Abstract
Silver diamine fluoride (SDF) has been long studied in laboratories, and its clinical effectiveness in the treatment and prevention of root caries has been reported. In the present study, we assessed the microbiological effects of SDF on dental biofilms grown on demineralized dentin in situ. Specifically, demineralized bovine root dentin slabs used as biofilm substrates were treated with 38% SDF, and the biofilms formed after this treatment were analyzed via real-time PCR, DEAD/LIVE cell staining, and SEM. Next, the viable cell count was determined, and microbial profiles were compared using 16S rRNA gene sequencing. Untreated slabs were used as controls. We observed significant decreases in viable cell counts (p < 0.05), number of biofilm-forming cells (p < 0.01), biofilm thickness (p < 0.01), and high proportion of dead cells with SDF treatment (p < 0.01). The microcolonies in the SDF-treated biofilms showed less complexity, and only a limited number of genera were differentially abundant between the groups. Microbial diversity index comparisons showed no significant differences between the groups with respect to treatments days (p = 0.362). Thus, SDF negatively influenced dental biofilm growth on demineralized root dentin in situ; however, its antimicrobial action did not target a specific oral taxon.
Similar content being viewed by others
Introduction
Oral biofilms are polymicrobial communities adhered to oral cavity surfaces and consist of multiple species of microorganisms embedded within a matrix containing extracellular polysaccharides. It is generally accepted that the interactions within and among oral biofilms, as well as with the host, are potentially responsible for oral health and disease states1,2,3. Dental caries is a prime example of oral infectious diseases that originate as a consequence of the interactions between microorganisms, hosts, and environmental conditions1. The etiology of dental caries has presumably been described as a shift to the predominance of some cariogenic species within symbiotic biofilms that eventually leads to an imbalance between demineralization and remineralization of tooth structures4,5,6.
Human life expectancy has increased, leading to increased awareness of dental health, including advanced oral therapeutic modalities. The number of remaining teeth with gingival recession has become significantly greater in the elderly. Once root surface/dentin is exposed to the oral environment, it is likely to be susceptible to demineralization owing to the acidic environment created by acid-producing bacteria in the supragingival biofilm. Compared to coronal enamel, root dentin and cementum are more susceptible to acidic pH due to their lower inorganic content and smaller size of hydroxyapatite crystallites7. In the elderly, decreased salivary flow resulting from medications or systemic diseases and difficulty in brushing the teeth regularly due to physical impairment can increase the risk of root caries development8,9, which presently is a crucial oral problem affecting them and worsening their quality of life.
Silver diamine fluoride (SDF) is an aqueous therapeutic agent containing silver and fluoride that is used to arrest caries and promote remineralization. Its clinical efficiency in the treatment of coronal caries of primary10,11 and permanent dentitions12,13, including root surface caries14, has been reported. SDF solution is also used for the treatment of dentinal hypersensitivity15. Further, a number of in vitro studies have demonstrated its efficacy in reducing mineral loss16,17, enhancing dentin hardening18, and ensuring collagen preservation, as inferred from an experiment involving acid challenge17. Furthermore, SDF can react with the inorganic components of the tooth and form fluorohydroxyapatite, which can promote tooth hardness and decrease the solubility of the affected dentin19. Additionally, the antimicrobial properties of SDF remarkably inhibit root caries progression. Specifically, it has been demonstrated that the growth rate of a monoculture of Streptococcus mutans is significantly lower under SDF treatment than under silver fluoride and potassium iodide treatment20. Moreover, it has also been observed that biofilm formation by Actinomyces naeslundii on dentin blocks decreases significantly after the direct application of SDF18. This is consistent with the results of a more recent study in which dual- and multi-species biofilm models were used21,22.
At present, 16S rRNA gene sequencing in combination with microbiological ecology methods has become a crucial part of studying oral microorganisms in health and diseases23. A recent clinical sample study on the root caries-related microbial profiles observed before and after SDF application on cavitated lesions of the root surface showed that even though there was no statistical difference in microbial diversity after SDF application, the relative abundance of cariogenic taxa (e.g., Scardovia, Bifidobacterium, and Actinomyces) decreased with time24. Therefore, it is presumed that the antimicrobial properties of SDF, as exhibited in the oral microbiome, play an important role in the prevention of the recurrence of root caries. However, no clear evidence that accurately validates these assumptions, particularly in relation to actual dental biofilms, has been reported. Hence, a combination of comprehensive quantitative data acquisition and gene-based characterization of microorganisms in the biofilm formed on SDF-applied tooth surfaces would provide a better understanding of the antibacterial effects of SDF.
The advancement of in vitro biofilm models has mitigated the complexity in studying oral biofilm nowadays25,26. Nevertheless, there are some challenging aspects that limit the capability of in vitro models in mimicking the natural oral condition, including the species diversity and the complex environment of the oral cavity26. Therefore, an in situ biofilm model could be considered as an alternative for the investigation of natural oral biofilms and various therapeutic modalities. To extend the microbiological perspective of using SDF as a topical medication for arresting and preventing dental caries, we therefore assessed the microbiological effect of 38% SDF on dental biofilms grown on demineralized dentin in situ in terms of bacterial viability and biomass. Furthermore, based on 16S rRNA gene sequencing technique, we analyzed bacterial profiles and changes in biofilm biodiversity, to observe the changes in the dental biofilm community presumably influenced by treatment with SDF. The null hypotheses of this study were that (i) the application of SDF on demineralized dentin does not inhibit the growth and viability of biofilms and (ii) there is no difference in the microbial community structure between the SDF and control groups.
Results
Quantity of bacteria in biofilm
Under both aerobic and anaerobic conditions, cultivable bacteria cell count revealed that the log CFU counts corresponding to the SDF-treated biofilms were significantly lower than those corresponding to the controls (Repeated Measure ANOVA, p < 0.01) (Fig. 1a). Consistent with the viable cell count, quantification via real-time PCR showed the same tendency, i.e., the SDF treatment group showed a lower bacterial load (Repeated Measure ANOVA, p < 0.001) (Fig. 1b).
(a) Bacterial viability (log colony-forming unit (CFU) per unit area) under aerobic and anaerobic conditions. ***p < 0.001, aerobic condition; †p < 0.05, ††p < 0.01, †††p < 0.001, anaerobic condition. (b) Real-time PCR quantification of biofilm-forming cells (log CFU per unit area). △△p < 0.01, △△△p < 0.001.
Biofilm biomass, thickness, and proportion of dead and live cells based on confocal laser scanning microscopy
The representative images corresponding to LIVE/DEAD bacterial cell staining obtained via confocal microscopy (Fig. 2a) revealed a noticeably lesser biofilm mass in the SDF group, with dead cells labelled in red enclosed within microcolonies. Although the biofilm biomass in both groups tended to increase with time, as demonstrated by the thickness of biofilm in Fig. 2b (p < 0.01), the dead to live cell volume ratio of the SDF-treated biofilm was significantly greater than that of the corresponding control group throughout the experimental period (p < 0.01) (Fig. 2c).
Confocal laser scanning microscopy (CLSM) analysis of biofilms grown on dentin slabs in the control and SDF groups using the Imaris imaging software. (a) Representative LIVE/DEAD staining images comparing biofilms formed in each group over the experiment days. Live and dead cells are labelled in green and red, respectively (scale bar, 20 μm). (b) Thickness of biofilms p** < 0.01. (c) Ratio of live to dead cells. p** < 0.01. The letters a, b, and c denote significant differences over different experiment days in the same group (p < 0.05).
Microscopic structure of biofilms based on SEM
High-magnification SEM images were captured to observe the characteristics of the ultrastructure of in situ biofilms of both groups. Figure 3 shows the co-aggregation of rod-shaped and columnar microcolonies, which penetrated the dentinal tubules of bovine root dentin in the control group on the first day. Further, the biofilm structure became more complex owing to the presence of substantial filamentous and coccal bacteria that grew in the subsequent days. In contrast, the microcolonies in the biofilms corresponding to the SDF group were sparsely detectable and were less complex. We also observed that small silver particles precipitated on the dentin surface in the SDF group.
Microbial profile based on 16S rRNA gene sequencing
With respect to the results of 16S rRNA gene sequencing involving a total of 80 samples (40 each from the control and SDF groups), 14 phyla, 21 classes, 33 orders, 58 families, and 90 genera were detected across all samples. Based on the relative abundances of the different taxa, the most abundant phyla in the control and SDF groups were Proteobacteria (31.54%) and Firmicutes (37.20%), respectively. Further, at the genus level, the three most abundant phyla in the control group were Neisseria (18.79%), Streptococcus (15.33%), and Porphyromonas (13.19%), while Streptococcus (22.17%), Haemophilus (11%), and Neisseria (10.96%) were the most abundant phyla in the SDF group. Furthermore, we examined the extent of changes in the relative abundances of the different genera. Table 1 shows the differential abundances of the taxa detected between the groups, compared on each experiment day. Among the more abundant taxa (average relative abundance > 3%), only Actinomyces (Log2FoldChange = − 2.68) was found to be significantly more abundant in the SDF group on day 3 (adjusted p-value < 0.001). However, Porphyromonas was in the control group (Log2FoldChange ranging from 1.36 to 2.42). Additionally, considering the pooled data between the groups, Granulicatella and unclassified genera in the family Gemellaceae were observed more frequently in the SDF treatment group.
Bacterial diversity of biofilm community
To further assess the effect of SDF application on community structure, alpha diversity parameters, including Chao1 and Shannon indices, were calculated. The calculated Chao1 indices showed significant differences between the control and SDF groups, regardless of the experiment day (control, 183.25 ± 46.82; SDF, 235.03 ± 65.09; adjusted p-value < 0.001). Similarly, the two groups also showed significant differences in Shannon indices (control, 3.99 ± 0.33; SDF, 4.38 ± 0.40; adjusted p-value < 0.001) (Fig. 4a). In contrast, when both indices were compared based on the experiment day, no significant differences were observed, except on day 2 for Shannon index. The PERMANOVA test showed significant differences in beta-diversity based on the Bray–Curtis distances between the biofilm communities when the groups were compared (adjusted p-value < 0.001). However, groups showed no significant differences with respect to the experiment day. Further, the PCoA plots did not show any clear clustering pattern for any of the factors considered (Fig. 4b,c).
Alpha and beta diversities of the bacterial community on biofilms. (a) Shannon and Chao1 diversity indices. (b) Principal coordinates analysis (PCoA) plot for pooled biofilm samples from the control and SDF groups based on Bray–Curtis distance. (c) PCoA plot based on Bray–Curtis distance when group and day factors were considered together. Statistical comparisons were done using the PERMANOVA test.
Discussion
In this study, we extensively assessed the effect of SDF treatment on dental biofilms from a microbiological perspective, combining conventional cultivable and microbial community profile analyses using current gene sequencing methods. Our first null hypothesis was rejected based on the results as a remarkable biofilm-inhibitory effect was observed with SDF treatment. In contrast, our findings supported the second null hypothesis, as the results did not demonstrate significant differences in microbial diversity between the biofilms of SDF and control groups on any experiment day.
To overcome the limitations associated with in vitro dental biofilms, we used a newly designed dentin slab-holding oral device to enable the growth of dental biofilms under actual oral conditions. Reportedly, bovine root dentin, which was used as a substrate in our in situ model instead of human root dentin, is suitable for investigating anticariogenic agents27, although its microhardness was found to be lower in another in situ study28. Furthermore, bovine dentin being more readily available and easy to manipulate than human dentin makes it more suitable for studies like ours, in which a large number of dentin slabs are required29. However, using sodium hypochlorite for dissolving the remaining organic tissues on root dentin after preparation might alter its surface characteristics, affecting the ability of the substrate to completely mimic the actual oral conditions. Hence, the results should be interpreted with caution.
Dental caries is caused by the acidic environment created by acidogenic and aciduric bacteria that leads to an imbalance between demineralization and remineralization on the tooth surface7. Carious lesions on the root surface are initiated by a polymicrobial biofilm covering the root surface and its metabolic products. The microbiota of root caries has been extensively studied, using both culture-based30,31,32 and non-culture-based33,34 approaches, and reportedly, it includes a variety of related bacterial species. Moreover, biodental engineering factors, such as chewing and parafunctional habits, capable of generating electrochemical transfer from the tooth surface to saliva and eventually leading to surface demineralization are involved in root caries etiology35. It has been reported that exposed root surface is a risk indicator associated with root caries in the elderly36. Traumatic tooth brushing is an etiological factor for gingival recession that exposes the root surface37. Consequently, improper brushing might increase the risk of root caries in older people.
For several decades, SDF has been used to prevent and arrest carious lesions in primary teeth11,38 and the root surface, particularly in the elderly13. However, the specific mechanism by which it exerts this effect has not yet been fully elucidated. Several in vitro studies have been conducted to investigate the underlying mechanisms extensively. Apart from its efficacy in preventing the degradation of dentin collagen17 and increasing microhardness as well as mineral contents after application18,39, it has also been demonstrated that SDF exerts various effects on cariogenic bacteria. Silver ions bind strongly to sulfhydryl groups and proteins on bacterial cell membranes, inhibiting intracellular enzyme activity as well as DNA replication40, eventually causing bacterial cell death and inhibiting biofilm formation41. A monoculture study of S. mutans and A. naeslundii showed significantly fewer CFU counts in the SDF treatment group than in the control18. This is also consistent with the results of another study that was performed using a co-culture model of S. mutans and Lactobacillus acidophilus on demineralized dentin blocks, which showed significantly lower CFU counts in SDF-treated blocks than water-applied blocks22. Similarly, the application of 38% SDF on cariogenic biofilms consisting of S. mutans, S. sorbrinus, L. acidophilus, Lactobacillus rhamnosus, and A. naeslundii, could inhibit the growth of this mixed-species biofilm21. Data from our present study on viable cell counts are consistent with the results of these aforementioned studies, demonstrating decreased CFU in the SDF group under both aerobic and anaerobic culture conditions. Similarly, real-time PCR quantification revealed a significantly lower total number of bacterial cells in the SDF group than in the control group, indicating that SDF reduces viable cell count in the biofilms formed on dentin slabs. Further, the viable cell count corresponding to the control group from both analyses exhibited a similar trend as that observed in a previous in situ study using hydroxyapatite (HA) disks42, suggesting the potential for the application of HA disks as a practical alternative to the bovine root dentin substrate for dental biofilm cultivation.
Regarding the biomass of the dental biofilms, CLSM images showed noticeably lesser biofilm formation, with dead cells, on the SDF-treated dentin slabs than in the control group. These findings are in accordance with previous in vitro studies18,21,22, although the SDF application sequences were different, and our experimental period was shorter. Similarly, the lower thickness of the biofilms following SDF treatment and their lower bacterial cell viability, as demonstrated by the dead to live cell ratio, indicated the anti-biofilm efficacy of SDF after application on demineralized root dentin. The CLSM results were further corroborated by the ultrastructural characterization of the biofilms based on SEM. While the actual maturation of biofilms could be observed in the control group as previously demonstrated in vivo43 and in situ using HA disks42, which showed the co-aggregation of various bacteria and the presence of matrix-like structures after 48 h of experimentation, biofilms on the dentin surfaces treated with SDF were scarce, and the bacterial composition was less complex. Additionally, consistent with earlier in vitro studies18,21, precipitated silver particles were also observed on the SDF-treated dentin surfaces in the present study. It has also been suggested that silver and fluoride ions in SDF interact with HA in the tooth and subsequently form CaF2 and Ag3PO444. In particular, CaF2 is a crucial fluoride reservoir that can eventually react with HA and form fluoroapatite, which reportedly is a favorable acid-resistant crystalline matrix that reduces the susceptibility of the tooth surface to demineralization10,19,45. Additionally, Ag3PO4 reacts with alkali chlorides to form AgCl, which is the major precipitate found on SDF-treated surfaces17,19, and owing to its low solubility, it has been affirmed that it has a slow-release effect with respect to silver ions that is responsible for its antibacterial properties46. Furthermore, it has been considered that the pronounced antibacterial effect of SDF is derived from the metallic silver nanoparticles that are also produced following the reaction between SDF and HA17. These particles are inert, but after coming in contact with moisture in the oral cavity, they release silver ions47. In a recent study, it was demonstrated that SDF-coated root dentin significantly attenuates lactate production in S. mutans48. For these reasons, SDF is considered an efficient agent for arresting caries.
In this study, biodiversity analyses using 16 s rRNA sequencing were performed to investigate the changes in the biofilm community on root dentin following SDF application. The alpha diversity indices (both Shannon and Chao1 indices) corresponding to the SDF group when data were pooled together regardless of the experiment day, were significantly greater than those corresponding to the control, suggesting that the bacterial taxa detected in the SDF group were richer and more equally distributed than those in the control group. However, most of the comparisons with respect to the experiment day showed no significant differences between the groups. These results are consistent with those of previous studies on gene sequencing in plaques, in which the effect of SDF application on bacterial profiles was investigated using plaque samples24,49. Moreover, performing the PERMANOVA test of Bray–Curtis distances revealed that SDF application had a significant effect on the bacterial communities between the groups, within the groups, and with respect to time. The PERMANOVA test also showed that the community structure corresponding to the SDF-treated biofilms was different from that corresponding to the control biofilms; this is inconsistent with the results obtained in a previous study24. This could be attributed to the differences in the experimental design; the method for normalization, including the statistical analyses conducted50; delayed colonization, which possibly influenced the number of bacterial populations observed in the community analysis; and the type of bacterial taxa in the initial biofilm.
Our differential abundance analysis revealed only four significant differences among the most frequently encountered taxa when collective data between groups were compared. Granulicatella and an unclassified genus in the Gemellaceae family were frequently detected in the SDF biofilms, whereas Porphyromonas and Neisseria were found to be significantly subtle. Further, Granulicatella and Gemella species are gram-positive facultative anaerobes, whereas Neisseria is gram-negative and predominantly colonizes aerobic environments. These microbes are considered commensal among oral flora and are frequently observed at all sites of the oral mucosal tissue51,52,53. Furthermore, oral Porphyromonas, such as P. gingivalis, is a gram-negative anaerobe that plays a key role in many oral infections, including periodontitis54. The results of this study possibly indicate a non-specific effect of SDF on the oral microbiome, given that changes in the relative abundances of both commensal and pathogenic bacteria were observed. Actinomyces were more abundant in the SDF group; however, based on the experiment day we observed that significant abundance could be detected only on days 2 and 3. This is consistent with the findings of Mei et al., who demonstrated that Actinomyces numbers tended to increase in arrested coronal caries in children after treatment with SDF49.
An evidence-based clinical practice guideline has been proposed by the American Dental Association for Evidence-Based Dentistry. According to it, biannual application of SDF is recommended to arrest advanced cavitated carious lesions on coronal surfaces, based on the highest rate of caries arrest observed in a clinical trial. It is also recommended to use SDF annually to arrest non-cavitated or cavitated root caries lesions55. Though we could not find guidelines for clinical use to prevent the onset of root caries by applying SDF to the exposed root surfaces, it has been reported that applying SDF on the exposed root surfaces may reduce the initiation of root caries56. This is also supported by the present study that shows a significant reduction in biofilm formation on demineralized root dentin in situ.
One of the limitations of this study is the short experimental period. The cultivation of biofilms was performed within four consecutive days of the start of the study owing to the practical difficulties associated with the availability of volunteers agreeing to wear the device along with their concerns regarding oral hygiene. Furthermore, since our 16S rRNA gene sequence analysis did not identify microbiota at the species level, our findings are restricted to the genus level, which can be elusive; thus, there is a necessity for careful consideration of the microbial abundance data interpretation. In conclusion, the application of SDF reduced the growth of dental biofilms grown on demineralized dentin in terms of bacterial cell viability and biofilm biomass under in situ conditions. Furthermore, even though SDF did not clearly alter the microbiome diversity and the differential abundances of specific bacterial genera, its potential antimicrobial effect is still recognized as a significant mode of action for root caries therapeutic interventions. Furthermore, its substantivity on tooth structures, particularly on the root dentin surface, would be valuable for future investigations, with a focus on establishing a protocol for SDF therapy and estimating the appropriate interval for re-application.
Conclusion
The application of SDF solution on demineralized dentin negatively affected the growth of dental biofilm by reducing bacterial cell viability and total biomass. However, bacterial composition analysis revealed that the antimicrobial effect of SDF did not directly influence the community structure of the dental biofilm. The efficiency of SDF in preventing and treating root caries may potentially arise from the biofilm inhibitory effect rather than driving the change in bacterial composition.
Methods
Volunteer recruitment
The study design, which included the usage of bovine dentin slabs inside the volunteer’s mouth, was approved by the Ethics Committee of the Osaka University Graduate School of Dentistry (approval number: H29-E17-2, approval date: 2017/9/27) and was conducted according to the guidelines of the Declaration of Helsinki. Ten healthy volunteers (students and staff of the Osaka University Graduate School of Dentistry), aged 26 to 31 years, were recruited for this study. The inclusion criteria were as follows: (1) no clinical signs of caries, gingivitis, or periodontitis, (2) no history of antibiotics usage for 3 months, (3) no history of smoking, and (4) no history of orthodontic treatment and denture use. All the volunteers were informed about the study protocols before signing an informed consent form.
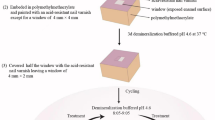
Dentin slab preparation and the in situ model
The customized in situ device used in this study was a modification of the biofilm model developed in previous studies42,57. Briefly, each intraoral device, which had left and right pieces, was designed to have eight rectangular slots for positioning the substrate used for biofilm cultivation. The slots were perforated to create a 1 mm × 3 mm opening at the buccal surface using a round diamond bur (Supplementary Fig. S1). Further, the root dentin of bovine anterior incisors was prepared for the study by cutting them into rectangular slabs. First, the bovine roots were cut along the first plane (thickness) using a cutting machine (IsoMet™; Buehler, Lake Bluff, IL, USA). Thereafter, the pieces of the dentin were further cut along the second plane (width and length) to match the designated dimensions (3 × 5 × 1 mm3) using a diamond wire saw (Well Diamond Wire Saws; Norcross, GA, USA). The remaining organic tissues in the dentin slabs were dissolved by ultrasonication with an aqueous solution of 2.5% sodium hypochlorite for 60 min (replenished every 10 min) prior to demineralizing the surface via ultrasonication with 20% citric acid for 30 min. The prepared dentin slabs were assigned to the control and SDF groups. The slabs assigned to the SDF group were treated with 38% SDF for 4 min before washing with distilled water for 30 s. Subsequently, the slabs were fit in the intraoral devices and sterilized using ethylene oxide gas (Supplementary Fig. S2).
Experimental design
The study volunteers were asked to wear the left and right pieces of the devices, each of which contained eight dentin slabs, for a total of 96 h to enable biofilm growth. Four dentin slabs with biofilms were extracted at 24, 48, 72, and 96 h, respectively, from both groups and subjected to different biofilm analyses. Throughout the experiment, all the sample collection procedures were performed at the same period for all the volunteers, who were allowed to remove the devices only during meals and when drinking. Tooth brushing was allowed but without toothpaste.
Viable cell count
After extraction, the dentin slabs were gently washed twice, vortexed for 30 s, and ultrasonicated in distilled water at 4 °C for 15 min to detach the biofilms. The resulting bacterial suspension was serially diluted by tenfold. Thereafter, droplets from each dilution were cultured on Columbia blood agar plates with 5% sheep blood (Nippon Becton Dickinson, Tokyo, Japan) and incubated aerobically and anaerobically for 48 h. After incubation, colonies were counted and expressed as log10 CFU/slab.
Quantification of biofilm-forming cells by real-time PCR
To quantify the number of biofilm-forming cells, real-time PCR was performed as previously described57. Briefly, bacterial genomic DNA was extracted from each biofilm sample using a DNA extraction kit (DNeasy PowerSoil Kit; Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at − 20 °C until further analysis. The assays (20 μL) were prepared by mixing 1 μL of extracted DNA, 10 μL of SYBR Select Master Mix (Applied Biosystems, Carlsbad, CA, USA), 0.5 μL each of bacterial universal primers 27F (5′- AGRGTTTGATCMTGGCTCAG -3′) and 338R (5′- TGCTGCCTCCCGTAGGAGT -3′). Real-time PCR (Applied Biosystems™ 7500 Fast Real-Time PCR System; Thermo Fisher Scientific, Tokyo, Japan) was performed, and standard curves were generated using S. mutans ATCC 25175 genomic DNA to amplify serial dilutions. The experiments were repeated three times per sample. Data were acquired and analyzed using the software provided by the manufacturer (Applied Biosystems 7500 System SDS software version 2.0.2; Thermo Fisher Scientific, Tokyo, Japan).
Confocal microscopy
The dentin slabs were washed in distilled water and stained using the LIVE/DEAD® Bacterial Viability Kit (BacLight™; Invitrogen, Carlsbad, CA, USA) for 30 min in the dark at room temperature. Thereafter, the slabs were observed using a confocal laser scanning microscope (LSM700; Carl Zeiss, Oberkochem, Germany), and five areas on the slabs were randomly selected and captured. The images obtained were reconstructed using an image analysis software (Imaris 9.2.1; Bitplane, Zurich, Switzerland), and the thickness of the biofilm as well as the volume of the dead and the live bacteria were assessed.
SEM observation
Biofilm samples were prepared according to a previously described protocol58. The samples were immersed in 50% Karnovsky’s solution for 30 min before they were dehydrated in a series of aqueous ethanol solutions (50, 70, 80, 90, 95, and 100%) and transferred into t-butyl alcohol. Thereafter, the specimens were freeze-dried (JFD-320; JEOL, Tokyo, Japan) and sputter-coated with platinum (Sputter Coater SC7620; Quorum Technologies, East Sussex, UK) before observation under an SEM (JSM-6390LV; JEOL, Tokyo, Japan) using the secondary electron emission mode and at an accelerating voltage of 20 kV. Magnification of 7500 was used.
16S rRNA gene amplicon sequencing
PCR amplification targeting the V1–V2 hypervariable region of the bacterial 16S rRNA was performed separately using forward and reverse primers containing the adapter sequence with barcode index; 27F (5′-ACACTCTTTCCCTACACGACGCTCTTCCG ATCT-NNNNN-AGRGTTTGATYMTGG.CTCAG-3′) and 338R (5′-GTGACTGGA GTTCAGACGTGTGCTCTTCCGATCT-NNNNN-TGCTGCCTCCCGTAGGAGT-3′). Paired-end sequencing (2 × 300 bp) of the amplicons was performed using an Illumina MiSeq platform (Illumina, San Diego, CA, USA) according to standard protocols. Further, raw sequences obtained from all samples were de-multiplexed using the Fastq barcode splitter tool from the Fastx toolkit (version 0.0.13). Only the sequences that exactly matched the start of the reading sequence or primer were extracted. After removing the primer sequence, the QIIME pipeline (version 2.0) was applied to denoise and cluster quality-checked sequences into operational taxonomic units (OTUs) at a similarity cut-off of 97% using UCLUST (version 1.2.22q). Representative sequences for each OTU were aligned using the Ribosomal Database Project Classifier (http://rdp.cme.msu.edu/) against the Greengenes database, and each sequence was assigned to the genus level. The relative abundances of the taxa and the alpha and beta diversities were computed using the QIIME script (version 2.0).
Statistical analysis
Statistical analyses were computed in R (RStudio version 1.2.1335; RStudio, Boston, MA, USA) for the Mac OS X development environment. Using the Shapiro–Wilk test, all sets of data were tested for normal distribution. The Friedman test was used to compare the mean CFU counts in aerobic and anaerobic conditions, including real-time PCR bacterial loads between groups and across days, followed by the Wilcoxon signed-rank test. Repeated measures ANOVA and multiple pairwise comparisons using Bonferroni’s correction were performed to compare the means of biofilm thickness in both groups and over different days. Further, dead and live cell volumes were calculated as ratios before analysis by the Friedman test and the Wilcoxon signed-rank test with Bonferroni’s correction. Based on the 16S rRNA sequencing data, Phyloseq and DESeq2 packages were used to perform bacterial community analysis59,60. For the alpha diversity index, Chao1 and Shannon indices, indicating taxa richness and abundance plus evenness, respectively, were determined using rarefied OTU tables. The Wilcoxon rank-sum test was used to compare the alpha diversity indices corresponding to each experiment day's control and SDF groups. Additionally, the Bray–Curtis distance matrix, a beta-diversity index, was measured from raw sequences that had been normalized based on the DESeq2 calculation. Using SHAMAN, a web application for metagenomic analysis61, a principal coordinates analysis (PCoA) plot was generated and a PERMANOVA test, which demonstrates the compositional difference across the groups of data, was performed. Comparative analysis of bacterial abundance was also performed using the DESeq2 method with a negative binomial model. The results were quantified and visualized as log2 fold change. Statistical differences (p-values) were corrected for multiple comparisons using the Benjamin-Hochberg method. For all tests, p-values < 0.05 were considered statistically significant.
Data availability
The data underlying this article cannot be shared publicly due to ethical concerns (the contents of agreements of the study). The data will be shared on reasonable request to the corresponding author.
References
Marsh, P. D., Moter, A. & Devine, D. A. Dental plaque biofilms: Communities, conflict and control. Periodontology 2000(55), 16–35. https://doi.org/10.1111/j.1600-0757.2009.00339.x (2011).
Mosaddad, S. A. et al. Oral microbial biofilms: An update. Eur. J. Clin. Microbiol. Infect. Dis. 38, 2005–2019. https://doi.org/10.1007/s10096-019-03641-9 (2019).
Mira, A., Simon-Soro, A. & Curtis, M. A. Role of microbial communities in the pathogenesis of periodontal diseases and caries. J. Clin. Periodontol. 44(Suppl 18), S23–S38. https://doi.org/10.1111/jcpe.12671 (2017).
Marsh, P. D. Are dental diseases examples of ecological catastrophes?. Microbiology 149, 279–294. https://doi.org/10.1099/mic.0.26082-0 (2003).
Marsh, P. D. Dental plaque as a biofilm and a microbial community: Implications for health and disease. BMC Oral Health 6, 1–7. https://doi.org/10.1186/1472-6831-6-S1-S14 (2006).
Bowen, W. H., Burne, R. A., Wu, H. & Koo, H. Oral Biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242. https://doi.org/10.1016/j.tim.2017.09.008 (2018).
Takahashi, N. & Nyvad, B. ecological hypothesis of dentin and root caries. Caries Res. 50, 422–431. https://doi.org/10.1159/000447309 (2016).
Vieira, A. R. & Gati, D. Elderly at greater risk for root caries: A look at the multifactorial risks with emphasis on genetics susceptibility. Int. J. Dent. 2011, 1–6. https://doi.org/10.1155/2011/647168 (2011).
Steele, J. G., Sheiham, A., Marcenes, W., Fay, N. & Walls, A. W. Clinical and behavioural risk indicators for root caries in older people. Gerodontology 18, 95–101. https://doi.org/10.1111/j.1741-2358.2001.00095.x (2001).
Yamaga, R., Nishino, M., Yoshida, S. & Yokomizo, I. Diammine silver fluoride and its clinical application. J. Osaka Univ. Dent. Sch. 12, 1–20 (1972).
Chu, C. H., Lo, E. C. M. & Lin, H. C. Effectiveness of silver diamine fluoride and sodium fluoride varnish in arresting dentin caries in Chinese pre-school children. J. Dent. Res. 81, 767–770. https://doi.org/10.1177/0810767 (2002).
Chu, C. H., Lee, A. H. C., Zheng, L., Mei, M. L. & Chan, G. C. F. Arresting rampant dental caries with silver diamine fluoride in a young teenager suffering from chronic oral graft versus host disease post-bone marrow transplantation: A case report. BMC Res. Notes 7, 3. https://doi.org/10.1186/1756-0500-7-3 (2014).
Hendre, A. D., Taylor, G. W., Chávez, E. M. & Hyde, S. A systematic review of silver diamine fluoride: Effectiveness and application in older adults. Gerodontology 34, 411–419. https://doi.org/10.1111/ger.12294 (2017).
Tan, H. P., Lo, E. C. M., Dyson, J. E., Luo, Y. & Corbet, E. F. A randomized trial on root caries prevention in elders. J. Dent. Res. 89, 1086–1090. https://doi.org/10.1177/0022034510375825 (2010).
Craig, G. G., Knight, G. M. & McIntyre, J. M. Clinical evaluation of diamine silver fluoride/potassium iodide as a dentine desensitizing agent. A pilot study. Aust. Dent. J. 57, 308–311. https://doi.org/10.1111/j.1834-7819.2012.01700.x (2012).
Knight, G. M. et al. Inability to form a biofilm of Streptococcus mutans on silver fluoride- and potassium iodide-treated demineralized dentin. Quintessence Int. 40, 155–161 (2009).
Mei, M. L. et al. Inhibitory effect of silver diamine fluoride on dentine demineralisation and collagen degradation. J. Dent. 41, 809–817. https://doi.org/10.1016/j.jdent.2013.06.009 (2013).
Chu, C. H., Mei, L. E. I., Seneviratne, C. J. & Lo, E. C. M. Effects of silver diamine fluoride on dentine carious lesions induced by Streptococcus mutans and Actinomyces naeslundii biofilms. Int. J. Paediatr. Dent. 22, 2–10. https://doi.org/10.1111/j.1365-263X.2011.01149.x (2012).
Mei, M. L. et al. Formation of fluorohydroxyapatite with silver diamine fluoride. J. Dent. Res. 96, 1122–1128. https://doi.org/10.1177/0022034517709738 (2017).
Knight, G. M. et al. An in vitro model to measure the effect of a silver fluoride and potassium iodide treatment on the permeability of demineralized dentine to Streptococcus mutans. Aust. Dent. J. 50, 242–245. https://doi.org/10.1111/j.1834-7819.2005.tb00367.x (2005).
Mei, M. L., Li, Q. L., Chu, C. H., Lo, E. C. M. & Samaranayake, L. P. Antibacterial effects of silver diamine fluoride on multi-species cariogenic biofilm on caries. Ann. Clin. Microbiol. Antimicrob. 12, 1. https://doi.org/10.1186/1476-0711-12-4 (2013).
Mei, M. L., Chu, C. H., Low, K. H., Che, C. M. & Lo, E. C. M. Caries arresting effect of silver diamine fluoride on dentine carious lesion with S mutans and L. acidophilus dual-species cariogenic biofilm. Med. Oral Patol. Oral Cir. Bucal 18, e824–e831. https://doi.org/10.4317/medoral.18831 (2013).
Xu, P. & Gunsolley, J. Application of metagenomics in understanding oral health and disease. Virulence 5, 424–432. https://doi.org/10.4161/viru.28532 (2014).
Mitwalli, H. et al. Effect of silver diamine fluoride treatment on microbial profiles of plaque biofilms from root/cervical caries lesions. Caries Res. 53, 555–566. https://doi.org/10.1159/000499578 (2019).
Watson, P. S. et al. Penetration of fluoride into natural plaque biofilms. J. Dent. Res. 84, 451–455. https://doi.org/10.1177/154405910508400510 (2005).
Prada-López, I., Quintas, V., Vilaboa, C., Suárez-Quintanilla, D. & Tomás, I. Devices for in situ development of non-disturbed oral biofilm A systematic review. Front. Microbiol. 7, 1055. https://doi.org/10.3389/fmicb.2016.01055 (2016).
Hara, A. T., Queiroz, C. S., Paes Leme, A. F., Serra, M. C. & Cury, J. A. Caries progression and inhibition in human and bovine root dentine in situ. Caries Res. 37, 339–344. https://doi.org/10.1159/000072165 (2003).
Turssi, C. P., Messias, D. F., Corona, S. M. & Serra, M. C. Viability of using enamel and dentin from bovine origin as a substitute for human counterparts in an intraoral erosion model. Braz. Dent. J. 21, 332–336. https://doi.org/10.1590/S0103-64402010000400008 (2010).
Mellberg, J. R. Hard-tissue substrates for evaluation of cariogenic and anti-cariogenic activity in situ. J. Dent. Res. 71, 913–919. https://doi.org/10.1177/002203459207100S25 (1992).
Papone, V. Y. Actinomyces. Its relation to dental plaque–root caries–actinomycosis. Odontoestomatologia 2, 47–53 (1989).
Schüpbach, P., Osterwalder, V. & Guggenheim, B. Human root caries: Microbiota in plaque covering sound, carious and arrested carious root surfaces. Caries Res. 29, 382–395. https://doi.org/10.1159/000262097 (1995).
Ellen, R. P., Banting, D. W. & Fillery, E. D. Longitudinal microbiological investigation of a hospitalized population of older adults with a high root surface caries risk. J. Dent. Res. 64, 1377–1381 (1985).
Preza, D. et al. Bacterial profiles of root caries in elderly patients. J. Clin. Microbiol. 46, 2015–2021. https://doi.org/10.1177/00220345850640121001 (2008).
Chen, L. et al. Extensive description and comparison of human supra-gingival microbiome in root caries and health. PLoS ONE 10, e0117064. https://doi.org/10.1371/journal.pone.0117064 (2015).
Grippo, J. O. & Masi, J. V. Role of biodental engineering factors (BEF) in the etiology of root caries. J. Esthet. Restor. Dent. 3, 71–76. https://doi.org/10.1111/j.1708-8240.1991.tb00813.x (1991).
Hayes, M. et al. Risk indicators associated with root caries in independently living older adults. J. Dent. 51, 8–14. https://doi.org/10.1016/j.jdent.2016.05.006 (2016).
Maroso, F. B., Gaio, E. J., Rösing, C. K. & Fernandes, M. I. Correlation between gingival thickness and gingival recession in humans. Acta Odontol. Latinoam. 28, 162–166. https://doi.org/10.1590/S1852-48342015000200011 (2015).
Mei, M. L., Lo, E. C. & Chu, C. H. Clinical use of silver diamine fluoride in dental treatment. Compend. Contin. Educ. Dent. 37, 93–98 (2016).
Mei, M. L. et al. An ex vivo study of arrested primary teeth caries with silver diamine fluoride therapy. J. Dent. 42, 395–402. https://doi.org/10.1016/j.jdent.2013.12.007 (2014).
Lansdown, A. B. G. Silver in health care: Antimicrobial effects and safety in use. Biofunct. Text. Skin. 33, 17–34. https://doi.org/10.1159/000093928 (2006).
Wu, M.-Y., Suryanarayanan, K., Van Ooij, W. J. & Oerther, D. B. Using microbial genomics to evaluate the effectiveness of silver to prevent biofilm formation. Water Sci. Technol. 55, 413–419. https://doi.org/10.2166/wst.2007.285 (2007).
Wake, N. et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ Biofilms Microbiomes https://doi.org/10.1038/npjbiofilms.2016.18 (2016).
Theilade, E., Wright, W. H., Jensen, S. B. & Löe, H. Experimental gingivitis in man: II. A longitudinal clinical and bacteriological investigation. J. Periodontal Res. 1, 1–13. https://doi.org/10.1111/j.1600-0765.1966.tb01842.x (1966).
Rosenblatt, A., Stamford, T. C. M. & Niederman, R. Silver diamine fluoride: A caries “silver-fluoride bullet”. J. Dent. Res. 88, 116–125. https://doi.org/10.1177/0022034508329406 (2009).
Ten Cate, J. M. & Featherstone, J. D. B. Mechanistic aspects of the interactions between fluoride and dental enamel. Crit. Rev. Oral Biol. Med. 2, 283–296. https://doi.org/10.1177/10454411910020030101 (1991).
Li, X. et al. Silver chloride loaded hollow mesoporous aluminosilica spheres and their application in antibacterial coatings. Mater. Lett. 105, 159–161. https://doi.org/10.1016/j.matlet.2013.04.077 (2013).
Peng, J. J. Y., Botelho, M. G. & Matinlinna, J. P. Silver compounds used in dentistry for caries management: A review. J. Dent. 40, 531–541. https://doi.org/10.1016/j.jdent.2012.03.009 (2012).
Ishiguro, T. et al. Sodium fluoride and silver diamine fluoride-coated tooth surfaces inhibit bacterial acid production at the bacteria/tooth interface. J. Dent. 84, 30–35. https://doi.org/10.1016/j.jdent.2018.12.017 (2019).
Mei, M. L. et al. Effect of silver diamine fluoride on plaque microbiome in children. J. Dent. 102, 103479. https://doi.org/10.1016/j.jdent.2020.103479 (2020).
Lin, H. & Peddada, S. D. Analysis of microbial compositions: A review of normalization and differential abundance analysis. NPJ Biofilms Microbiomes https://doi.org/10.1038/s41522-020-00160-w (2020).
Cargill, J. S., Scott, K. S., Gascoyne-Binzi, D. & Sandoe, J. A. T. Granulicatella infection: Diagnosis and management. J. Med. Microbiol. 61, 755–761. https://doi.org/10.1099/jmm.0.039693-0 (2012).
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I. & Dewhirst, F. E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. https://doi.org/10.1128/JCM.43.11.5721-5732.2005 (2005).
Liu, G., Tang, C. M. & Exley, R. M. Non-pathogenic Neisseria: Members of an abundant, multi-habitat, diverse genus. Microbiology 161, 1297–1312. https://doi.org/10.1099/mic.0.000086 (2015).
How, K. Y., Song, K. P. & Chan, K. G. Porphyromonas gingivalis: An overview of periodontopathic pathogen below the gum line. Front. Microbiol. 7, 1–14. https://doi.org/10.3389/fmicb.2016.00053 (2016).
Slayton, R. L. et al. Evidence-based clinical practice guideline on nonrestorative treatments for carious lesions: A report from the American Dental Association. J. Am. Dent. Assoc. 149, 837-849.e19. https://doi.org/10.1016/j.adaj.2018.07.002 (2018).
Wierichs, R. J. & Meyer-Lueckel, H. Systematic review on noninvasive treatment of root caries lesions. J. Dent. Res. 94, 261–271. https://doi.org/10.1177/0022034514557330 (2015).
Sotozono, M. et al. Impacts of sleep on the characteristics of dental biofilm. Sci. Rep. 11, 1–12. https://doi.org/10.1038/s41598-020-80541-5 (2021).
Asahi, Y. et al. Synergistic effects of antibiotics and an N-acyl homoserine lactone analog on Porphyromonas gingivalis biofilms. J. Appl. Microbiol. 112, 404–411. https://doi.org/10.1111/j.1365-2672.2011.05194.x (2012).
McMurdie, P. J. & Holmes, S. phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. https://doi.org/10.1371/journal.pone.0061217 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Volant, S. et al. SHAMAN: A user-friendly website for metataxonomic analysis from raw reads to statistical analysis. BMC Bioinform. 21, 345. https://doi.org/10.1186/s12859-020-03666-4 (2020).
Acknowledgements
This study was supported by JSPS KAKENHI (Grant numbers 17K11703 and 20K09953) and also partly supported by the SECOM Science and Technology Foundation.
Author information
Authors and Affiliations
Contributions
K.K.: Conceptualization, investigation, formal analysis, writing—original draft, and visualization. Y.A.: Conceptualization, methodology, formal analysis, writing—review and editing, and supervision. M.S.: Formal analysis and writing—review and editing. HMaezono, N.K., S.E., and M.H.: Writing—review and editing. HMachi: Methodology.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Klanliang, K., Asahi, Y., Maezono, H. et al. An extensive description of the microbiological effects of silver diamine fluoride on dental biofilms using an oral in situ model. Sci Rep 12, 7435 (2022). https://doi.org/10.1038/s41598-022-11477-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-11477-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.