Abstract
Prey spectra (the number and composition of captured arthropods) represent a crucial aspect of carnivorous plant ecology, yet remain poorly studied. Traditional morphology-based approaches for prey identification are time-intensive, require specialists with considerable knowledge of arthropod taxonomy, and are hampered by high numbers of unidentifiable (i.e., heavily digested) prey items. We examined prey spectra of three species of closely-related annual Drosera (Droseraceae, sundews) from tropical northern Australia using a novel DNA metabarcoding approach with in-situ macro photography as a plausibility control and to facilitate prey quantity estimations. This new method facilitated accurate analyses of carnivorous plant prey spectra (even of heavily digested prey lacking characteristic morphological features) at a taxonomic resolution and level of completeness far exceeding morphology-based methods and approaching the 100% mark at arthropod order level. Although the three studied species exhibited significant differences in detected prey spectra, little prey specialisation was observed and habitat or plant population density variations were likely the main drivers of prey spectra dissimilarity.
Similar content being viewed by others
Introduction
Carnivorous plants are characterised by adaptations to trap, kill and derive nutritional benefit from animal prey1. Since they typically grow in soils where nitrogen and phosphorus are limiting, obtaining these macronutrients by means of arthropod prey capture forms an essential component of their survival strategy2. This ecological distinction has made them a popular subject of research since the early work of Darwin3, yet studies focussing on the number and composition of captured prey (i.e., their prey spectra) remain surprisingly scarce. Only five such studies have been done in Australia, the global centre of carnivorous plant diversity with approx. 250 species4,5,6,7,8. Characterising the prey spectra of carnivorous plants is crucial for understanding their requirements for survival.
Where prey spectra of carnivorous plants have been examined, they have traditionally been analysed by collecting samples of their trapping leaves (dry or in alcohol) before identifying captured prey items by morphological features, usually under a stereo microscope (e.g.4,5,6,7,9,10,11,12,13,14). This method is extremely time-intensive and requires considerable taxonomic identification skills covering a wide range of arthropod taxa, or help of insect specialists to identify prey items. Identification may also become impossible for heavily digested prey items that have lost crucial diagnostic morphological features8. Collecting trapping leaves can also be logistically challenging in remote regions that may only be accessible by air travel during the main growing season of most carnivorous plant taxa, as such travel precludes the usage of alcohol for conserving samples. An alternative approach using in-situ macro photography of prey items was tested by Krueger et al.8. While this method allowed rapid and non-invasive documentation of carnivorous plant prey spectra data even under extreme field conditions and provided highly accurate data on prey quantity (prey count), a significant proportion (up to 80%8) of prey items remained unidentifiable. Additionally, identification below the taxonomic level of arthropod order using only in-situ macro photographs proved to be extremely difficult, and was often impossible8.
With recent advances in technologies such as high throughput DNA sequencing, DNA metabarcoding has become a promising tool for analysing environmental samples containing diverse and complex arthropod assemblages15,16,17,18,19,20,21,22. Here, DNA of all specimens contained in a sample is extracted holistically and amplified using universal barcode primers targeting the mitochondrial cytochrome c oxidase subunit I (CO1-5P) gene16,23. After sequencing, each DNA barcode sequence is subsequently compared with reference libraries on curated databases such as BOLD24,25 and NCBI GenBank (https://www.ncbi.nlm.nih.gov) to obtain taxonomic identifications. Metabarcoding thus promises to allow for taxonomic resolution at much finer scale and much higher completeness for environmental bulk samples, such as carnivorous plant prey spectra, provided reference sequences from reliably determined voucher specimens of the targeted species are available on these databases. However, metabarcoding usually does not allow for accurate prey quantity estimations (neither total count of individuals nor biomass), as the CO1 marker is not capable of distinguishing between individuals and obtained read count data are affected by the amount of DNA extracted from prey items which is likely highly variable among different prey taxa, sizes and digestion stages26,27. Indeed, the only available study analysing prey spectra of carnivorous plants using metabarcoding did not attempt to obtain prey quantity estimates for these reasons27. Thus, metabarcoding of carnivorous plant prey assemblages allows for analysis of what is captured (i.e., taxonomic analysis, such as prey composition), but not how much is captured (i.e., prey quantity or biomass or even relative prey abundances). Crucially, metabarcoding approaches also require plausibility controls as they are extremely sensitive and therefore prone to false positive identifications by even minuscule DNA contamination15,28.
To study the prey spectra of three closely related Western Australian carnivorous plant species, we developed and evaluated a novel approach combining DNA metabarcoding and in-situ macro photography. All three species (Drosera finlaysoniana, D. hartmeyerorum and D. margaritacea) belong to Drosera sect. Arachnopus and have similar, linear adhesive trapping leaves. The prey spectra of D. finlaysoniana and D. margaritacea have not been characterised previously, that of D. hartmeyerorum was studied by Krueger et al.8. This group of sundews is of particular interest for prey spectra research as they are annuals which appear to depend heavily on supplementary nutrition derived from prey capture29 and furthermore have evolved highly specific morphological features which have been hypothesised to function as prey attractants, such as trap scent and specialised leaf trichomes8,30,31,32. By using in-situ macro photography (as established by Krueger et al.8) as a plausibility control to detect false positive identifications or contaminations in the metabarcoding data and to enable the calculation of total prey quantity, we aimed to obtain, for the first time, carnivorous plant prey spectra data of unprecedented taxonomic resolution and completeness. At coarse taxonomic levels, we expected that this new approach would yield prey spectra similar to that previously observed in D. sect. Arachnopus8. We further expected to confirm significant prey spectra differences between study sites8,27 and among species with different trapping leaf sizes8,33.
Materials and methods
Study sites
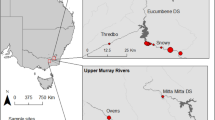
Plants were sampled at three study sites in Western Australia in July 2020 (Table 1). Sites 1 and 3 featured large plant populations in freshwater lake margin habitats (especially at Site 1 which consisted of an extremely large and high-density population of D. finlaysoniana, comprising millions of individuals), while only ca. 100 plants of D. margaritacea were found in a small and dry artificial drainage channel at Site 2 (Supplementary Fig. S1). All studied plants were identified by T.K. and voucher specimens (Krueger 6, Krueger 7 and Krueger 8) were deposited in the Western Australian Herbarium (PERTH). Collection of plant material complied with relevant institutional, national, and international guidelines and legislation. A flora taking licence (FT61000038-2) was obtained from the Western Australian Department of Biodiversity, Conservation and Attractions. Plant material was exported to Germany for scientific study under export permit WT2020-001235 issued by the Australian Department of Agriculture, Water and the Environment.
Leaf sampling
Ten plants from each population were randomly selected for study, and five randomly chosen leaves per plant were removed with forceps (scientific collection license FT61000038-2). Each sample thus constituted of five leaves belonging to a single individual and there were ten individuals (replicates) per species. Only fully developed, mucilage-secreting (i.e., “active”) leaves were collected as the heavily digested prey items found on old leaves would complicate both quantitative analysis (e.g., by counting fragmented prey items multiple times) and metabarcoding (due to heavily degraded prey DNA). Three species of D. sect. Arachnopus were studied (N = 30), consisting of ten replicates per species and a total of 150 collected leaves (Table 1).
In-situ macro photography
Detached sampled leaves were carefully placed on paper sheets (with the tentacle-bearing, sticky, prey-containing side facing upwards so that no prey items would be lost by adhering to the sheets) with a scale (ruler) to record leaf length. Paper sheets were not re-used to prevent cross-contamination among samples. 447 macro photographs of the collected leaves were taken using a digital camera with a macro lens, and total prey was counted for each sample based on these images. All visible prey items were counted, regardless of digestion state. Other plant material sticking to the leaves (such as seed) did not trigger any tentacle motion and was thus easy to distinguish from prey items. While some prey items may have originated from the same captured prey animal (which was subsequently disintegrated), such cases were often uncertain or difficult to reconstruct and therefore always counted as if they were separate prey. The prey count value per sample was defined as the total number of observed prey items on five randomly selected leaves of a single Drosera individual. Finally, the strong effect of leaf size on prey counts8 was mitigated by calculating prey count values per cm of leaf length (because even within a single individual of D. sect. Arachnopus leaf size can be highly variable8). All three studied Drosera species have a narrowly linear-lanceolate leaf shape and prey counts per cm of leaf length thus closely approximate prey counts per leaf area. For leaf length, the arithmetic mean of the five collected leaves was used for each sample.
Sample preparation, lysis and DNA extraction
After all leaves were measured and photographed, the five leaves belonging to each individual plant were pooled in 15 ml sterile sample tubes containing 96% denatured ethanol and stored at ~ 5 °C. The ethanol supernatant of all 30 samples was carefully removed immediately before shipment to the Botanische Staatssammlung Munich (SNSB-BSM, Germany) for further processing (export permit WT2020-001235), where 96% denatured ethanol was re-added to the samples. Prey items were separated from the leaves in order to reduce the amount of plant tissue per sample relative to the amount of insect tissue (Drosera leaf tissue is rich in polyphenols and polysaccharides which are known to interfere with DNA extraction and amplification34); for this, prey items still attached to the leaves were carefully detached from the leaves using forceps under a stereomicroscope, and prey items were transferred into 2 ml lysis cups that were filled with 96% denatured ethanol. Therefore, most of the leaf tissue (except for the tentacles) was removed before lysis.
For better lysis and DNA extraction of the insect tissue, samples were subsequently homogenised for 30–60 s using a FastPrep96 (MP Biomedicals) with addition of sterile steel beads. DNA extraction was conducted following the protocol of Ivanova et al.35. Briefly, 200 µl of insect lysis buffer with proteinase K in a 1:20 ratio was added to the sample. Polyvinylpyrrolidone was added (until 2% concentration in the solution) to block inhibiting substances such as polyphenols. Samples were incubated overnight at a temperature of 56 °C and lysates were frozen before extraction.
DNA amplification and metabarcoding
DNA metabarcoding was conducted at the AIM Lab (AIM—Advanced Identification Methods GmbH, Leipzig, Germany), following the methodology of Morinière et al.16, Hardulak et al.21, and Hausmann et al.22,36.
From each sample, 5 µl of extracted total DNA was used for PCR, along with Plant MyTAQ (Bioline, Luckenwalde, Germany) and High Throughput Sequencing (HTS) adapted mini-barcode primers targeting the mitochondrial cytochrome c oxidase subunit I (CO1-5P) (primers and amplification following Morinière et al.16). Amplification success and fragment lengths were verified by gel electrophoresis. Amplified DNA was cleaned up using a 1% sodium acetate and 70% ethanol precipitation method37 and resuspended in 50 µl purified water for each sample before proceeding. Illumina Nextera XT (Illumina Inc., San Diego, USA) indices were ligated to the samples in a second PCR reaction applying the same annealing temperature as for the first PCR reaction but with only seven cycles, and ligation success was confirmed by gel electrophoresis. DNA concentrations were measured using a Qubit fluorometer, and adjusted to 40 µl pools containing equimolar concentrations of 100 ng/µl DNA template each. Pools were purified using MagSi-NGSprep Plus (Steinbrenner Laborsysteme GmbH, Wiesenbach, Germany) beads. A final elution volume of 20 µl was used. High Throughput Sequencing (HTS) was performed on an Illumina MiSeq (Illumina Inc., San Diego, USA) using v3 chemistry (2 × 300 basepairs, 600 cycles, maximum of 25 million paired-end reads).
Barcode sequence analysis, processing and OTU identification
FASTQ files were combined and sequence processing was performed with the VSEARCH v2.4.3 suite38 and cutadapt v1.1439. Since not all of the sequenced samples yielded reverse reads of high enough quality to enable paired-end merging, only forward reads were utilised. Reads were removed in cases where either the number of mismatches was > 5, where the alignment was shorter than 16 basepairs, or where identity percentage of the alignment was < 90%. Forward primers were removed with cutadapt. Quality filtering was done with the fastq_filter program of VSEARCH (fastq_maxee 2, minimum length of 100 bp). Sequences were dereplicated with derep_fulllength, first at the sample level, and then concatenated into one fasta file, which was then dereplicated. Chimeric sequences were filtered out from the large fasta file using uchime_denovo. Remaining sequences were clustered into Operational Taxonomic Units (OTUs) at 97% identity with cluster_size, and an OTU table was created with usearch_global. To reduce false positives, a cleaning step was employed which excluded read counts in the OTU table of less than 0.01% of the total. OTUs were blasted against a custom database downloaded from BOLD (on 03 February 2021) and NCBI GenBank (February 2020), including taxonomy and BIN (Barcode Index Number) information (contained in the BOLD database), by using Geneious (v.10.2.5; Biomatters, Auckland, New Zealand) and following the methods described in Morinière et al.16. The resulting csv file which included the OTU ID, BOLD Process ID, BIN, Hit-%-ID value (percentage of overlap similarity (identical base pairs) of an OTU query sequence with its closest counterpart in the database), length of the top BLAST hit sequence, and phylum, class, order, family, genus, and species information for each detected OTU was exported from Geneious and combined with the OTU table generated by the bioinformatic pipeline (Supplementary Data S1). A consensus taxonomy of the two BLASTs (i.e., only showing the highest taxonomic level of agreement among BOLD and NCBI GenBank for each OTU) was used in subsequent analyses (Supplementary Data S1).
Sample pooling, data exclusion and plausibility control
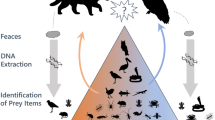
OTUs were first pooled to the level of arthropod family, as prey spectra analysis was not conducted below this taxonomic level and only 303 of the 739 retrieved OTUs could be identified to genus or below by metabarcoding (Supplementary Data S1). This also resulted in the exclusion of 87 OTUs above the taxonomic level of organismic order. In addition, microorganisms (such as the arthropod intracellular bacteria of the genus Wolbachia), marine taxa, fungi and other obvious contaminants (such as Homo sapiens–referring to contamination during human handling and processing of samples) were excluded from analysis. The rather ubiquitous phytophagous mealybugs and mites of Pseudococcidae, Trombidiformes and Mesostigmata were not considered to have been captured as prey, but rather because they parasitised the collected plant tissues, and were thus also excluded. The in-situ macro photographs obtained during sampling were used as a plausibility control of the prey spectra data generated by metabarcoding. Each taxon in each sample was carefully attempted to be matched with one or several of the prey items visible in the photographs (Fig. 1). This pictorial plausibility control was conducted conservatively, as taxa were only excluded from further analysis if they consisted of large prey animals (such as, for example, wasps, beetles or moths, documented for each case in Supplementary Data S2) which would have been clearly visible in the pictures if they were truly present. Families mostly consisting of small prey animals were generally impossible to confirm or exclude by pictorial plausibility control, as small unidentifiable “crumbs” of prey material were present on most leaves (see8). Data on prey spectra composition was compiled and analysed as presence/absence only, because metabarcoding does not allow for accurate estimations of prey quantity16,26,27. Finally, the number of samples in which each prey taxon was present was counted for each Drosera species, as well as across all three species.
Example of pictorial plausibility control of DNA metabarcoding using in-situ macro photography. Left image is a macro photograph of two of the five leaves in sample 2 of Drosera margaritacea, on the right is a table of prey families detected by metabarcoding for the same sample showing their read counts in the right-bound column. Only the four prey groups with highest read counts are shown. Colours match detected prey families with visible prey items in the macro photograph (pictorial plausibility control). Picture by T. Krueger.
Statistical analysis
Prey spectra composition was compared between all three species (including all pairwise comparisons) by using analysis of similarity (ANOSIM) in PRIMER 740. After creating Bray–Curtis resemblance matrices, prey spectra dissimilarity was quantified using the ANOSIM R-statistic which ranges from 0 (100% similarity) to 1 (0% similarity)40. No data transformations were required, as metabarcoding data were treated as presence/absence only. Subsequently, similarity percentages (SIMPER) were calculated in PRIMER 7 to identify prey groups contributing most to dissimilarity (more than 15% for arthropod orders and the five taxa contributing most to dissimilarity for arthropod families8).
Total numbers of captured prey per cm of leaf length (as determined by analysis of in-situ prey pictures) were compared between all three species using non-parametric Kruskal–Wallis tests with Dunn-Bonferroni post-hoc pairwise comparisons.
Results
Prey spectra detected by DNA metabarcoding
DNA metabarcoding confirmed 92 arthropod families belonging to 12 orders caught as prey across all 30 Drosera samples (Supplementary Table S1; Figs. 2, 3). Hits from an additional 25 arthropod families were excluded by pictorial plausibility control, most of them detected in D. hartmeyerorum samples 1, 4 and 9 (Supplementary Data S2). We found no instance of any prey item being clearly identifiable in the macro photographs but not present in the barcoding data. Of the 739 retrieved OTUs, 71%, 41% and 17% were identified to family-, genus- and species-level, respectively (Supplementary Data S1).
Examples of captured arthropod prey detected and correctly identified by DNA metabarcoding in three Western Australian species of Drosera sect. Arachnopus. The lowest taxonomic level determined by metabarcoding and the corresponding family, order and BOLD Barcode Index Number (BIN) is indicated. (a) Symplecta sp. (Limoniidae, Diptera, BOLD:AAF8963) captured by D. finlaysoniana (Sample 5). (b) Praxis marmarinopa (Erebidae, Lepidoptera, BOLD:AAC9474) captured by D. finlaysoniana (Sample 9). (c) 2 individuals of Utetheisa lotrix (Erebidae, Lepidoptera, BOLD:AAA4528) captured by D. margaritacea (Sample 2). (d) Cecidomyiidae (Diptera, BOLD:ACK2565) captured by D. margaritacea (Sample 9). (e) Early instar nymph of Gryllotalpa pluvialis (Gryllotalpidae, Orthoptera, BOLD:AAF7358) captured by D. hartmeyerorum (Sample 1). (f) Nysius plebeius (Lygaeidae, Hemiptera, BOLD:AAI3382) captured by D. hartmeyerorum (Sample 7). All pictures by T. Krueger.
Curculionidae was the family most commonly excluded by pictorial plausibility control as these characteristic weevil beetles were clearly not present, at least as prey (but possibly as eggs in the plant tissue in the case of phytophagous species), in nine of the thirteen samples where they were detected by metabarcoding (in the remaining four samples they were either confirmed by the pictorial plausibility control or not excluded with certainty; see Supplementary Data S2).
Ten of the twelve detected arthropod orders were insects, with only Araneae (spiders, Arachnida, present in 30% of total samples) and Entomobryomorpha (springtails, Collembola, present in 10% of total samples) not belonging to this class (Fig. 3). These two orders were also the only orders exclusively consisting of non-flying prey. Although some of the captured insect families such as Formicidae (ants, Hymenoptera, present in 17% of samples) and larvae of Gryllotalpidae (mole crickets, Orthoptera, larvae only present in sample 1 of D. hartmeyerorum; Fig. 2e) include non-flying prey taxa, in the majority of samples only flying adult insects were detected as prey.
The prey orders Diptera and Hemiptera were confirmed to be present in all 30 samples (100%), while Hymenoptera (87%), Lepidoptera (77%) and Thysanoptera (57%) were detected in more than half of samples (Fig. 3). The most commonly (≥ 50%) detected prey groups were “Other Hemiptera” (i.e., hemipterans which could not be assigned by metabarcoding to any family; present in 97% of samples), Hemiptera–Cicadellidae (83%), “Other Diptera” (73%), Diptera–Cecidomyiidae (70%) and Hemiptera–Lygaeidae (70%; Fig. 2f; Supplementary Table S1).
Prey families detected in more than 50% of samples in each of the three species were Diptera–Cecidomyiidae (60–90%; Fig. 2d) and Hemiptera–Cicadellidae (50–90%; Supplementary Table S1). “Other Hemiptera” were present in 90–100% of samples of each species, but the data did not allow for exact identification to family-level.
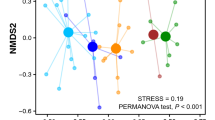
ANOSIM indicated that differences in the prey spectra between the three species were highly significant at prey family-level (R = 0.784, P < 0.001) but non-significant at the level of order (R = 0.079, P = 0.063). Additionally, all three species-pairwise comparisons at prey family-level were significant, with the highest R value observed in the comparison between D. margaritacea and D. finlaysoniana (ANOSIM R = 0.918, P < 0.001; Table 2). The only significant pairwise comparison at prey order-level was D. margaritacea–D. finlaysoniana (ANOSIM R = 0.134, P = 0.046; Table 2). SIMPER analysis indicated that no single prey family contributed more than 5% to prey spectra dissimilarity in any of the three pairwise comparisons (Table 2). Aleyrodidae (Hemiptera) contributed most to dissimilarity in both pairwise comparisons involving D. margaritacea (this prey family was detected in much more samples of this species), while Lygaeidae had the highest contribution in the SIMPER comparison of D. finlaysoniana and D. hartmeyerorum (where it was more commonly detected in the latter species; Table 2). However, the individual contributions to dissimilarity of most prey families were generally very similar within the pairwise species comparisons (Table 2). When analysed at order-level, Lepidoptera contributed most to prey dissimilarity in both the D. margaritacea–D. finlaysoniana and D. margaritacea–D. hartmeyerorum comparisons (in both cases detected much less commonly in the D. margaritacea samples) but did not contribute more than 15% to dissimilarity in the D. finlaysoniana–D. hartmeyerorum comparison (Table 2). SIMPER analysis further indicated that all pairwise comparisons among species showed higher average dissimilarity than samples of the same species.
Observed total numbers of captured prey
Total prey capture per cm of leaf length, as observed by counting prey items in the in-situ macro photographs, did differ significantly among all three studied Drosera species (Kruskal–Wallis test, H = 19.19, P < 0.001) and in the two pairwise comparisons D. margaritacea–D. finlaysoniana (P < 0.001) and D. finlaysoniana–D. hartmeyerorum (P = 0.004). Prey numbers did not differ in the comparison D. margaritacea–D. hartmeyerorum (P = 0.966). Among the three species, D. margaritacea featured the highest average number (2.25 ± 0.65) of prey items per cm of leaf length (Fig. 4). The average measured leaf length of this species was 7.1 ± 1.3 cm (Supplementary Table S2). For D. hartmeyerorum, the average number of prey items per cm of leaf length was 1.80 ± 0.50, with an average leaf length in this species of 5.3 ± 1.1 cm (Fig. 4; Supplementary Table S2). Despite D. finlaysoniana having by far the largest leaves (average leaf length of 10.4 ± 0.6 cm; Supplementary Table S2), this species had the lowest observed number of prey items per cm of leaf length of the three species (0.81 ± 0.29; Fig. 4).
Discussion
A combined DNA metabarcoding/in-situ macro photography approach to reliably analyse carnivorous plant prey spectra
Results indicate that DNA metabarcoding allows for reliable analysis of prey spectra composition in carnivorous plants at a taxonomic resolution and level of completeness unachievable by traditional morphology-based approaches (as performed, for example, by4,5,6,7,9,10,11). Even in remote tropical northern Western Australia, where many (if not most) arthropod species have not yet been accessioned into the BOLD or GenBank barcode reference libraries, this method identified over 90% of obtained OTUs from our sample set; most of them at family-level, but 41% to genus-level, and 17% even down to species rank (Supplementary Data S1). Lekesyte et al.27 were able to identify 80% of the analysed prey items found on D. rotundifolia in England to species-level. However, their sampling was performed in western Europe, whose entomofauna is comparatively well studied taxonomically and has an excellent coverage in the BOLD reference library of DNA barcodes41. New insect barcodes are regularly added to the BOLD library through large-scale initiatives such as the international Barcode of Life Project (iBOL; https://ibol.org/) and its Australian node Australian Barcode of Life Network (ABOLN), hence accuracy of future metabarcoding research performed in Australia can be expected to increase to similar levels soon.
In-situ macro photography was found to provide a valuable plausibility control tool for the prey taxa identified by metabarcoding. While many of the smaller prey taxa detected by metabarcoding were impossible to identify in the in-situ macro photographs due to their tendency to quickly degenerate after digestion into small, shapeless “crumbs”8, this control method considerably reduced the amount of prey taxa detected which were not actually present as prey in the Drosera samples. This flaw of metabarcoding is most commonly a consequence of procedural errors resulting in cross-contamination within the DNA extraction procedure27, usually resulting in low read numbers. However, in-situ macro photographs may also fail to detect species if prey captured by the sundew escaped from the trap33,42, or was stolen by larger animals. In both cases, a DNA imprint left on the Drosera leaves as excretions, detached scales, hairs or, frequently, as autotomised (shedded) body parts42 could have been detected by metabarcoding. Additionally, some barcoding-detected taxa may not constitute prey if they were associated with another captured prey taxon (either as part of its diet, or as a parasite). The latter may explain some barcode hits for taxa not immediately apparent from the in-situ macro photographs, as they are (endo)parasites of captured prey taxa. This was likely the case in the detected Strepsiptera (stylops) which are frequently contained as larvae and adult females in their hymenopteran and orthopteran hosts43. However, insect endoparasites and other non-obvious prey taxa were by default not excluded by the very conservative approach of pictorial plausibility control. Additionally, in the case of endoparasites, these organisms would also contribute to plant nutrition as “bycatch” after being digested together with their host, despite not having been actively attracted to the carnivorous traps. Finally, the control method tested in this study showed that even heavily digested prey items in the samples had sufficient amounts of intact (mitochondrial) DNA present to be detected by metabarcoding, as we found no instance of any prey item being clearly identifiable in the macro photographs but not present in the barcoding data.
Prey spectra composition of the studied Drosera species
The analysed prey spectra of the three studied species from D. sect. Arachnopus most commonly contained flying insects (especially of the orders Diptera and Hemiptera, both present in 100% of the samples; Fig. 3), thus confirming earlier in-situ macro photography-based studies of closely-related D. sect. Arachnopus species by Krueger et al.8. All members of D. sect. Arachnopus are characterised by a large, erect growth habit and thread-like aerial leaves which usually do not contact the ground8,32, thereby excluding most ground-dwelling arthropods as prey. This result is also similar to other prey spectra studies of erect-leaved Drosera from different geographic areas, where flying insects (particularly Diptera) unanimously comprised almost the entire recorded prey5,11,44. Furthermore, this study confirmed the result of Krueger et al.8 that Hemiptera—and within this order especially the Cicadellidae—are exceptionally common in the prey spectra of D. sect. Arachnopus compared with all other, previously studied Drosera. A possible explanation for this may be the relatively high abundance of Cicadellidae in tropical habitats45 compared to subtropical or temperate habitats where the above-mentioned previous Drosera prey spectra studies were conducted.
Of the five most commonly detected orders, Lepidoptera generally comprised the largest prey items in terms of body size or wingspan, respectively. This prey order was exceptionally common in D. finlaysoniana, being present in 100% of samples and also visually conspicuous in the in-situ photographs. Since this Drosera species had by far the largest trapping leaves among the three species studied with an average leaf length of 10.4 ± 0.6 cm (Suppl Appendix S7), and exhibits the largest leaves in D. section Arachnopus32, this may represent an example of large prey items being more easily captured by species with larger trapping leaves33. Additionally, the sampled population of D. finlaysoniana was huge and dense (see Supplementary Figure S1), probably attracting larger prey and enabling capture of larger prey items by “collective” trapping46. Alternatively, Fleischmann30 suggested that captured Lepidoptera themselves could attract further individuals of the same species by pheromone release, potentially explaining the very high numbers of this insect order observed in D. finlaysoniana.
Differences among observed prey spectra
Comparison of prey spectra between the three studied Drosera species revealed significant differences at arthropod family-level but not at the higher level of arthropod orders, indicating that at a coarse taxonomic resolution, the same five arthropod orders (Diptera, Hemiptera, Hymenoptera, Lepidoptera and Thysanoptera) generally comprise most of the prey in D. sect. Arachnopus, regardless of given Drosera species or habitat. However, as strong differences were discovered in the ANOSIM comparison at family-level, it can be concluded that differences might likely increase with finer taxonomic resolution of prey taxa, a conclusion also reached by the carnivorous plant prey spectra meta-analysis of Ellison & Gotelli47. While these differences may be partially attributed to different morphological traits of the three species such as leaf scent8,30 or eglandular appendages31, the very high ANOSIM R-values returned and the large number of prey families contributing nearly equally to dissimilarity (Table 2) indicate that the most likely explanation is very different available prey spectra at the three study sites. Indeed, significant differences among different study sites, even within the same species, were previously reported for Drosera rotundifolia by Lekesyte et al.27 and for four species from D. sect. Arachnopus by Krueger et al.8. Notably, the three study sites feature different habitat types and climate regimes (Supplementary Fig. S1).
Analyses indicate that there is likely little specialisation in prey capture by the three studied Drosera species. For example, the relatively high detection rate of Lepidoptera in the samples of D. finlaysoniana and D. hartmeyerorum compared to D. margaritacea may be explained by the lake margin habitats of the former two species, while the latter species was found in a completely dry drainage channel lacking any nearby waterbodies (Supplementary Fig. S1). Lepidoptera are likely to occur in much higher concentrations near water sources, especially during the dry season (May to November) when the surrounding areas are lacking other water sources (G. Bourke in Fleischmann30).
Estimating prey quantity
In addition to providing a plausibility control for the compositional prey analysis by metabarcoding, the in-situ macro photography method facilitated an estimation of prey quantity per sample. Metabarcoding by itself is currently not a reliable tool for prey quantification due to the lack of a linear relationship between the number of sequence reads and organism biomass26,27.
In contrast to Krueger et al.8, who generally found more prey items on larger trapping leaves in species of D. sect. Arachnopus (even when values were compared as per cm of trapping leaf length), the species with the largest leaves studied here (D. finlaysoniana) captured significantly less prey items than the smaller-leaved species D. margaritacea and D. hartmeyerorum (Fig. 4). However, while Krueger et al.8 was able to compare sympatric species (thus minimising any potential effects of the habitat or region on prey spectra), the three species in this study were studied at three different, geographically distant sites. While it is possible that overall prey abundance in the habitat was much lower at the D. finlaysoniana study site (Site 1), it can be hypothesised that the low total prey capture observed in this species may be due to the very large and extremely dense population resulting in strong intraspecific competition for prey (see Supplementary Fig. S1). This effect of population structure on prey capture has also been observed by Gibson48 and Tagawa and Watanabe46 who found a significant negative correlation between total prey capture and population density in different species of Drosera.
Conclusions and outlook
Our study is the first to employ a DNA metabarcoding approach supported by controls for species presence to analyse carnivorous plant prey spectra. When combined with in-situ macro photography, this method is clearly superior in terms of taxonomic resolution and completeness for analysis of environmental bulk samples (containing different organisms in highly variable states of preservation), as used here for the reconstruction of prey spectra of carnivorous plants. The capability of this method increases with new reference barcodes being regularly added to DNA barcode libraries (such as BOLD and NCBI GenBank) and it thus has the potential to become the standard methodology for future carnivorous plant prey spectra research.
Additional studies are needed to test this method for other carnivorous plant species and genera, especially those possessing different trap types. Within Western Australia, three additional trap types occur: snap traps (Aldrovanda), suction traps (Utricularia) and pitfall traps (Cephalotus). In particular, it might be expected that in-situ macro photography will not work as well for the extremely small, typically submerged traps of Aldrovanda and Utricularia (which also completely enclose their captured, microscopic prey items49), potentially necessitating usage of alternative control methods for metabarcoding data. Furthermore, even within Drosera (adhesive traps) some species may require adjustments to the methodology presented here as they accumulate captured prey in a central point via tentacle movement (e.g., many climbing tuberous Drosera) or their leaves may be very difficult to place on paper sheets with the sticky side facing upwards (e.g., all pygmy Drosera). The latter problem may be solved by using reverse action forceps and photographing the leaves while held in place by the forceps.
Extensive sampling of sites with co-occurring species from D. sect. Arachnopus is clearly required to better understand the ecological role of trap scent and eglandular appendages in this section. For example, manipulation experiments involving the removal of all yellow blackberry-shaped appendages of D. hartmeyerorum (which have been hypothesised to function as visual prey attractants31) and subsequent metabarcoding prey spectra comparisons of mutilated plants lacking emergences with control plants are proposed. Potential effects of population density on prey spectra (as hypothesised here for D. finlaysoniana) could be studied by comparing prey spectra of individual plants from within mass populations with more exposed-growing individuals of the same population.
References
Juniper, B. E., Robins, R. J. & Joel, D. M. The Carnivorous Plants. (Academic Press, 1989).
Adamec, L. & Pavlovič, A. Mineral nutrition of terrestrial carnivorous plants. In Carnivorous Plants: Physiology, Ecology, and Evolution (eds. Ellison, A. & Adamec, L.) 285–293 (Oxford University Press, 2018).
Darwin, C. Insectivorous Plants. (John Murray, 1875).
Watson, A. P., Matthiessen, J. N. & Springett, B. P. Arthropod associates and macronutrient status of the red-ink sundew (Drosera erythrorhiza Lindl.). Aust. J. Ecol. 7(1), 13–22. https://doi.org/10.1111/j.1442-9993.1982.tb01296.x (1982).
Verbeek, N. A. M. & Boasson, R. Relationship between types of prey captured and growth form in Drosera in southwestern Australia. Aust. J. Ecol. 18(2), 203–207. https://doi.org/10.1111/j.1442-9993.1993.tb00444.x (1993).
Jobson, R. W. & Morris, E. C. Feeding ecology of a carnivorous bladderwort (Utricularia uliginosa, Lentibulariaceae). Aust. Ecol. 26(6), 680–691. https://doi.org/10.1046/j.1442-9993.2001.01149.x (2001).
Płachno, B. J., Wołowski, K., Fleischmann, A., Lowrie, A. & Łukaszek, M. Algae and prey associated with traps of the Australian carnivorous plant Utricularia volubilis (Lentibulariaceae: Utricularia subgenus Polypompholyx) in natural habitat and in cultivation. Aust. J. Bot. 62(6), 528–536. https://doi.org/10.1071/BT14176 (2014).
Krueger, T., Cross, A. T. & Fleischmann, A. Size matters: trap size primarily determines prey spectra differences among sympatric species of carnivorous sundews. Ecosphere 11(7), e03179. https://doi.org/10.1002/ecs2.3179 (2020).
Zamora, R. The feeding ecology of a carnivorous plant (Pinguicula nevadense): prey analysis and capture constraints. Oecologia 84(3), 376–379. https://doi.org/10.1007/BF00329762 (1990).
Chin, L., Chung, A. Y. & Clarke, C. Interspecific variation in prey capture behavior by co-occurring Nepenthes pitcher plants: Evidence for resource partitioning or sampling-scheme artifacts?. Plant Signal. Behav. 9(1), e27930. https://doi.org/10.4161/psb.27930 (2014).
Costa, J. et al. Arthropods associated with the carnivorous plant Drosera latifolia (Droseraceae) in an area of Atlantic Forest (southeastern Brazil). Acta Biol. Paranaense 43(1–2), 61–68. https://doi.org/10.5380/abpr.v43i0.38097 (2014).
Bertol, N., Paniw, M. & Ojeda, F. Effective prey attraction in the rare Drosophyllum lusitanicum, a flypaper-trap carnivorous plant. Am. J. Bot. 102(5), 689–694. https://doi.org/10.3732/ajb.1400544 (2015).
Annis, J., Coons, J., Helm, C. & Molano-Flores, B. The role of red leaf coloration in prey capture for Pinguicula planifolia. Southeast. Nat. 17(3), 433–437. https://doi.org/10.1656/058.017.0308 (2018).
Horstmann, M., Fleischmann, A., Tollrian, R. & Poppinga, S. Snapshot prey spectrum analysis of the phylogenetically early-diverging carnivorous Utricularia multifida from U. section Polypompholyx (Lentibulariaceae). PLoS ONE 16(4), e0249976. https://doi.org/10.1371/journal.pone.0249976 (2021).
Ji, Y. et al. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecol. Lett. 16(10), 1245–1257. https://doi.org/10.1111/ele.12162 (2013).
Morinière, J. et al. Species identification in malaise trap samples by DNA barcoding based on NGS technologies and a scoring matrix. PLoS ONE 11(5), e0155497. https://doi.org/10.1371/journal.pone.0155497 (2016).
Morinière, J. et al. A DNA barcode library for 5200 German flies and midges (Insecta: Diptera) and its implications for metabarcoding-based biomonitoring. Mol. Ecol. Resour. 19(4), 900–928. https://doi.org/10.1371/journal.pone.0155497 (2019).
Bittleston, L. S., Baker, C. C., Strominger, L. B., Pringle, A. & Pierce, N. E. Metabarcoding as a tool for investigating arthropod diversity in Nepenthes pitcher plants. Austral Ecol. 41(2), 120–132. https://doi.org/10.1111/aec.12271 (2016).
Fernandes, K. et al. Invertebrate DNA metabarcoding reveals changes in communities across mine site restoration chronosequences. Restor. Ecol. 27(5), 1177–1186. https://doi.org/10.1111/rec.12976 (2019).
Hardulak, L. A. Development and improvement of next generation sequencing pipelines for mixed and bulk samples of German fauna. Doctoral dissertation. Ludwig-Maximilians-University Munich, Germany. Available from https://edoc.ub.uni-muenchen.de/27000/ (2020).
Hardulak, L. A. et al. DNA metabarcoding for biodiversity monitoring in a national park: Screening for invasive and pest species. Mol. Ecol. Resour. 20(6), 1542–1557. https://doi.org/10.1111/1755-0998.13212 (2020).
Hausmann, A. et al. Toward a standardized quantitative and qualitative insect monitoring scheme. Ecol. Evol. 10(9), 4009–4020. https://doi.org/10.1002/ece3.6166 (2020).
Hebert, P. D., Ratnasingham, S. & De Waard, J. R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Ser. B 270, S96–S99. https://doi.org/10.1098/rsbl.2003.0025 (2003).
Ratnasingham, S. & Hebert, P. D. BOLD: The barcode of life data system. Mol. Ecol. Notes 7(3), 355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x (2007).
Ratnasingham, S. & Hebert, P. D. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE 8(7), e66213. https://doi.org/10.1371/journal.pone.0066213 (2013).
Deagle, B. E., Thomas, A. C., Shaffer, A. K., Trites, A. W. & Jarman, S. N. Quantifying sequence proportions in a DNA-based diet study using Ion Torrent amplicon sequencing: which counts count?. Mol. Ecol. Resour. 13(4), 620–633. https://doi.org/10.1111/1755-0998.12103 (2013).
Lekesyte, B., Kett, S. & Timmermans, M. J. What’s on the menu: Drosera rotundifolia diet determination using DNA data. J. Lundy Field Soc. 6, 55–64 (2018).
Creedy, T. J., Ng, W. S. & Vogler, A. P. Toward accurate species-level metabarcoding of arthropod communities from the tropical forest canopy. Ecol. Evol. 9(6), 3105–3116. https://doi.org/10.1002/ece3.4839 (2019).
Karlsson, P. S. & Pate, J. S. Contrasting effects of supplementary feeding of insects or mineral nutrients on the growth and nitrogen and phosphorous economy of pygmy species of Drosera. Oecologia 92(1), 8–13. https://doi.org/10.1007/BF00317256 (1992).
Fleischmann, A. Olfactory prey attraction in Drosera?. Carniv. Plant Newsl. 45(1), 19–25 (2016).
Hartmeyer, I. & Hartmeyer, S. Drosera hartmeyerorum—Der Sonnentau mit Lichtreflektoren. Das Taublatt 56, 4–8 (2006).
Krueger, T. & Fleischmann, A. A new species of Drosera section Arachnopus (Droseraceae) from the western Kimberley, Australia, and amendments to the range and circumscription of Drosera finlaysoniana. Phytotaxa 501(1), 56–84 (2021).
Gibson, T. C. Differential escape of insects from carnivorous plant traps. Am. Midl. Nat. 125(1), 55–62. https://doi.org/10.2307/2426369 (1991).
Fleischmann, A. & Heubl, G. Overcoming DNA extraction problems from carnivorous plants. An. Jard. Bot. Madr. 66(2), 209–215. https://doi.org/10.3989/ajbm.2198 (2009).
Ivanova, N. V., Dewaard, J. R. & Hebert, P. D. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 6(4), 998–1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x (2006).
Hausmann, A. et al. DNA barcoding of fogged caterpillars in Peru: A novel approach for unveiling host-plant relationships of tropical moths (Insecta, Lepidoptera). PLoS ONE 15(1), e0224188. https://doi.org/10.1371/journal.pone.0224188 (2020).
Green, M. R. & Sambrook, J. Precipitation of DNA with ethanol. Cold Spring Harb. Protoc. 12, 2016. https://doi.org/10.1101/pdb.prot093377 (2016).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. https://doi.org/10.7717/peerj.2584 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17(1), 10–12 (2011).
Clarke, K. R. & Gorley, R. N. PRIMER v7: User manual/tutorial. PRIMER-E (Plymouth, 2015).
Gaytán, Á. et al. DNA barcoding and geographical scale effect: the problems of undersampling genetic diversity hotspots. Ecol. Evol. 10(19), 10754–10772. https://doi.org/10.1002/ece3.6733 (2020).
Cross, A. T. & Bateman, P. W. How dangerous is a Drosera? Limb autotomy increases passive predation risk in crickets. J. Zool. 306(4), 217–222. https://doi.org/10.1111/jzo.12609 (2018).
Kathirithamby, J., Ross, L. D. & Johnston, J. S. Masquerading as self? Endoparasitic Strepsiptera (Insecta) enclose themselves in host-derived epidermal bag. PNAS 100(13), 7655–7659. https://doi.org/10.1073/pnas.1131999100 (2003).
Thum, M. Segregation of habitat and prey in two sympatric carnivorous plant species Drosera rotundifolia and Drosera intermedia. Oecologia 70(4), 601–605. https://doi.org/10.1007/BF00379912 (1986).
Nielson, M. W. & Knight, W. J. Distributional patterns and possible origin of leafhoppers (Homoptera, Cicadellidae). Rev. Bras. Zool. 17(1), 81–156. https://doi.org/10.1590/S0101-81752000000100010 (2000).
Tagawa, K. & Watanabe, M. Group foraging in carnivorous plants: Carnivorous plant Drosera makinoi (Droseraceae) is more effective at trapping larger prey in large groups. Plant Species Biol. 36(1), 114–118. https://doi.org/10.1111/1442-1984.12290 (2021).
Ellison, A. M. & Gotelli, N. J. Energetics and the evolution of carnivorous plants—Darwin’s ‘most wonderful plants in the world’. J. Exp. Bot. 60(1), 19–42. https://doi.org/10.1093/jxb/ern179 (2009).
Gibson, T. C. Competition among threadleaf sundews for limited insect resources. Am. Nat. 138(3), 785–789 (1991).
Poppinga, S. et al. Prey capture analyses in the carnivorous aquatic waterwheel plant (Aldrovanda vesiculosa L., Droseraceae). Sci. Rep. 9(1), 1–13. https://doi.org/10.1038/s41598-019-54857-w (2019).
Acknowledgements
Funding for metabarcoding was provided by the project “SNSB-Innovativ 2020” of the Staatliche Naturwissenschaftliche Sammlungen Bayerns SNSB (Bavarian Natural History Collections, Munich, Germany) to Andreas Fleischmann and Axel Hausmann. Vedran Bozicevic is thanked for providing helpful comments about the barcode sequence analysis and processing method. A flora taking licence (FT61000038-2) was obtained for this project from the Western Australian Department of Biodiversity, Conservation and Attractions. Plant material was exported to Germany under an export permit (WT2020-001235) from the Australian Department of Agriculture, Water and the Environment. This study was supported by a Postgraduate Research Stipend Scholarship from Curtin University to Thilo Krueger.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
T.K. designed the macro photography procedure, conducted fieldwork and initial sample processing, analysed and evaluated data, wrote the first manuscript draft and prepared all figures. T.K., A.T.C. and A.F. had the initial idea for this research, designed the leaf sampling procedure and contributed most to manuscript revision. A.F., J.H., J.M. and A.H. performed sample processing, DNA extraction, amplification and metabarcoding. J.H., J.M. and A.H. contributed to writing the methods section of the manuscript. All authors critically revised the manuscript and approved its final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krueger, T., Cross, A.T., Hübner, J. et al. A novel approach for reliable qualitative and quantitative prey spectra identification of carnivorous plants combining DNA metabarcoding and macro photography. Sci Rep 12, 4778 (2022). https://doi.org/10.1038/s41598-022-08580-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-08580-8
This article is cited by
-
Better to risk limb than life: some insects use autotomy to escape passive predation by carnivorous plants
Arthropod-Plant Interactions (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.