152–156 (2009).
Broehan, G., Kroeger, T., Lorenzen, M. & Merzendorfer, H. Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genom. 14, 6 (2013).
Grubbs, N., Haas, S., Beeman, R. W. & Lorenzen, M. D. The ABCs of Eye Color in Tribolium castaneum: Orthologs of the Drosophila white, scarlet, and brown Genes. Genetics 199, 749–759 (2015).
Khan, S. A., Reichelt, M. & Heckel, D. G. Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci. Rep. 7, 40025 (2017).
Xue, W.-H. et al. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 93, 19–26 (2018).
Tsuji, T. et al. Molecular characterization of eye pigmentation-related ABC transporter genes in the ladybird beetle Harmonia axyridis reveals striking gene duplication of the white gene. Zool. Sci. 35, 260–267 (2018).
Jiang, Y. & Lin, X. Role of ABC transporters White, Scarlet and Brown in brown planthopper eye pigmentation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 221–222, 1–10 (2018).
Brent, C. S. & Hull, J. J. RNA interference-mediated knockdown of eye coloration genes in the western tarnished plant bug (Lygus hesperus Knight). Arch. Insect Biochem. Physiol. 100, e21527 (2019).
Reding, K. & Pick, L. High-efficiency CRISPR/Cas9 mutagenesis of the white gene in the milkweed bug Oncopeltus fasciatus. Genetics 215, 1027–1037 (2020).
Heu, C. C., McCullough, F. M., Luan, J. & Rasgon, J. L. CRISPR-Cas9-based genome editing in the silverleaf whitefly (Bemisia tabaci). CRISPR J. 3, 89–96 (2020).
Panizzi, A. R. Wild hosts of pentatomids: Ecological significance and role in their pest status on crops. Annu. Rev. Entomol. 42, 99–122 (1997).
McPherson, J. E. & McPherson, R. Stink Bugs of Economic Importance in America North of Mexico (CRC Press, 2000).
Nilakhe, S. S., Chalfant, R. B. & Singh, S. V. Evaluation of southern green stink bug damage to cowpeas. J. Econ. Entomol. 74, 589–592 (1981).
Hall, D. G. & Teetes, G. L. Yield loss-density relationships of four species of panicle-feeding bugs in sorghum. Environ. Entomol. 11, 738–741 (1982).
Barbour, K. S., Bradley, J. R. & Bacheler, J. S. Reduction in yield and quality of cotton damaged by green stink bug (Hemiptera: Pentatomidae). J. Econ. Entomol. 83, 842–845 (1990).
Yates, I. E., Tedders, W. L. & Sparks, D. Diagnostic evidence of damage on pecan shells by stink bugs and coreid bugs. J. Am. Soc. Hortic. Sci. 116, 42–46 (1991).
Greene, J. K., Turnipseed, S. G., Sullivan, M. J. & May, O. L. Treatment thresholds for stink bugs (Hemiptera: Pentatomidae) in cotton. J. Econ. Entomol. 94, 403–409 (2001).
Tindall, K. V. et al. Yield components and quality of rice in response to graminaceous weed density and rice stink bug populations. Crop Prot. 24, 991–998 (2005).
Reay-Jones, F. P. F. Spatial and temporal patterns of stink bugs (Hemiptera: Pentatomidae) in wheat. Environ. Entomol. 39, 944–955 (2010).
Ni, X. et al. Impact of brown stink bug (Heteroptera: Pentatomidae) feeding on corn grain yield components and quality. J. Econ. Entomol. 103, 2072–2079 (2010).
Musser, F. R., Catchot, A. L., Gibson, B. K. & Knighten, K. S. Economic injury levels for southern green stink bugs (Hemiptera: Pentatomidae) in R7 growth stage soybeans. Crop Prot. 30, 63–69 (2011).
Leskey, T. C. et al. Pest status of the brown marmorated stink bug, Halyomorpha halys in the USA. Outlooks Pest Manag. 23, 218–226 (2012).
Venugopal, P. D., Coffey, P. L., Dively, G. P. & Lamp, W. O. Adjacent habitat influence on stink Bug (Hemiptera: Pentatomidae) densities and the associated damage at field corn and soybean edges. PLoS ONE 9, e109917 (2014).
Rijal, J. P., Joyce, A. L. & Gyawaly, S. Biology, ecology, and management of hemipteran pests in almond orchards in the United States. J. Integr. Pest Manag. 12, 24 (2021).
Vyavhare, S. S., Way, M. O., Pearson, R. A. & Medina, R. F. Redbanded stink bug (Hemiptera: Pentatomidae) infestation and occurrence of delayed maturity in soybean. J. Econ. Entomol. 108, 1516–1525 (2015).
Naranjo, S. E. Impacts of Bt transgenic cotton on integrated pest management. J. Agric. Food Chem. 59, 5842–5851 (2011).
Zeilinger, A. R., Olson, D. M. & Andow, D. A. Competition between stink bug and heliothine caterpillar pests on cotton at within-plant spatial scales. Entomol. Exp. Appl. 141, 59–70 (2011).
Zeilinger, A. R., Olson, D. M. & Andow, D. A. Competitive release and outbreaks of non-target pests associated with transgenic Bt cotton. Ecol. Appl. 26, 1047–1054 (2016).
Douris, V. et al. Using CRISPR/Cas9 genome modification to understand the genetic basis of insecticide resistance: Drosophila and beyond. Pestic. Biochem. Physiol. 167, 104595 (2020).
Liu, S. H., Yang, B. J., Wang, A. Y., Luo, J. & Tang, J. The white gene in Nilaparvata lugens and its expression pattern under two different survival stresses. J. Asia-Pac. Entomol. 21, 701–707 (2018).
Klobasa, W. et al. Microinjection of corn planthopper, Peregrinus maidis, embryos for CRISPR/Cas9 genome editing. J. Vis. Exp. 169, e62417. https://doi.org/10.3791/62417 (2021).
Fishilevich, E. et al. Use of chromatin remodeling ATPases as RNAi targets for parental control of western corn rootworm (Diabrotica virgifera virgifera) and Neotropical brown stink bug (Euschistus heros). Insect Biochem. Mol. Biol. 71, 58–71 (2016).
Lu, Y., Chen, M., Reding, K. & Pick, L. Establishment of molecular genetic approaches to study gene expression and function in an invasive hemipteran, Halyomorpha halys. EvoDevo 8, 15 (2017).
Cagliari, D. et al. First transcriptome of the Neotropical pest Euschistus heros (Hemiptera: Pentatomidae) with dissection of its siRNA machinery. Sci. Rep. 10, 4856 (2020).
Sharma, R., Christiaens, O., Taning, C. N. T. & Smagghe, G. RNAi-mediated mortality in Southern green stinkbug Nezara viridula by oral delivery of dsRNA. Pest Manag. Sci. 77(1), 77–84 (2021).
Riga, M. et al. Development of efficient RNAi in Nezara viridula for use in insecticide target discovery. Arch. Insect Biochem. Physiol. 103, e21650 (2020).
Cantón, P. E. & Bonning, B. C. Transcription and activity of digestive enzymes of Nezara viridula maintained on different plant diets. Front. Physiol. 10, 1553 (2020).
Owczarzy, R. et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 36, W163–W169 (2008).
Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR: Methods and Applications (eds Meuer, S. et al.) 21–34 (Springer, 2001). https://doi.org/10.1007/978-3-642-59524-0_3.
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1 (2002).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33 (2006).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001).
SAS Institute Inc. SAS 9.4® Statements: Reference (SAS Institute Inc., 2013).
Ferrari, S. & Cribari-Neto, F. Beta regression for modelling rates and proportions. J. Appl. Stat. 31, 799–815 (2004).
Stroup, W. W. Rethinking the analysis of non-normal data in plant and soil science. Agron. J. 107, 811–827 (2015).
Christensen, S. A., Santana, E. A., Alborn, H. T., Block, A. K. & Chamberlain, C. A. Metabolomics by UHPLC-HRMS reveals the impact of heat stress on pathogen-elicited immunity in maize. Metabolomics 17, 6 (2021).
Pluskal, T., Castillo, S., Villar-Briones, A. & Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 11, 395 (2010).
Lavore, A., Pagola, L., Esponda-Behrens, N. & Rivera-Pomar, R. The gap gene giant of Rhodnius prolixus is maternally expressed and required for proper head and abdomen formation. Dev. Biol. 361, 147–155 (2012).
Paim, R. M. M., Araujo, R. N., Lehane, M. J., Gontijo, N. F. & Pereira, M. H. Long-term effects and parental RNAi in the blood feeder Rhodnius prolixus (Hemiptera; Reduviidae). Insect Biochem. Mol. Biol. 43, 1015–1020 (2013).
Vargas-Lowman, A. et al. Cooption of the pteridine biosynthesis pathway underlies the diversification of embryonic colors in water striders. Proc. Natl. Acad. Sci. 116, 19046–19054 (2019).
Khila, A., Abouheif, E. & Rowe, L. Evolution of a novel appendage ground plan in water striders is driven by changes in the Hox gene ultrabithorax. PLOS Genet. 5, e1000583 (2009).
Liu, P. Z. & Kaufman, T. C. Hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 131, 1515–1527 (2004).
O’Hare, K., Murphy, C., Levis, R. & Rubin, G. M. DNA sequence of the white locus of Drosophila melanogaster. J. Mol. Biol. 180, 437–455 (1984).
Howells, A. J., Summers, K. M. & Ryall, R. L. Developmental patterns of 3-hydroxykynurenine accumulation in white and various other eye color mutants of Drosophila melanogaster. Biochem. Genet. 15, 1049–1059 (1977).
Yagi, S. & Ogawa, H. Effect of tryptophan metabolites on fluorescent granules in the malpighian tubules of eye color mutants of Drosophila melanogaster. Zool. Sci. 13, 97–104 (1996).
Cölln, K. & Hedemann, E. 3-OH-Kynurenine content and ommochrome formation in the developing compound eye of Ephestia kuehniella. Experientia 40, 494–496 (1984).
Chiou, S.-J. et al. Purification of toxic compounds from larvae of the gray fleshfly: The identification of paralysins. Biochem. Biophys. Res. Commun. 246, 457–462 (1998).
Han, Q., Fang, J. & Li, J. 3-Hydroxykynurenine transaminase identity with alanine glyoxylate transaminase: A probable detoxification protein in Aedes aegypti. J. Biol. Chem. 277, 15781–15787 (2002).
Cerstiaens, A. et al. Neurotoxic and neurobehavioral effects of kynurenines in adult insects. Biochem. Biophys. Res. Commun. 312, 1171–1177 (2003).
Cromartie, R. I. T. Insect pigments. Annu. Rev. Entomol. 4, 59–76 (1959).
Ryall, R. L. & Howells, A. J. Ommochrome biosynthetic pathway of Drosophila melanogaster: Variations in levels of enzyme activities and intermediates during adult development. Insect Biochem. 4, 47–61 (1974).
Pfleiderer, W. Pteridines. Properties, reactivities and biological significance. J. Heterocycl. Chem. 29, 583–605 (1992).
Follett, P. A., Calvert, F. & Golden, M. Genetic studies using the orange body color type of Nezara viridula (Hemiptera: Pentatomidae): inheritance, sperm precedence, and disassortative mating. Ann. Entomol. Soc. Am. 100, 433–438 (2007).
Vivan, L. M. & Panizzi, A. R. Two New Morphs of the Southern Green Stink Bug, Nezara viridula (L.) (Heteroptera: Pentatomidae), Brazil. Neotrop. Entomol. 31, 475–476 (2002).
Kiritani, K. Studies on the adult polymorphism in the southern green stink bug, Nezara viridula (hemiptera: Pentatomidae). Popul. Ecol. 12, 19–34 (1970).
Harris, V. E., Todd, J. W. & Mullinix, G. Color change as an indicator of adult diapause in the southern green stink bug, Nezara viridula. J. Agric. Entomol. 1, 82–91 (1984).
Takeda, K., Musolin, D. L. & Fujisaki, K. Dissecting insect responses to climate warming: Overwintering and post-diapause performance in the southern green stink bug, Nezara viridula, under simulated climate-change conditions. Physiol. Entomol. 35, 343–353 (2010).
Niva, C. C. & Takeda, M. Color changes in Halyomorpha brevis (Heteroptera: Pentatomidae) correlated with distribution of pteridines: Regulation by environmental and physiological factors. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 132, 653–660 (2002).
Moraes, A. S., Pimentel, E. R., Rodrigues, V. L. C. C. & Mello, M. L. S. Eye pigments of the blood-sucking insect, Triatoma infestans klug (Hemiptera, Reduviidae). Braz. J. Biol. 65, 477–481 (2005).
Insausti, T. C., Gall, M. L. & Lazzari, C. R. Oxidative stress, photodamage and the role of screening pigments in insect eyes. J. Exp. Biol. 216, 3200–3207 (2013).
Busnel, R. G. & Drilhon, A. Action vitaminique de la riboflavine extraite des tubes de Malpighi des insectes. C. R. Seances Soc. Biol. 137, 752–753 (1943).
Metcalf, R. L. The storage and interaction of water soluble vitamins in the malpighian system of Periplaneta americana (L.). Arch Biochem 2, 55–62 (1943).
Vincentiis, M. D. Sulle sostanze fluorescenti del tubo malpighiano di Sarcophaga haemorrhoidalis Fll. Boll. Zool. 23, 1–11 (1956).
Caspari, E. & Blomstrand, I. A yellow pigment in the testis of Ephestia: Its development and its control by genes. Genetics 43, 679–694 (1958).
Wessing, A. & Eichelberg, D. Die fluoreszierenden Stoffe aus den Malpighischen-Gefäßen der Wildform und verschiedener Augenfarbenmutanten von Drosophila melanogaster. Z. Für Naturforschung B 23, 376–386 (1968).
Kikkawa, H. Biochemical Genetics of Bombyx mori (Silkworm). In Advances in Genetics Vol. 5 (ed. Demerec, M.) 107–140 (Academic Press, 1953).
Zhang, H., Kiuchi, T., Hirayama, C., Katsuma, S. & Shimada, T. Bombyx ortholog of the Drosophila eye color gene brown controls riboflavin transport in Malpighian tubules. Insect Biochem. Mol. Biol. 92, 65–72 (2018).
Zhang, H. et al. A reexamination on the deficiency of riboflavin accumulation in Malpighian tubules in larval translucent mutants of the silkworm, Bombyx mori. Genetica 146, 425–431 (2018).
Esquivel, J. F. Stages of gonadal development of the southern green stink bug (Hemiptera: Pentatomidae): Improved visualization. Ann. Entomol. Soc. Am. 102, 303–309 (2009).
Gilby, A. R. & Waterhouse, D. F. Secretions from the lateral scent glands of the green vegetable bug, Nezara viridula. Nature 216, 90–91 (1967).
Nakabachi, A. & Ishikawa, H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J. Insect Physiol. 45, 1–6 (1999).
Hosokawa, T., Koga, R., Kikuchi, Y., Meng, X.-Y. & Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. 107, 769–774 (2010).
Nikoh, N. et al. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. 111, 10257–10262 (2014).
Bustamante-Brito, R., Vera-Ponce de León, A., Rosenblueth, M., Martínez-Romero, J. C. & Martínez-Romero, E. Metatranscriptomic analysis of the bacterial symbiont Dactylopiibacterium carminicum from the carmine cochineal Dactylopius coccus (Hemiptera: Coccoidea: Dactylopiidae). Life 9, 4 (2019).
Noda, T. & Kamano, S. Artificial rearing of Nezara viridula (L.) and N. antennata Scott (Heteroptera: Pentatomidae) with semi-solid meridic diets. Appl. Entomol. Zool. 37, 43–50 (2002).
Miller, S. G. & Silhacek, D. L. Riboflavin binding proteins and flavin assimilation in insects. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 110, 467–475 (1995).
Dow, J. A., Maddrell, S. H., Davies, S. A., Skaer, N. J. & Kaiser, K. A novel role for the nitric oxide-cGMP signaling pathway: The control of epithelial function in Drosophila. Am. J. Physiol.-Regul. Integr. Comp. Physiol. https://doi.org/10.1152/ajpregu.1994.266.5.R1716 (1994).
Davies, S. A. et al. CAP2b, a cardioacceleratory peptide, is present in Drosophila and stimulates tubule fluid secretion via cGMP. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 269, R1321–R1326 (1995).
Quinlan, M. C., Tublitz, N. J. & O’Donnell, M. J. Anti-diuresis in the blood-feeding insect Rhodnius prolixus Stål: The peptide CAP2b and cyclic GMP inhibit Malpighian tubule fluid secretion. J. Exp. Biol. 200, 2363–2367 (1997).
Chintapalli, V. R., Wang, J. & Dow, J. A. T. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 (2007).
O’donnell, M. J., Maddrell, S. H. P. & Gardiner, B. O. C. Transport of uric acid by the malpighian tubules of Rhodnius prolixus and other insects. J. Exp. Biol. 103, 169–184 (1983).
Benedict, M. Q., Cohen, A., Cornel, A. J. & Brummett, D. L. Uric acid in anopheles mosquitoes (Diptera: Culicidae): Effects of collarless, stripe, and white mutations. Ann. Entomol. Soc. Am. 89, 261–265 (1996).
Wang, L. et al. Mutation of a novel ABC transporter gene is responsible for the failure to incorporate uric acid in the epidermis of ok mutants of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 43, 562–571 (2013).
Matsuo, T. & Ishikawa, Y. Protective role of uric acid against photooxidative stress in the silkworm, Bombyx mori (Lepidoptera : Bombycidae). Appl. Entomol. Zool. 34, 481–484 (1999).
Hu, Y.-G., Shen, Y.-H., Zhang, Z. & Shi, G.-Q. Melanin and urate act to prevent ultraviolet damage in the integument of the silkworm, Bombyx mori. Arch. Insect Biochem. Physiol. 83, 41–55 (2013).
Hackman, R. H. Green pigments of the hemolymph of insects. Arch. Biochem. Biophys. 41, 166–174 (1952).
Terwilliger, N. B. Hemolymph proteins and molting in crustaceans and insects. Integr. Comp. Biol. 39, 589–599 (1999).
Markl, J. & Decker, H. Molecular Structure of the Arthropod Hemocyanins. In Blood and Tissue Oxygen Carriers (ed. Mangum, C. P.) 325–376 (Springer, 1992). https://doi.org/10.1007/978-3-642-76418-9_12.
Hagner-Holler, S. et al. A respiratory hemocyanin from an insect. Proc. Natl. Acad. Sci. 101, 871–874 (2004).
Kuballa, A. V., Holton, T. A., Paterson, B. & Elizur, A. Moult cycle specific differential gene expression profiling of the crab Portunus pelagicus. BMC Genom. 12, 147 (2011).
Kuballa, A. V. & Elizur, A. Differential expression profiling of components associated with exoskeletal hardening in crustaceans. BMC Genom. 9, 575 (2008).
Glazer, L. et al. Hemocyanin with phenoloxidase activity in the chitin matrix of the crayfish gastrolith. J. Exp. Biol. 216, 1898–1904 (2013).
Jaenicke, E., Föll, R. & Decker, H. Spider hemocyanin binds ecdysone and 20-OH-ecdysone. J. Biol. Chem. 274, 34267–34271 (1999).
Chinzei, Y., Haruna, T., Miura, K., Numata, H. & Nakayama, S. Purification and characterization of biliverdin-associated cyanoprotein from eggs and hemolymph of the bean bug, Riptortus clavatus (Heteroptera: Alydidae). Insect Biochem. 20, 545–555 (1990).
Miura, K. et al. Two hexameric cyanoprotein subunits from an insect, Riptortus clavatus. Eur. J. Biochem. 258, 929–940 (1998).
Gäde, G., Auerswald, L., Šimek, P., Marco, H. G. & Kodrík, D. Red pigment-concentrating hormone is not limited to crustaceans. Biochem. Biophys. Res. Commun. 309, 967–973 (2003).
Josefsson, L. Chemical properties and physiological actions of crustacean chromatophorotropins. Integr. Comp. Biol. 23, 507–515 (1983).
Acknowledgements
This material is based upon work supported by the National Science Foundation I/UCRC, the Center for Arthropod Management Technologies under Grant No. IIP-1821914 and by industry partners. The authors would like to thank Dr. Pablo E. Cantón (Entomology and Nematology Department, University of Florida, Gainesville, FL) for donating a SGSB lab population, and Dr. Marcus Guest (Syngenta Crop Protection, Jealott’s Hill International Research Centre, Berkshire, UK) for providing SGSB gene sequences used in this study.
Author information
Authors and Affiliations
Contributions
D.S., K.W., P.S. and B.S. designed the study. P.S. and B.S. obtained the grant to support the research. D.S., S.C., K.W., K.K. and C.D. performed the experiments. D.S. and K.W. analyzed the data. D.S., K.W. and L.B. photo documented insect phenotypes generated. D.S. wrote the manuscript. All authors reviewed the manuscript and critically discussed the results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Souza, D., Christensen, S.A., Wu, K. et al. RNAi-induced knockdown of white gene in the southern green stink bug (Nezara viridula L.). Sci Rep 12, 10396 (2022). https://doi.org/10.1038/s41598-022-14620-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-14620-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.