Abstract
JAZ is a plant-specific protein family involved in the regulation of plant development, abiotic stresses, and responses to phytohormone treatments. In this study, we carried out a bioinformatics analysis of JAZ genes in turnip by determining the phylogenetic relationship, chromosomal location, gene structure and expression profiles analysis under stresses. The 36 JAZ genes were identified and classified into four subfamilies (ZML, JAZ, PPD and TIFY). The JAZ genes were located on 10 chromosomes. Two gene pairs were involved in tandem duplication events. We identified 44 collinear JAZ gene pairs in the turnip genome. Analysis of the Ka/Ks ratios indicated that the paralogs of the BrrJAZ family principally underwent purifying selection. Expression analysis suggested JAZ genes may be involved in the formation of turnip tuberous root, and they also participated in the response to ABA, SA, MeJA, salt stress and low-temperature stress. The results of this study provided valuable information for further exploration of the JAZ gene family in turnip.
Similar content being viewed by others
Introduction
Jasmonic acid and its biologically active derivatives are referred to as jasmonates (JAs), which participate in plant defenses to insects and contribute to developmental and growth controls1,2,3. JAs were first identified when plants suffered from biotic stresses (e.g., mechanical damage, pests, and diseases)4. Afterward, numerous studies revealed their significant role in plant responses to abiotic stresses such as low temperature, high temperature, drought, heavy metal, and salt stresses5. Under stress treatment, JAs induce the gene expression of signal transduction pathways, thereby regulating plant responses to adversity6. The interaction of JAs with other phytohormones and thus the regulation of plant stress resistance has also become a hot topic of research7.
JA signaling pathway, including the biosynthesis and metabolism of signal transduction molecules, JA signaling, and downstream gene response, is a complex process involving many genes and proteins5,8. When plants are stimulated by the external environment, they synthesize large amounts of jasmonic acid, which is formed into the highly biologically active JA-Ile by the action of the adenylate-forming enzyme JAR1. JA-Ile binds specifically to the jasmonic acid receptor F-box protein COI1 (coronatine insensitve1)9. The JAS domain is involved in the binding of COI1 and MYC2. The ZIM domain containing the TIFY motif is involved in binding NINJA (novel interactors of JAZ)10. JAZ is also considered to be a component of the JA co-receptor11 and acts as a "repressor" in the JA pathway12. In the absence of JA-Ile, JAZ proteins interact with NINJA proteins13 to recruit the co-repressor TPL (topless), which allows JAZ proteins to interact with downstream transcription factors, such as MYC2, to inhibit the transcriptional activation of JA-responsive genes by MYC2. In the presence of JA-Ile, the JA-Ile accumulated in response to stress binds to COI1 and promotes direct binding of the COI1-JAZs complex, forming a complex and causing ubiquitination of JAZ proteins by the E3 ubiquitin ligase SCFCOI1 (Skp/Cullin/F-box) complex, which eventually degrades the JAZ repressor through the 26S proteasome. The SCFCOI1 complex is formed by the binding of COI1 to ASK1/ASK2, Cullin1, and Rbx1, which are important components mediating the JA signaling response. Among them, MYC2 not only participates in the activation of jasmonic acid signaling, but also regulates the termination of jasmonic acid signaling, and can interoperate with MTB (MYC2-targeted bHLH) to regulate jasmonic acid signaling14,15.
JAZ gene family has many members that are involved in the regulation of plant development, abiotic stresses, and responses to phytohormone treatments, each with a different biological function16. For example, Overexpression of the OsJAZ9 gene improves rice (Oryza sativa) tolerance to potassium deficiency by changing JA level and JA signal transduction pathway17. Overexpression of the GsJAZ2 gene in soybean (Glycine max) significantly enhanced the resistance of transgenic lines to saline stress18. Overexpression of AtJAZ1 in Arabidopsis can enhance host resistance to Spodoptera exigua19. Overexpression of OsJAZs in rice can lead to malformations in floral organ development20,21.
Turnip (Brassica rapa L. subsp. rapa) is a crucial root vegetable belonging to the Brassica subspecies of the family Cruciferae. Turnips are very sensitive to environmental stress which seriously affect the quality and yield of the tuberous roots22,23,24,25. Despite extensive studies of the JAZ family in various plant species, including cotton, rice, tomato, soybean, and cabbage, JAZ family genes have not yet been identified in turnip18,26,27,28,29,30.
In this study, we used genomic resources to systematically identify members of the turnip JAZ gene family and investigated phylogeny, chromosome locations, evolutionary history, structural characteristics. Furthermore, we also analyzed expression patterns of JAZ genes after different abiotic stresses and phytohormone treatments. This study will be useful for functional studies of JAZs in turnip.
Materials and methods
Identification of the B. rapa JAZ family genes
The genome sequences and annotation files of Arabidopsis thaliana, B. oleracea var. Capitata, and Brassica rapa subsp. rapa were obtained from the TAIR database (http://www.arabidopsis.org/), CNGB database (http://db.cngb.org/search/project/CNP0000469/), and Turnip Genome Database in JBrowse website (https://www.bioinformatics.nl/brassica/index.html?data=bras_tp%2Fdata&loc=A01%3A11421217..17131178&tracks=DNA&highlight ), respectively21.
To find the JAZ family genes in turnip genome, we downloaded the Markov model (HMM) files corresponding to the TIFY domain (PF06200) and JAS domain (PF09425) from Pfam protein family database (http://pfam.sanger.ac.uk/)31. The former two HMM profiles were used to search the turnip protein database for target hits with the TIFY and JAS domain using HMMER 3.0 software. The candidate JAZ proteins with E-values < 1.0E−05 were selected.
The JAZ protein sequences of 18 A. thaliana and 48 B. oleracea obtained from previous studies were used as query sequences to blast against turnip protein sequences30,32. All non-redundant sequences with E < 1.0e−5 were selected as candidate JAZ proteins.
The candidate JAZ protein sequences obtained by the above two methods were combined and uploaded to NCBI CD-Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to confirm the conserved domain. The molecular weight (MW) and isoelectric point (pI) of each JAZ protein were analyzed with the online tool ExPASy (http://www.expasy.org). The subcellular locations were predicted using Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
Analysis of conserved motif and gene structure
The BrrJAZ proteins were used to create multiple protein sequence alignments using MEGA 7 software with the default parameter setting MUSCLE method33. The Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.cn) was employed to determine the exon/intron organization of turnip JAZ genes by comparing predicted coding sequences with their corresponding full-length sequences. The conserved motifs in the identified turnip JAZ proteins were identified by MEME (http://meme-suite.org/).
Sequence alignment and phylogenetic analysis
To infer the evolutionary relationship among A. thaliana, B. oleracea var. Capitata, and B. rapa, the phylogenetic analysis was performed. Multiple JAZ protein sequences were aligned using MEGA 7 software with the default parameter setting MUSCLE method. Based on this result, the neighbor-joining phylogenetic tree was constructed, with 1000 bootstrap values.
Gene location and collinearity analysis and gene replication analysis
The position information of BrrJAZ genes was acquired from the genomic sequence annotation. TBtools software was used for the mapping of JAZ genes in the corresponding chromosome34. MCscanX software was used to analyze the gene duplication events35. Ks (synonymous) and Ka (non-synonymous) substitution of each duplicated JAZ gene pairs were calculated using KaKs_Calculator 2.0. To exhibit the synteny relationship of the orthologous JAZ genes obtained from turnip and other selected species, the syntenic analysis maps were constructed using the Dual Systeny Plotter software.
Expression analysis from RNA-Seq data
The Illumina RNA-seq data were downloaded from the NCBI (Accession number: PRJNA273340) to study the expression patterns of BrrJAZ genes that participate in the tuberous root development. The turnip cultivar “Chang Huang Man Jing” was used as plant material. Samples consisting of tuberous root tissues were collected on day 18 (the early stage before cortex splitting, ES), day 28 (the stage of cortex splitting, CSS) and day 64 (the stage of root thickening, RTS) after sowing. Additionally, every stage had two independent biological replicates. The gene expression level was calculated using the Fragments Per Kilobase per Million reads (FPKM) method.
RT-qPCR analysis of JAZ genes of turnip under abiotic stress and exogenous phytohormone treatment
Plant growth and treatments
The seeds of turnip cultivar “Qiamagu” were purchased from Tian Di He Co., Ltd. (Urumqi, China). All the experimental research on plants were conducted according to the proper guidelines and legislation of national and international regulations. Seeds were sterilized using sodium hypochlorite (5%) for 15 min and then rewashed with distilled water for 15 min. Thereafter, seeds were placed on filter paper in 9-cm petri dishes filled with 5 mL distilled water to germinate. Germinated turnip seeds (1-mm radicle emerged from the seed coat) were planted in plastic pots (20 × 12 cm) with coconut fiber as the substrate. Every pot was planted with 3 seedlings. All pots were placed in the greenhouse where the temperature was maintained at 25℃ and the photoperiod was 16 h/8 h (day/night). Each pot was irrigated with 50 mL of 1/2 Hoagland nutrient solution every 3 days. Two-week-old (two leaves) turnip seedlings with uniform sizes were selected for different abiotic stresses and exogenous phytohormone treatments.
Phytohormone treatment The turnip seedlings were sprayed with 100 μmol/L salicylic acid, abscisic acid, and methyl jasmonate, respectively.
Abiotic stress treatment The turnip seedlings were irrigated with 100 mmol/L NaCl solution as salt stress treatment. The turnip seedlings were placed in the 4 °C incubators as low-temperature treatment.
There were three repetitions in every treatment, and each repetition consisted of 9 plants. After 24 h, the leaf samples of every treatment were taken and frozen in liquid nitrogen and stored at – 80 °C for RNA extraction.
Extraction of total RNA and analysis of gene expression
Total RNA of turnip leaves was extracted using Trizol Kit (Beyotime, China). The quantity and purity of RNA were estimated by nanodrop microspectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). First-strand cDNA synthesis was carried out by reverse transcription Kit (Takara, Japan) with gDNA eraser. The specific primers of BrrJAZ genes were designed using NCBI primer-blast tools.
The sequences, amplification length, and locations of each primer have been listed in Table S1, and the specificity of the amplification products was tested by agarose gel electrophoresis. Each reaction contained 1.0 μL of cDNA, 0.4 μL of forward and reverse primer (10 μM), 10.0 μL of 2× SYBR qPCR Master Mix (Biosharp, China), and 8.2 μL double-distilled H2O in a total reaction volume of 20 μL and was conducted in ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with 3 technical replicates by using hard-shell PCR plates. The reaction conditions were as follows: 95 °C for 3 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 20 s. The 2−ΔΔCT algorithm was used to analyze the relative gene expression levels. β-Actin of turnip was used as the internal control to normalize the expression of the target genes. Between phytohormone treated and control samples, statistical analysis to find significant differential expression was determined using a two-tailed Student’s t-test in SPSS version 19.0 (IBM, Chicago, IL, USA, https://www.ibm.com/analytics/spss-statistics-software).
Research involving plants
Experimental research and field studies on plants in this work comply with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Results
Identification and chromosome mapping of JAZ genes in turnip genome
Based on the genome data of turnip, HMM search was carried out using the HMM profiles of the TIFY domain (PF06200) and JAS domain (PF09425) as queries against the local protein database. By retrieving the database, we detected 35 non-redundant sequences. Then, 36 and 37 homologous proteins were obtained according to the BLASTP search using 18 A. thaliana JAZ proteins and 36 B. oleracea JAZ proteins, respectively. Subsequently, all the candidate JAZ proteins were merged and scanned using NCBI-CDD for the identification of their conserved domains. Finally, a total of 36 non-redundant JAZ genes were identified in turnip, including 26 JAZ, 2 PPD, 5 ZML, and 3 TIFY genes (Table S2).
Basic information of nucleotide and amino acid sequences of the BrrJAZ genes was summarized (Table 1). Based on the chromosomal location and the subfamily classification, the 36 JAZ genes in B. rapa were renamed. The length of these JAZ proteins ranged from 112 (BrrTIFY2) to 364 (BrrTIFY3) amino acid (aa) residues with an average length of 248.75 aa. The molecular weight ranged from 12.02 to 39.68 kDa, and the pI values varied from 4.56 to 10.02. Subcellular localization prediction showed that all JAZ proteins were in the nucleus.
All 36 JAZ genes were assigned to ten chromosomes of B. rapa (Fig. 1), and the distribution of the JAZ genes on each chromosome was uneven. Chromosome 8 contained the largest number of JAZ genes (6 genes), followed by chromosomes 1, 2, and 7, which contained 5 genes. Only one JAZ gene was located on chromosome 4.
Phylogenetic analysis of JAZ protein in turnip
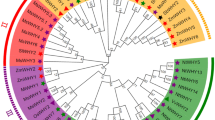
Based on the amino acid sequences of full-length JAZ proteins in A. thaliana (18), B. oleracea (36), and B. rapa (36), the phylogenetic tree was constructed using the neighbor-joining method in MEGA 7 software. The 90 JAZ proteins were grouped into eight clades (Fig. 2). Among these clades, Clade 1 was formed with 5 TIFY proteins (1 of A. thaliana, 2 of B. rapa, 2 of B. oleracea). Six PPD proteins (2 of A. thaliana, 2 of B. rapa, 2 of B. oleracea) were gathered together in Clade 2. Clades 3, 6, and 7 were three JAZ subfamily clades, including 7 (1 of A. thaliana, 3 of B. rapa, 3 of B. oleracea), 6 (2 of A. thaliana, 2 of B. rapa, 2 of B. oleracea), 12 (3 of A. thaliana, 5 of B. rapa, 4 of B. oleracea) members, respectively. Clade 4, 5, and 8 were mixed branches. Clade 4 was formed with TIFY (3 of B. oleracea) and JAZ proteins (2 of A. thaliana, 3 of B. oleracea, and 5 of B. rapa). Clade 5 was composed of 26 proteins, all of which were members of the JAZ except for the BoTIFY7 protein. 12 ZML (2 of A. thaliana, 5 of B. oleracea, and 5 of B. rapa) and 3 TIFY (each species possessed one TIFY protein) proteins were clustered in Clade 8.
Gene structure and conserved motif analysis of JAZ genes in turnip
The phylogenetic relationships of the 36 JAZ family genes in turnip were closely related to their gene structures and motif compositions (Figs. 3, 4). The TIFY subfamily proteins did not contain the JAS domain. Five genes of the ZML subfamily clustered in one group, which contained similar motif compositions and gene structures, and their protein sequences contained the GATA structural domain. The 26 members of the JAZ subfamily all possessed the TIFY and JAS domains. The TIFY domain corresponded to motif 1. Motif 2 constituted the JAS domain. The EAR domain corresponded to motif 3. The GATA domain of ZML subfamily consists of motif 4 (Fig. 3B).
Gene structure, conserved motif and genetic relationship of JAZ protein in turnip. (A) Phylogenetic analysis of BrrJAZ proteins. The phylogenetic tree was performed in MEGA 7 with the neighbor-joining method. (B) The distribution of conserved motifs in BrrJAZ proteins. Each motif was represented by a colored box. (C) Exon/intron structure and conserved domains of BrrJAZ genes. Exons and introns were represented by yellow boxes and black lines, respectively. Each conserved domain was represented by a colored box.
We compared the CDS sequences of turnip JAZ family genes and analyzed their exon–intron structures (Fig. 3C). The results showed that, among the JAZ family genes, BrrTIFY2 and BrrJAZ8 had the simplest gene structure, containing only one exon, whereas BrrZML2 contained the highest number (9) of exons. BrrZML2 had the highest number of introns (8).
Gene duplication and collinearity analysis of JAZ family genes in turnip
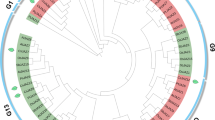
Gene duplication events can lead to the expansion of gene families and play a crucial role in the adaptation by acquiring new gene functions. Given the importance of gene duplication in the evolution of plant gene families, we analyzed the duplication patterns of 36 JAZ family genes in the turnip genome, and 44 homologous duplicated gene pairs were identified (Fig. 5). Among these homologous duplicated gene pairs, BrrJAZ14/BrrJAZ18 and BrrJAZ15/BrrJAZ16 are two tandem duplicated gene pairs, while the other homologous gene pairs are formed by segmental duplication or whole-genome duplication. To estimate the evolutionary rates and selective pressure of the JAZ gene family in turnip, Ka and Ks analysis was subsequently performed (Table 2).
In the turnip genome, the ka/ks values of 44 duplicated JAZ gene pairs were lower than 1, suggesting that JAZ family genes evolved mainly under the influence of purifying selection.
The turnip JAZ family genes were distributed on 10 chromosomes, of which chromosome 2 (14), chromosome 7 (16), and chromosome 8 (15) had the highest number of homologous genes. BrrJAZ6 of chromosome 2, BrrJAZ20 of chromosome 8, and BrrJAZ23 of chromosome 9 contained the highest number (5) of homologous genes in the turnip genome, while BrrTIFY2 had no homologous genes in the turnip genome.
To infer the evolutionary relationship of JAZ genes among different species, the genomes of A. thaliana, B. oleracea, and turnip were analyzed by collinearity (Fig. 6). We detected many collinear blocks between their genomes. A total of 54 homologous JAZ gene pairs existed between the A. thaliana and turnip genomes. The homologous fragments between the two species were mainly distributed on chromosome 1 of A. thaliana, with 31 JAZ gene pairs. Chromosome 8 of turnip contained 10 homologous gene pairs.
A total of 121 pairs of JAZ genes between the B. oleracea and turnip genomes were covalently related. Homologous segments containing more pairs between species were mainly found on chromosomes 7 and 8 of turnip, containing 22 and 21 homologous pairs, respectively. Correspondingly, on chromosomes 6 and 8 of B. oleracea, containing 20 and 24 JAZ homologous pairs, respectively.
Transcriptome analysis of JAZ family genes in turnip
To explore the expression of JAZ family genes of turnip involved in tuberous root development, we analyzed the transcriptomic data published by Li et al. (Fig. 7). The expression patterns of turnip JAZ family genes in the three developmental periods could be distinguished. A total of five genes, including BrrJAZ9, BrrJAZ10, BrrJAZ19, BrrJAZ22, and BrrTIFY2, had no detectable expression. Most members of the TIFY and ZML subfamilies have close gene expression patterns, suggesting similar functions in the processes involved in tuberous root development. The diverse expression patterns of JAZ family genes in the three periods suggest that these members play more enriched functions in participating in the development of turnip tuberous roots.
Expression analysis of JAZ family genes in turnip under abiotic stress and exogenous phytohormone treatment
To understand the expression pattern of JAZ family genes of turnip under different exogenous phytohormone and abiotic stress treatments, the leaves of turnip treated with ABA, SA, MeJA, salt stress, and low-temperature stress for 24 h were collected in this study, and the expression of JAZ family genes in each treatment was detected by qRT-PCR (Fig. 8). We found that BrrJAZ21 and BrrZML3 responded to all treatments. In all treatments, the expression of the above two genes was significantly different from the control.
The analysis of expression data showed that most of the JAZ family genes were up-regulated under exogenous ABA treatment. Among the 15 JAZ family genes tested, the expression of BrrJAZ1, 7, 11, 15, 17, 19, 21, 24, and BrrZML3 were significantly up-regulated.
After exogenous SA treatment, the 15 JAZ genes exhibited distinct expression patterns. The expression levels of BrrJAZ4, 9, 21, 25 and BrrZML3 were significantly up-regulated compared with the control group, while the expression of BrrJAZ2 and BrrJAZ18 were significantly down-regulated.
Exogenous MeJA treatment increased the expression of the 15 JAZ family genes detected in the leaves of turnip seedlings. Except for the gene expression of BrrJAZ11, which was not significantly different from the control group, the gene expression of the other 14 genes was significantly increased compared with the control group.
After salt stress treatment, the gene expression of BrrJAZ2 and BrrJAZ18 was significantly lower than that of the control, while BrrJAZ21 and BrrZML3 were significantly higher than that of the control.
We also analyzed the expression of JAZ genes in turnip under low temperature stress. The expression of BrrJAZ2, 7, 21 and BrrZML3 were significantly up-regulated after low temperature stress treatment, whereas BrrJAZ9 was significantly lower than the control group.
Discussion
JAZ is a plant-specific gene family with prominent roles in the regulation of many physiologic processes in plant growth and stress response through JA signalings, such as seed germination36, flower development37, response to salt, drought, high temperature, wound, and diseases18,38,39. However, few studies have been reported on the functional analysis of turnip JAZ gene family members. Therefore, in this study, we identified the JAZ family genes in the turnip genome and analyzed the sequence information of each member to investigate their expression patterns under abiotic stresses and exogenous phytohormone treatments.
JAZ family genes were widely identified in some Brassica crops. Previous research identified 36, 38, 36, and 36 JAZ genes in B. rapa L.40, B. juncea var. tumida41, B. napus L.42, and B. oleracea var. capitata30, respectively. In the present study, we identified 36 members of JAZ genes in the turnip genome. This result suggested that the number of JAZ family genes is conservative and has not changed significantly during the process of species formation. The composition of the turnip JAZ gene family members was more similar to that of other dicotyledons. The JAZ protein sequences of the tea plant43, Arabidopsis44, tomato45, and Brassica30,40,41,42 all contain members of the TIFY, JAZ, and PPD subfamilies, and members of these three subfamilies have been identified in the JAZ family protein sequence of turnip. JAZ family genes have numerous members and are likely to perform different functions in response to adversity stress.
Gene duplications contribute to the expansion of new gene family members and provide an opportunity for novel functions in the evolution of the plant genome. Therefore, investigating gene duplication can help us understand the evolution of genes and species. Whole-genome duplication, segmental duplication, and tandem duplication are the three main pathways of gene duplication44. Previous studies showed that no tandem duplication events of JAZ family genes were found in the B. rapa L. and B. juncea var. tumida40,41, whereas two pairs of tandem duplication genes were identified in Brassica oleracea var. capitata30. In concert with the findings in B. oleracea var. capitata, we also detected only two pairs of tandem duplication JAZ genes in the turnip genome. Our results indicate that whole-genome duplication or segmental duplication were predominant duplication events for JAZ genes.
To determine the selective evolutionary pressure for BrrJAZ genes differentiation after duplication, Ka and Ks values for duplicated BrrJAZ gene pairs were calculated using the Ka/Ks calculator. In general, Ka/Ks = 1 indicates neutral selection, Ka/Ks > 1 indicates positive selection, and Ka/Ks < 1 indicates purification selection46. Our results showed that the Ka/Ks value of each duplicated BrrJAZ gene pair was less than 1, which indicated the purification selection during evolution. Similarly, the Ka/Ks values of duplicated homologous gene pairs in the JAZ gene family of Solanum lycopersicum45, Phyllostachys edulis47, B. oleracea var. capitata30, and Petunia48 were less than 1, indicating that the JAZ genes of these species were subjected to strong purifying selection, which may have led to functional conservation or pseudogenization. In contrast, in the maize genome, three repetitive blocks had Ka/Ks > 1, indicating accelerated evolution under positive selection45.
JAZ proteins may be involved in the root development process in plants. Han et al. found that JAZ proteins interact with RHD6/RSL1, a transcription factor that regulates root growth, repressing the transcriptional function of RHD6 and interfering with the interaction between RHD6 and RSL1, suggesting that JAZ proteins play an important role in Arabidopsis root development49. In this study, after mining transcriptome data of Li et al. during the development of turnip tuberous roots, we found that JAZ family genes varied greatly during three periods of turnip tuberous root growth, indicating that JAZ genes are likely to be involved in the development of turnip tuberous roots, and this will be used as an entry point for validation in future studies50.
Plants regulate responses to growth, development and environmental stresses at the transcriptional level. Therefore, we analyzed the expression of JAZ family genes in turnips under different stress conditions. Our results showed that most BrrJAZs responded significantly to abiotic stress and/or exogenous phytohormone treatments, which is in agreement with the results obtained in other Brassica crops30,40,41,42.
Many studies have demonstrated that exogenous JAs treatment can strongly induce the expression of JAZ genes. Saha et al. found that the expression of JAZ genes was significantly up-regulated by exogenous JA treatment, increased 15-fold to 800-fold compared with the control40. Liu et al. found that all BoJAZ family genes were up-regulated after exogenous MeJA treatment, and the expression of 8 genes showed a highly significant increased, which was more than fivefold higher than the control30. Our results were in agreement with the findings above. We found that MeJA treatment increased the expression of JAZ family genes. The expression of BrrJAZ4 was elevated the most compared to the control group, reaching 36.4-fold.
Different expression patterns of JAZ family genes emerged after exogenous SA treatment. A total of seven genes showed significant differences in expression from the control. Among them, two genes were significantly down-regulated in expression, while five genes were significantly up-regulated in expression. Liu et al. found that the expression of JAZ family genes showed insignificant changes after induction by exogenous SA, and only 3 of the 22 JAZ genes were up-regulated30. This suggests that although closely related species have similar numbers of JAZ family genes and relatively close phylogenetic relationships, they may have different functions.
JAZ genes are transcriptional repressors of jasmonate-responsive genes, which contain two highly conserved sequence regions: N-terminal ZIM/TIFY structural domain mediates homomeric and heteromeric interactions between most JAZ proteins. C-terminal JAS domain plays a key role in destabilizing JA-Ile response repressors51. Abiotic stresses such as low-temperature, drought, and salt stress can induce up-regulation of JAZ gene expression in rice. Moreover, overexpression of OsTIFY11a significantly increased tolerance to salt and dehydration stresses26. In grapes, 11 TIFY genes were found to be responsive to osmotic stress and low-temperature stress28. Our findings were slightly different from the above studies. We found that most of the turnip JAZ genes were not significantly changed under salt stress treatment. Among the 15 genes tested, only two genes were significantly up-regulated and two genes were significantly down-regulated. Moreover, the qPCR data revealed that only a small number of genes were up-regulated in expression under low-temperature stress, while most JAZ family genes did not show significant differences in expression compared to the control. Taken together, the above qPCR data analysis showed that the BrrZML3 gene responded positively to all exogenous plant hormone treatments and abiotic stress treatments. This is most likely related to its gene structure.
Conclusions
In this study, we identified 36 JAZ genes from the turnip genome and classified them into four subfamilies. They were unevenly distributed among 10 chromosomes. Gene structure and conserved motifs of BrrJAZs were similar within the subfamilies, but the differences between the subfamilies were large. Although the proteins varied in length, MW, and pI, all contained a conserved TIFY or JAS domain. Phylogenetic and collinearity analysis provided some valuable clues to the evolutionary characteristics of BrrJAZ genes. Expression analysis suggested JAZ genes may be involved in the formation of turnip tuberous root, and they also participated in the response to salt and low-temperature stress. Several BrrJAZ genes were also responsive to ABA, SA and MeJA treatment. Overall, our findings will help understand the biological functions of the BrrJAZ genes in turnip.
References
Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 4. https://doi.org/10.1093/aob/mcm079 (2007).
Wasternack, C. & Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. https://doi.org/10.1093/jxb/erw443 (2017).
Yu, X. et al. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 46, 197–212. https://doi.org/10.1071/fp18106 (2019).
Santino, A. et al. Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 32, 1085–1098. https://doi.org/10.1007/s00299-013-1441-2 (2013).
Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. https://doi.org/10.1016/j.tplants.2015.02.001 (2015).
Goossens, J., Fernandez-Calvo, P., Schweizer, F. & Goossens, A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689. https://doi.org/10.1007/s11103-016-0480-9 (2016).
Per, T. S. et al. Jasmonates in plants under abiotic stresses: Crosstalk with other phytohormones matters. Environ. Exp. Bot. 145, 104–120. https://doi.org/10.1016/j.envexpbot.2017.11.004 (2018).
Kazan, K. & Manners, J. M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31. https://doi.org/10.1016/j.tplants.2011.10.006 (2012).
Yan, J. et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. https://doi.org/10.1105/tpc.109.065730 (2009).
Thines, B. et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 (2007).
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. https://doi.org/10.1038/nature09430 (2010).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010).
Katsir, L., Chung, H. S., Koo, A. J. & Howe, G. A. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435 (2008).
Wang, C., Liu, Y., Li, S. S. & Han, G. Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167, 872–886. https://doi.org/10.1104/pp.114.247403 (2015).
Pieterse, C. M. J., Pierik, R. & van Wees, S. C. M. Different shades of JAZ during plant growth and defense. New Phytol. 204, 261–264. https://doi.org/10.1111/nph.13029 (2014).
Singh, A. P., Pandey, B. K., Mehra, P., Heitz, T. & Giri, J. OsJAZ9 overexpression modulates jasmonic acid biosynthesis and potassium deficiency responses in rice. Plant Mol. Biol. 104, 397–410. https://doi.org/10.1007/s11103-020-01047-2 (2020).
Zhao, C. et al. Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean. Mol. Breed. 40, 1–13. https://doi.org/10.1007/s11032-020-01113-z (2020).
Chung, H. S. et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964. https://doi.org/10.1104/pp.107.115691 (2008).
Hori, Y., Kurotani, K., Toda, Y., Hattori, T. & Takeda, S. Overexpression of the JAZ factors with mutated jas domains causes pleiotropic defects in rice spikelet development. Plant Signal. Behav. 9, e970414. https://doi.org/10.4161/15592316.2014.970414 (2014).
Lin, K. et al. Beyond genomic variation–comparison and functional annotation of three Brassica rapa genomes: A turnip, a rapid cycling and a Chinese cabbage. BMC Genomics 15, 250. https://doi.org/10.1186/1471-2164-15-250 (2014).
Parveen, T., Hussain, A. & SomeshwarRao, M. Growth and accumulation of heavy metals in turnip (Brassica rapa L.) irrigated with different concentrations of treated municipal wastewater. Hydrol. Res. 46, 60–71. https://doi.org/10.2166/nh.2014.140 (2015).
Wu, Y. et al. Comparative expression analysis of heavy metal ATPase subfamily genes between Cd-tolerant and Cd-sensitive turnip landraces. Plant Divers. 41, 275–283. https://doi.org/10.1016/j.pld.2019.02.001 (2019).
Yang, Y. et al. A splice variant of BrrWSD1 in turnip (Brassica rapa var. rapa) and its possible role in wax ester synthesis under drought stress. J. Agric. Food Chem. 67, 11077–11088. https://doi.org/10.1021/acs.jafc.9b04069 (2019).
Jia, K., Yan, C., Yan, H. & Gao, J. Physiological responses of turnip (Brassica rapa L. subsp rapa) seedlings to salt stress. HortScience 55, 1567–1574. https://doi.org/10.21273/hortsci15187-20 (2020).
Ye, H., Du, H., Tang, N., Li, X. & Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71, 291–305. https://doi.org/10.1007/s11103-009-9524-8 (2009).
Chini, A., Ben-Romdhane, W., Hassairi, A. & Aboul-Soud, M. A. M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 12, e0177381. https://doi.org/10.1371/journal.pone.0177381 (2017).
Zhang, Y. et al. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 7, e44465. https://doi.org/10.1371/journal.pone.0044465 (2012).
Sun, Q. et al. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci. Rep. 7, 1–9. https://doi.org/10.1038/srep42418 (2017).
Liu, X. et al. Genome-wide identification, expression profile of the TIFY gene family in Brassica oleracea var. capitata, and their divergent response to various pathogen infections and phytohormone treatments. Genes 11, 127. https://doi.org/10.3390/genes11020127 (2020).
Finn, R. D. et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 34, 247–251. https://doi.org/10.1093/nar/gkj149 (2006).
Nishii, A. et al. Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 64, 1402–1409. https://doi.org/10.1271/bbb.64.1402 (2000).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Chen, C. et al. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 (2020).
Wang, Y. et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. https://doi.org/10.1093/nar/gkr1293 (2012).
Ju, L. et al. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 223, 246–260. https://doi.org/10.1111/nph.15757 (2019).
Yu, X. et al. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 267, 65–73. https://doi.org/10.1016/j.plantsci.2017.11.008 (2018).
He, X. et al. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 19, 896–908. https://doi.org/10.1111/mpp.12575 (2018).
Liu, S., Zhang, P., Li, C. & Xia, G. The moss jasmonate ZIM-domain protein PnJAZ1 confers salinity tolerance via crosstalk with the abscisic acid signalling pathway. Plant Sci. 280, 1–11. https://doi.org/10.1016/j.plantsci.2018.11.004 (2019).
Saha, G., Park, J. I., Kayum, M. A. & Nou, I. S. A genome-wide analysis reveals stress and hormone responsive patterns of TIFY family genes in Brassica rapa. Front. plant sci. 7, 936. https://doi.org/10.3389/fpls.2016.00936 (2016).
Cai, Z., Chen, Y., Liao, J. & Wang, D. Genome-wide identification and expression analysis of jasmonate ZIM domain gene family in tuber mustard (Brassica juncea var. tumida). PLoS ONE 15, e0234738. https://doi.org/10.1371/journal.pone.0234738 (2020).
He, X. et al. Genome-wide identification and functional analysis of the TIFY gene family in the response to multiple stresses in Brassica napus L. BMC Genomics 21, 736. https://doi.org/10.1186/s12864-020-07128-2 (2020).
Shen, J. et al. Genome-wide analysis reveals stress and hormone responsive patterns of JAZ family genes in Camellia sinensis. Int. J. Mol. Sci. 21, 2433. https://doi.org/10.3390/ijms21072433 (2020).
Howe, G. A. & Yoshida, Y. Evolutionary origin of JAZ proteins and jasmonate signaling. Mol. Plant 12, 153–155. https://doi.org/10.1016/j.molp.2019.01.015 (2019).
Heidari, P., Faraji, S., Ahmadizadeh, M., Ahmar, S. & Mora-Poblete, F. New insights into structure and function of TIFY genes in Zea mays and Solanum lycopersicum: A genome-wide comprehensive analysis. Front. Genet. 12, 657970. https://doi.org/10.3389/fgene.2021.657970 (2021).
Hurst, L. D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 18, 486–487. https://doi.org/10.1016/s0168-9525(02)02722-1 (2002).
Huang, Z. et al. Genome-wide identification and characterization of TIFY family genes in Moso Bamboo (Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses. PeerJ 4, e2620. https://doi.org/10.7717/peerj.2620 (2016).
Tian, S. et al. Genome-wide identification and characterization of JAZ protein family in two Petunia progenitors. Plants 8, 203. https://doi.org/10.3390/plants8070203 (2019).
Han, X., Zhang, M., Yang, M. & Hu, Y. Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 32, 1049–1062. https://doi.org/10.1105/tpc.19.00617 (2020).
Li, J. et al. Integrative analysis of mRNA and miRNA expression profiles of the tuberous root development at seedling stages in turnips. PLoS ONE 10, e0137983. https://doi.org/10.1371/journal.pone.0137983 (2015).
Pauwels, L. & Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089. https://doi.org/10.1105/tpc.111.089300 (2011).
Funding
This research was supported by the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China (IVF201701); Natural Science Project of University Scientific Research Plan of Xinjiang Autonomous Region (XJEDU2017S017); Postgraduate Innovation Project of Xinjiang Agricultural University (XJAUGRI2017001); and Xinjiang Uyghur Autonomous Region Key Discipline Fund of Horticulture Science (2016-10758-3).
Author information
Authors and Affiliations
Contributions
K.J., H.Z.Y., and J.G. conceived the study. Y.C.Y., J.Z., and Y.X.C. contributed to the sampling. W.W.L. collected and analyzed the data. K.J. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jia, K., Yan, C., Zhang, J. et al. Genome-wide identification and expression analysis of the JAZ gene family in turnip. Sci Rep 11, 21330 (2021). https://doi.org/10.1038/s41598-021-99593-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99593-2
This article is cited by
-
Genome-wide identification and expression analysis of PtJAZ gene family in poplar (Populus trichocarpa)
BMC Genomic Data (2023)
-
Identification and expression analysis of SQUAMOSA promoter-binding protein (SBP) genes in mungbean
Plant Biotechnology Reports (2023)
-
MdARF8: An Auxin Response Factor Involved in Jasmonate Signaling Pathway in Malus domestica
Journal of Plant Growth Regulation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.