Abstract
Sepsis is a potentially fatal condition caused by infection. It is frequently difficult to distinguish sepsis from systemic inflammatory response syndrome (SIRS), often resulting in poor prognoses and the misuse of antibiotics. Hence, highly sensitive and specific biomarkers are needed to differentiate sepsis from SIRS. Urine samples were collected and segregated by group (a sepsis group, a SIRS group, and a healthy control group). iTRAQ was used to identify the differentially expressed proteins among the three groups. The identified proteins were measured by ELISA in urine samples. Finally, all the acquired data were analyzed in SPSS. C-reactive protein, leucine-rich alpha glycoprotein-1 and serum amyloid A (SAA) protein were differentially expressed among the three groups. The adjusted median concentrations of urinary C-reactive protein were 1337.6, 358.7, and 2.4 in the sepsis, SIRS, and healthy control groups, respectively. The urinary leucine-rich alpha glycoprotein-1 levels in these three groups were 1614.4, 644.5, and 13.6, respectively, and the levels of SAA were 6.3, 2.9, and 0.07, respectively. For all three of these measures, the sepsis group had higher levels than the SIRS group (P < 0.001), and the SIRS group had higher levels than the healthy control group. When combined, the three biomarkers had a sensitivity of 0.906 and a specificity of 0.896 in distinguishing sepsis from SIRS. Urinary C-reactive protein, urinary leucine-rich alpha glycoprotein-1 and urinary SAA have diagnostic value in cases of sepsis. This initial study suggests the possibility of improved differential diagnosis between sepsis and systemic inflammatory response syndrome; additional confirmation is necessary to corroborate the findings.
Similar content being viewed by others
Introduction
Sepsis was first defined in 19921 as a systemic inflammatory response syndrome (SIRS) caused by confirmed infection (Sepsis 1.0). The signs and symptoms of sepsis were expanded in Sepsis 2.02. Sepsis is now defined as organ dysfunction caused by a systemic inflammatory response to pathogenic microorganisms, which can be fatal (Sepsis 3.0)3. In the last decade, the incidence rate of sepsis was 437 per 100,000 person-years, and the incidence of severe sepsis (as defined by Sepsis 2.02) was 270 per 100,000 person-years in developed countries4. Fleischmann et al.4 also inferred that there were 31.5 million sepsis cases per year and 19.4 million severe sepsis cases per year worldwide, with 5.3 million deaths annually. The case-fatality rate can reach up to 30% in sepsis, 50% in severe sepsis and 80% in septic shock5. In addition, the prevalence of sepsis and the contribution of sepsis to all-cause mortality rates have been increasing in the last several years6. Severe sepsis in elderly patients was an independent risk factor for substantial and persistent new cognitive impairment and functional disability among survivors7. Delays in treatment and inappropriate antibiotic therapy dramatically reduce survival rates in septic shock8,9. Hence, the early diagnosis of sepsis is particularly important. At present, the diagnosis of sepsis is difficult and complicated. Although there are clinical guidelines and many laboratory tests to diagnose sepsis, e.g., the C-reactive protein (CRP) test, procalcitonin (PCT) test, and white blood cell (WBC) count, their specificity is unsatisfactory. Blood culture provides conclusive evidence for the diagnosis of sepsis, but the sensitivity of blood culture is very low, and the process is time consuming, usually delaying diagnosis. On the other hand, SIRS patients can be misdiagnosed with sepsis because the symptoms and signs of SIRS are very similar to those of sepsis when the blood culture is negative; the misdiagnosis of SIRS as sepsis leads to antibiotic misuse and possible selection for drug resistance. The major differences between sepsis and SIRS include organ failure, which is assessed mainly by the Sequential (sepsis-related) Organ Failure Assessment (SOFA) score, and infectious states, measured by biological phenotype and clinical symptoms3. Biomarkers are defined as measurable and quantifiable biological parameters, which can be molecules, genes, proteins or other variables. There are hundreds of biomarkers, but only a small fraction of these are useful biomarkers for sepsis10, e.g., CRP, PCT, serum amyloid A (SAA), and triggering receptors expressed on myeloid cells-1 (TREM-1). Biomarkers are a promising way to diagnose sepsis, and this approach can facilitate early and accurate diagnosis, forecast organ dysfunction and assist in defining appropriate therapeutic plans11. Human urine contains thousands of proteins12,13 and extracellular vesicles14, which could be good resources to use as biomarkers. The method of isobaric tags for relative and absolute quantification (iTRAQ) is a mass spectrometry (MS)-based relative proteomic quantification method utilizing the derivatization of primary amino groups in intact proteins and isobaric tags for different peptide fragments15,16. iTRAQ can be used to screen the differentially expressed proteins among eight samples simultaneously and is highly sensitive and specific17. In this study, we sought to identify new diagnostic biomarkers of sepsis in urine by utilizing iTRAQ and to verify the biomarkers using enzyme-linked immunosorbent assays (ELISAs). In addition, we explored the diagnostic value of the combined biomarkers through logistic regression to enhance the diagnostic sensitivity and specificity to sepsis and help clinicians determine the appropriate therapeutic strategy.
Methods and materials
Urine samples and data collection
This study was performed at the Second Affiliated Hospital of Chongqing Medical University. The procedures used in this study were in accordance with the 2008 Declaration of Helsinki and approved by the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University (Grant no. 201916). The urine samples and patient data came from the Adult Multidisciplinary Intensive Care Unit (ICU) and Infection Department. All patients or their bailors were informed about the study procedures, risk and privacy policy, and written consent was signed by each participant. Urine samples were collected from patients in the sepsis group, SIRS group and healthy control group. Consecutive patients with sepsis or SIRS were enrolled, and the healthy control group was enrolled by recruitment. The sepsis inclusion criteria3 were as follows: (1) age 18 years or older, (2) proven infection or suspected infection as adjudicated retrospectively by three physicians, and (3) a SOFA score of 2 or more points. The specific criteria for the SOFA score are listed in Table 13,18, and the effectiveness of the SOFA has been assessed19. The SIRS inclusion criteria1 were as follows: (1) temperature > 38 °C or < 36 °C; (2) heart rate > 90/min; (3) respiratory rate > 20/min or PaCO2 < 32 mmHg; (4) white blood cell count > 12 × 109/L or < 4 × 109/L or > 10% immature bands in peripheral blood; (5) proven lack of infection, or no suspected infection as adjudicated retrospectively by three physicians; and (6) age 18 years or older. The patients who were diagnosed with SIRS must have presented with two or more of the first four criteria. Healthy control inclusion criteria were that the healthy volunteer did not suffer from infectious disease or take antibiotics. The exclusion criteria were as follows: (1) immune deficiency, (2) autoimmune diseases, (3) pregnancy; (4) use of any antibiotics before hospitalization; and (5) refusal to take part in the study or refusal to provide written and signed consent. Urine samples were collected as soon as the clinical diagnosis was made, and the interval time was more than 4 h from the patients’ last micturition. The urine was collected in a 50 mL centrifuge tube directly from the bladder, and no protease inhibitor was added to the samples. The collected samples were transported in ice water mixture. These samples were centrifuged at 2000×g for 10 min to separate the cellular or tissue debris. Finally, the samples were divided into five 2.0 mL Eppendorf tubes and stored at −80 °C until analysis20. The patient data were collected through the electronic medical system of the Second Affiliated Hospital of Chongqing Medical University.
Materials and measurement procedure
Protein extraction from urine and iTRAQ procedure
We collected 10 samples from each of the groups to perform a preliminary analysis of the differentially expressed proteins among the three groups. The samples were pooled by group (sepsis, SIRS, and healthy control), and the proteins were precipitated using two volumes of cooled acetone (2 h)13. Finally, the three samples were centrifuged at 25,000×g at 4 °C for 15 min, and the supernatants were discarded. This procedure was repeated twice.
The precipitates were air-dried and dissolved at room temperature in an appropriate amount of lysis buffer using an ultrasonic homogenizer. Finally, the solutions were centrifuged at 25,000×g at 4 °C for 15 min, and the supernatants were collected. The Bradford method was used to measure the protein concentrations.
An iTRAQ 8-Plex Reagent Kit (Applied Biosystems, Foster City, CA, USA) was used to label the protein. The prepared protein was precipitated, redissolved, alkylated, cysteine blocked and digested following the iTRAQ Kit instructions and the study by Wisniewski22. The sepsis group was labeled with tag 113, the SIRS group was labeled with tag 119, and the healthy control group was labeled with tag 121. The labeled peptide solutions were pooled and freeze-dried under vacuum prior to further analysis.
The peptides were fractioned as previously described23,24. The prepared peptides were analyzed on the TripleTOF 5600 system coupled to an Eksigent NanoLC-2D system (AB Sciex, Framingham, MA, USA), and each of the samples was analyzed twice. The data were processed by ProteinPilot V2.0 (Applied Biosystems) and searched against the UniProt human proteome database (https://www.uniprot.org/). Proteins with an Unused Protscore > 1.3, a fold change > 2 and a P value < 0.05 were defined as differentially expressed.
ELISA measurement procedure
We employed the Human AACT-a1 (ab217779, Abcam, UK), Human C Reactive Protein ELISA Kit (ab9995, Abcam, UK), Human PEGC ELISA Kit (ab275552, Abcam, UK), Human Glutaredoxin-1 (abx151683, Abbexa, UK), Human Haptoglobin ELISA (ab219048, Abcam, UK), Human HLA-II histocompatibility antigen (abx387810, Abbexa, UK), Human LRG1 ELISA Kit (NBP2-60577, Novus Biologicals, Centennial, CO, USA), Human Resistin ELISA Kit (ab183364, Abcam, UK) and Human Serum Amyloid A ELISA Kit (KT-547, Kamiya Biomedical Company, Seattle, WA, USA) to measure the concentrations of target proteins. Urine samples from each patient were measured by ELISA, and the measurement protocol was performed according to the instructions of the three respective ELISA kits. The specifics of each ELISA kit are listed in Table 2. Duplicate standards or samples were set, and the mean absorbance of duplicate wells was calculated. The concentration of urinary proteins (CRP, LRG1, SAA) was normalized to urinary creatinine (u-Cr) and expressed as protein/u-Cr in μg/mmol to adjust for individual differences and potential kidney injury25,26. All statistical analyses were performed using Microsoft Excel 2019 (Microsoft Corporation, Redmond, WA, USA) and SPSS 25 (International Business Machines Corporation, Armonk, NY, USA).
Results
Subject characteristics
There were a total of 151 subjects included in our study: 53 sepsis patients, 48 SIRS patients and 50 healthy volunteers. The average ages of the three groups were 58.4, 62.0 and 51.0 years. The percentages of males were 64.2%, 75%, and 56%, respectively. In the sepsis group, there were 24 patients whose blood cultures were positive. The detected bacteria consisted of Escherichia coli (10, 41.7%), Staphylococcus aureus (6, 25%), Enterococcus faecium (3, 12.5%), Acinetobacter baumannii (2, 8.3%), Streptococcus (1, 4.2%), Lactobacillus brevis (1, 4.2%) and Klebsiella pneumoniae (1, 4.2%). There were no statistically significant differences in the distributions of age or gender among the three groups. The distribution of WBC count and neutrophil percentage were statistically similar and could not distinguish the sepsis and SIRS patients in this study. In addition, the sepsis patients presented higher urinary creatinine levels and pulse rates than the healthy control group. Patient data are summarized in Table 3.
iTRAQ identification of differentially expressed proteins
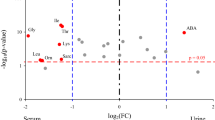
iTRAQ identified 11278 and 14904 proteins in duplicate analyses. There were 1970 and 1985 proteins with Unused ProtScore values > 1.3. Among these proteins, there were 114 and 118 with fold change > 2 between the sepsis group and healthy control group. There were 69 proteins that were detected in both iTRAQ procedures. After eliminating the proteins with P values > 0.05, 45 proteins met the screening criteria. The flow chart is presented in Fig. 1. Nine differentially expressed proteins that might be related to sepsis and SIRS were tested further by ELISA, and CRP, LRG1, and SAA were chosen as the target biomarkers (Fig. 2). Their iTRAQ information are listed in Table 4. The blood levels of these three proteins have previously been found to be related to sepsis and SIRS27,28,29.
Quantitative and statistical analysis
The concentrations of uCRP, uLRG1 and uSAA are shown in Table 3. Considering the variability of glomerular filtration rate (GFR), 24-h urinary volume and glomerular filtration barrier among the different subjects, the concentrations normalized to urinary creatinine levels were also listed as uCRP/u-Cr, uLRG1/u-Cr and uSAA/u-Cr in μg/mmol and were used for further statistical analyses. The normality of acquired data was tested, and we found that not all the data followed a Gaussian distribution. Although efforts were made to transform the data to a Gaussian distribution by converting initial data to the square root or logarithm, the data did not conform. Therefore, the data are presented as the median (25th percentile, 75th percentile), and nonparametric tests were used to analyze the statistical characteristics. Spearman's rank correlation coefficient was calculated (Table 5). Serum CRP was strongly correlated with urine CRP, which may indicate that urinary CRP came from the blood. In addition, the three biomarkers (uCRP, uLRG1, uSAA) were pairwise correlated, which may indicate that they had similar status during the infection process. The median values were highest in the sepsis patients, intermediate in the SIRS patients and lowest in the healthy control group (P < 0.001).
When these biomarkers were adjusted to urine creatinine, the same conclusion could be drawn; their values are presented in Fig. 2. For the adjusted concentrations and s-CRP, receiver operating characteristic curves (ROC) were drawn by SPSS 25 (Fig. 3). The area under the curve values were 0.878 (uCRP/u-Cr) and 0.874 (uLRG1/u-Cr) and 0.849 (uSAA/u-Cr) and 0.891 (s-CRP). The Youden index (sensitivity + specificity – 1) was used to determine the best cutoff value. The sensitivity, specificity, cutoff value and area under the curve values are listed in Table 6. s-CRP had the highest diagnostic efficiency. The diagnostic difference was small among uCRP/u-Cr, uLRG1/u-Cr and uSAA/u-Cr. The adjusted concentrations (uCRP/u-Cr, uLRG1/u-Cr and uSAA/u-Cr) of the sepsis and SIRS groups were transformed to binary variables based on whether they were greater than the cutoff value and further analyzed by logistic regression. The forward stepwise (likelihood ratio) method was employed to process the data. The omnibus test was used to verify the statistical significance (P < 0.001), and the Hosmer and Lemeshow test showed that the model had taken full advantage of the acquired data (P = 0.712). The regression coefficient and P values are listed in Table 7. According to the regression results, all three biomarkers were included in the regression model. If the patients’ parameters exceeded the cutoff value, the corresponding risk of sepsis would increase 9.913-fold (uCRP/u-Cr), 15.936-fold (uLRG1/u-Cr) and 12.793-fold (uSAA/u-Cr) compared to SIRS patients. The sepsis risk score was logit(P) = 2.294*X1 + 2.734*X2 + 2.549*X3-3.714 (X1 = 1, if uCRP/u-Cr > 746.1 μg/mmol; X1 = 0, if uCRP/u-Cr > 746.1 μg/mmol; X2 = 1, if uLRG1/u-Cr > 1174.8 μg/mmol; X2 = 0, if uLRG1/u-Cr < 1174.8 μg/mmol; X3 = 1, if uSAA/u-Cr > 4.4 μg/mmol; X3 = 0, if uSAA/u-Cr < 4.4 μg/mmol). According to logit(P), the prediction probabilities were calculated by P = elogit(P)/(1 + elogit(P)), and its ROC curve was also drawn (Fig. 3). The area under the curve was 0.937. The cutoff of logit(P) was 0.514. Its sensitivity and specificity were 0.906 and 0.896, respectively. This indicated that the sensitivity and specificity of diagnosis would be greatly enhanced by combining the panel of biomarkers.
Discussion
Because the symptoms of sepsis and SIRS are quite similar, it is frequently difficult to distinguish a septic infection from SIRS. After Sepsis 3.0 was issued, the situation became more difficult, in part because patients suffering from infection-related SIRS, but without organ dysfunction, can be confused with patients who have SIRS without sepsis. Biomarkers, especially acute-phase proteins, could be key to distinguishing sepsis from SIRS, but their specificity has been unsatisfactory, making it necessary to combine multiple biomarkers to enhance overall specificity28,29. The three biomarkers identified in the present study were all acute-phase reaction proteins30,31,32,33,34,35. CRP was identified in the 1930s and is synthesized in the liver and released in response to infection, trauma and immune disorders. The mean s-CRP levels were 98 mg/L in sepsis patients and 70 mg/L in SIRS patients36. Most recently, bacterial infection is suspected when s-CRP is greater than 100 mg/L in the clinical context. Conversely, the absence of severe bacterial infection is indicated when s-CRP is less than 20 mg/L33. In the present study, we first verified the existence of CRP in urine, and uCRP/u-Cr was strongly related to s-CRP. Although the diagnostic efficiency of uCRP/u-Cr was slightly lower than that of s-CRP, uCRP/u-Cr could still be a good substitute for s-CRP, considering the convenience and noninvasiveness of specimen collection and the strong relationship between the urinary and serum concentrations. Leucine-rich α-2 glycoprotein 1 (LRG1), first identified in 197737, is synthesized by hepatocytes, stored in neutrophils38 or myeloid cells39, and released into the serum when the body is in the acute phase of stimulation by bacterial or viral infection40. LRG1 belongs to the leucine-rich repeat (LRR) protein family and plays a role in protein interactions, innate immunity, platelet aggregation and angiogenesis. LRG1 is usually used as a biomarker of tumors, appendicitis, diabetes complications, and inflammatory disease41,42,43,44. According to our measurement results, uLRG1/u-Cr of sepsis patients was obviously higher in sepsis patients than in either SIRS patients or healthy volunteers (P < 0.001), indicating that urinary LRG1 is a promising biomarker in diagnosing sepsis and distinguishing sepsis from SIRS. Considering the lower sensitivity (0.774) and higher specificity (0.854), LRG1 would be a good choice to combine with other biomarkers. Serum amyloid A protein is also a well-known acute-phase protein produced by the liver. Its concentration is very low (1 μg/mL) in healthy people but can increase dramatically (100-fold or more) when the human body is in the acute phase45. Human SAA consists of 4 isotypes (SAA1, SAA2, SAA3, SAA4)46, but the sequence identity of SAA1 and SAA2 is more than 93%, and SAA1 is the main isotype in serum47. The SAA ELISA kits we employed in the current study were not sensitive to the SAA isotypes, and the concentration represented the concentration of total SAA. SAA is regulated by cytokines (interleukin-1β, interleukin-6, tumor necrosis factor, etc.) and regulate cytokines (interleukin-23, interleukin-33, interleukin-10, etc.) which is summarized as the cytokine-SAA-chemokine network48. Considering its interaction with cytokines, it is believed that SAA takes part in inflammatory diseases, angiogenesis and tumor growth, as do other acute-phase proteins49,50,51. In our study, the sensitivity and specificity of uSAA/u-Cr were between those of uCRP/u-Cr and uLRG1/u-Cr. This was also the first time the value of uSAA for sepsis diagnosis and differential diagnosis was verified.
In the current study, a range of inflammatory biomarkers were measured in patients with sepsis and SIRS. CRP is a nonspecific indicator of systemic inflammation, and patients with noninfectious causes of SIRS may also have markedly increased serum concentrations of this biomarker. The sensitivity and specificity of CRP for differential diagnosis between sepsis and SIRS were lower than those of the three combined biomarkers in our study52. PCT has been used to differentiate sepsis from SIRS53, but patients with systemic inflammation of noninfectious etiology may have increased levels of PCT in some cases54. Therefore, the diagnostic utility of measurement of CRP and PCT should be considered combined with other clinical and laboratory information in the clinical setting. In addition to CRP and PCT, the concentrations of the circulating anti-inflammatory cytokines IL-Ra and IL-10 were found to be significant in discriminating between sepsis and SIRS55,56,57. The diagnostic accuracy of urinary orosomucoid (u-ORM) for sepsis (AUC ROC: 0.954) was similar to that of PCT58. Few studies have been performed regarding urinary GSN (u-GSN) levels in sepsis. Maddens et al. reported increased u-GSN levels in septic mice but did not predict the diagnostic value of u-GSN in sepsis59. The diagnostic value of these biomarkers was lower than that of the three parameters in our study. However, in different studies, different experimental methods and populations have an impact on the experimental results. Therefore, larger samples and more in-depth studies are needed to compare the diagnostic utility of these indicators.
Although the three biomarkers were normalized to urinary creatinine in the statistical process, adjusting the concentration had only a small influence on the statistical results and no influence on the final conclusion. When the panel of biomarkers was combined, the sensitivity to sepsis was increased to 0.906, and the specificity was increased to 0.896, which greatly exceeded any single biomarker. The area under the ROC curve was 0.937, which indicated favorable diagnostic efficiency. Serum CRP data were acquired through the medical electronic system, but serum LRG1 and SAA were not available by routine examination at the Second Affiliated Hospital of Chongqing Medical University. Hence, serum LRG1 and SAA data were lacking in our study. The correlation between serum LRG1 and urinary LRG1 was not verified, nor was the correlation of serum SAA and urinary LRG1. According to the correlation between serum CRP and urinary CRP, we inferred that urine CRP came from serum CRP. Considering the similar mechanism of protein excretion by the kidney, we hypothesized that urinary LRG1 and SAA also came from the blood. There were only 101 patients included in our study, and it would be necessary to recruit more patients to further verify the efficiency of the combined biomarkers. The logistic regression model represented only a “discovery” finding, and we did not use K-fold cross-validation or other forms of cross-validation; thus, unfortunately, we must regard it as preliminary. In addition, the diagnostic value of LRG1 for sepsis was not fully investigated, and additional research is necessary to further understand the diagnostic significance of LRG1.
Conclusions
C-reactive protein, serum amyloid A protein and leucine-rich alpha glycoprotein-1 are present in urine. Urinary C-reactive protein comes from blood and can substitute for serum C-reactive protein in sepsis diagnosis. Urine SAA and leucine-rich alpha glycoprotein-1 were also useful biomarkers for sepsis diagnosis. This initial study suggests the possibility of improved differential diagnosis between sepsis and systemic inflammatory response syndrome, and further confirmation is necessary to further confirm the findings (Suppl. information).
Abbreviations
- SIRS:
-
Systemic inflammatory response syndrome
- CRP:
-
C-reactive protein
- uCRP:
-
Urinary CRP
- PCT:
-
Procalcitonin
- SAA:
-
Serum amyloid A protein
- uSAA:
-
Urinary SAA
- TREM-1:
-
Triggering receptors expressed on myeloid cells-1
- iTRAQ:
-
Isobaric tags for relative and absolute quantification
- ELISA:
-
Enzyme linked immunosorbent assay
- ICU:
-
Intensive care unit
- SOFA:
-
Sequential (sepsis-related) organ failure assessment
- LRG1:
-
Leucine-rich alpha glycoprotein-1
- uLRG1:
-
Urinary LRG1
- u-Cr:
-
Urinary creatinine
- CV:
-
Coefficient of variation
- SPSS:
-
Statistical product and service solutions
- GFR:
-
Glomerular filtration rate
- ROC:
-
Receiver operating characteristic curve
- LRR:
-
Leucine rich repeat
References
Bone, R., Balk, R. & Cerra, F. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit. Care Med. 20(6), 864–874 (1992).
Levy, M. M. et al. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit. Care Med. 31(4), 1250–1256 (2003).
Singer, M. et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315(8), 801–810 (2016).
Fleischmann, C. et al. Assessment of global incidence and mortality of hospital-treated sepsis: Current estimates and limitations. Am. J. Respir. Crit. Care Med. 193(3), 259–272 (2016).
Jawad, I., Luksic, I. & Rafnsson, S. B. Assessing available information on the burden of sepsis: Global estimates of incidence, prevalence and mortality. J. Glob. Health 2(1), 010404 (2012).
Gobatto, A. L., Besen, B. A. & Azevedo, L. C. How can we estimate sepsis incidence and mortality?. Shock 47(1S Suppl 1), 6–11 (2017).
Iwashyna, T. J. et al. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA 304(16), 1787–1794 (2010).
Kumar, A. et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34(6), 1589–1596 (2006).
Kumar, A. et al. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 136(5), 1237–1248 (2009).
Prucha, M., Bellingan, G. & Zazula, R. Sepsis biomarkers. Clin. Chim. Acta 440, 97–103 (2015).
Singer, M. Biomarkers in sepsis. Curr. Opin. Pulm. Med. 19(3), 305–309 (2013).
Chen, Y. Variations of human urinary proteome. Adv. Exp. Med. Biol. 845, 91–94 (2015).
Thongboonkerd, V. et al. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 62(4), 1461–1469 (2002).
Pisitkun, T., Shen, R. F. & Knepper, M. A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 101(36), 13368–13373 (2004).
Wiese, S. et al. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 7(3), 340–350 (2007).
Ross, P. L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteom. 3(12), 1154–1169 (2004).
Unwin, R. D. Quantification of proteins by iTRAQ. Methods Mol. Biol. 658, 205–215 (2010).
Vincent, J. L. et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 22(7), 707–710 (1996).
Vincent, J. L. et al. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: Results of a multicenter, prospective study. Working group on “sepsis-related problems” of the European Society of Intensive Care Medicine. Crit. Care Med. 26(11), 1793–1800 (1998).
Zhou, H. et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 69(8), 1471–1476 (2006).
Teasdale, G. & Jennett, B. Assessment of coma and impaired consciousness. A practical scale. Lancet 2(7872), 81–84 (1974).
Wisniewski, J. R. et al. Universal sample preparation method for proteome analysis. Nat. Methods 6(5), 359–362 (2009).
She, S. et al. Proteomics based identification of autotaxin as an anti-hepatitis B virus factor and a promoter of hepatoma cell invasion and migration. Cell Physiol. Biochem. 45(2), 744–760 (2018).
Yang, Y. et al. Discovery of SLC3A2 cell membrane protein as a potential gastric cancer biomarker: Implications in molecular imaging. J. Proteome Res. 11(12), 5736–5747 (2012).
Hou, L. N. et al. Excretion of urinary orosomucoid 1 protein is elevated in patients with chronic heart failure. PLoS ONE 9(9), e107550 (2014).
Kustan, P. et al. Urinary orosomucoid: A novel, early biomarker of sepsis with promising diagnostic performance. Clin. Chem. Lab. Med. 55(2), 299–307 (2017).
Mei-Hong, Yu. et al. Prognostic value of the biomarkers serum amyloid A and nitric oxide in patients with sepsis. Int. Immunopharmacol. 62, 287–292 (2018).
Fish-Low, C.-Y., Than, L. T. L., Ling, K.-H., Lin, Q. & Sekawi, Z. Plasma proteome profiling reveals differentially expressed lipopolysaccharide-binding protein among leptospirosis patients. J. Microbiol. Immunol. Infect. 53(1), 157–162 (2020).
Kumar, V. Toll-like receptors in sepsis-associated cytokine storm and their endogenous negative regulators as future immunomodulatory targets. Int. Immunopharmacol. 89, 107087 (2020).
Kargaltseva, N. M. et al. Inflammation markers and bloodstream infection (review of literature). Klin. Lab Diagn. 64(7), 435–442 (2019).
Di Somma, S. et al. Opinion paper on innovative approach of biomarkers for infectious diseases and sepsis management in the emergency department. Clin. Chem. Lab Med. 51(6), 1167–1175 (2013).
Okamura, J. M. et al. Potential clinical applications of C-reactive protein. J. Clin. Lab Anal. 4(3), 231–235 (1990).
Knudtzen, F. C. et al. Characteristics of patients with community-acquired bacteremia who have low levels of C-reactive protein (</=20 mg/L). J. Infect. 68(2), 149–155 (2014).
Perez, L. Acute phase protein response to viral infection and vaccination. Arch. Biochem. Biophys. 671, 196–202 (2019).
Shirai, R. et al. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem. Biophys. Res. Commun. 382(4), 776–779 (2009).
Povoa, P. C-reactive protein: A valuable marker of sepsis. Intensive Care Med. 28(3), 235–243 (2002).
Heinz, H. & Siegfried, B. Isolierung und Charakterisierung eines bisher unbekannten leucinreichen 3.1S-α2-Glykoproteins aus Humanserum, in Hoppe-Seyler’s. Z. Physiol. Chem. 77, 639 (1977).
Druhan, L. J. et al. Leucine rich alpha-2 glycoprotein: A novel neutrophil granule protein and modulator of myelopoiesis. PLoS ONE 12(1), e0170261 (2017).
O’Donnell, L. C., Druhan, L. J. & Avalos, B. R. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J. Leukoc. Biol. 72(3), 478–485 (2002).
Bini, L. et al. Two-dimensional electrophoretic patterns of acute-phase human serum proteins in the course of bacterial and viral diseases. Electrophoresis 17(3), 612–616 (1996).
Yamamoto, M. et al. Overexpression of leucine-rich alpha2-glycoprotein-1 is a prognostic marker and enhances tumor migration in gastric cancer. Cancer Sci. 108(10), 2052–2060 (2017).
Kharbanda, A. B. et al. Novel serum and urine markers for pediatric appendicitis. Acad. Emerg. Med. 19(1), 56–62 (2012).
Hong, Q. et al. LRG1 promotes diabetic kidney disease progression by enhancing TGF-beta-induced angiogenesis. J. Am. Soc. Nephrol. 30(4), 546–562 (2019).
Shinzaki, S. et al. Leucine-rich alpha-2 glycoprotein is a serum biomarker of mucosal healing in ulcerative colitis. J. Crohns Colitis 11(1), 84–91 (2017).
Shainkin-Kestenbaum, R., Winikoff, Y. & Cristal, N. Serum amyloid A concentrations during the course of acute ischaemic heart disease. J. Clin. Pathol. 39(6), 635–637 (1986).
De Buck, M. et al. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr. Med. Chem. 23(17), 1725–1755 (2016).
Yamada, T. et al. Serum amyloid A1 alleles and plasma concentrations of serum amyloid A. Amyloid 6(3), 199–204 (1999).
De Buck, M. et al. The cytokine-serum amyloid A-chemokine network. Cytokine Growth Factor Rev. 30, 55–69 (2016).
Yarur, A. J. et al. Serum amyloid A as a surrogate marker for mucosal and histologic inflammation in patients with Crohn’s disease. Inflamm. Bowel Dis. 23(1), 158–164 (2017).
Connolly, M. et al. Acute serum amyloid A is an endogenous TLR2 ligand that mediates inflammatory and angiogenic mechanisms. Ann. Rheum. Dis. 75(7), 1392–1398 (2016).
Shiels, M. S. et al. Circulating inflammation markers, risk of lung cancer, and utility for risk stratification. J. Natl. Cancer Inst. 107(10), 199 (2015).
Faix, J. D. Biomarkers of sepsis. Crit. Rev. Clin. Lab. Sci. 50(1), 23–36 (2013).
Biron, B. M., Ayala, A. & Lomas-Neira, J. L. Biomarkers for sepsis: What is and what might be?. Biomark Insights 10(Suppl 4), 7–17 (2015).
Henriquez-Camacho, C. & Losa, J. Biomarkers for sepsis. BioMed. Res. Int. 9, 547818 (2014).
Arend, W. P. & Guthridge, C. J. Biological role of interleukin 1 receptor antagonist isoforms. Ann. Rheum. Dis. 59(Suppl. 1), i60–i64 (2000).
Duque, G. A. & Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 5, 491 (2014).
Janson, R. W., Hance, K. R. & Arend, W. P. Production of IL-1 receptor antagonist by human in vitro-derived macrophages: Effects of lipopolysaccharide and granulocyte macrophage colony-stimulating factor. J. Immunol. 147(12), 4218–4223 (1991).
Kustán, P. Corresponding, Zoltán Horváth-Szalai, Diána Mühl. Nonconventional markers of sepsis. EJIFCC. 28(2), 122–133 (2017).
Maddens, B. et al. Chitinase-like proteins are candidate biomarkers for sepsis-induced acute kidney injury. Mol. Cell Proteom. 11(6), M111.013094 (2012).
Acknowledgements
The study was supported by the Natural Science Foundation of China (grant no. 81171560), the “Par-Eu Scholars Program” of Chongqing City, the National Science and Technology Major Project of China (grant no. 2012ZX10002007001), and the High-Level Medical Reserved Personnel Training Project of Chongqing (grant no. 2017HBRC005).
Author information
Authors and Affiliations
Contributions
X.H.W. conceived and designed the study, and X.H.W. is the guarantor of the study. Y.T., N.L. and S.Y.L. collected the urine samples and patient data. Y.T. and W.Y.Z. performed the iTRAQ and ELISA procedures. J.H. was responsible for the statistical analysis. Y.T. and N.L. wrote the paper. H.R. and Y.X.Y. edited the clinical theses. All the authors have approved the final manuscript and declare that they have no conflicts of interest.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tang, Y., Ling, N., Li, S. et al. A panel of urine-derived biomarkers to identify sepsis and distinguish it from systemic inflammatory response syndrome. Sci Rep 11, 20794 (2021). https://doi.org/10.1038/s41598-021-99595-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99595-0
This article is cited by
-
Deficiency of circadian clock gene Bmal1 exacerbates noncanonical inflammasome-mediated pyroptosis and lethality via Rev-erbα-C/EBPβ-SAA1 axis
Experimental & Molecular Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.