Abstract
A vertically transmitted microsporidian, Microsporidia MB, with the ability to disrupt Plasmodium development was reported in Anopheles arabiensis from Kenya, East Africa. To demonstrate its range of incidence, archived DNA samples from 7575 Anopheles mosquitoes collected from Ghana were screened. MB prevalence was observed at 1.8%. An. gambiae s.s constituted 87% of positive mosquitoes while the remaining were from An. coluzzii. Both sibling species had similar positivity rates (24% and 19%; p = 0.42) despite the significantly higher number of An. gambiae s.s analysed (An. gambiae s.s = 487; An. coluzzii = 94; p = 0.0005). The microsporidian was also more prevalent in emerged adults from field-collected larvae than field-caught adults (p < 0.0001) suggestive of an efficient vertical transmission and/or horizontal transfer among larvae. This is the first report of Microsporidia MB in Anopheles mosquitoes in West Africa. It indicates possible widespread among malaria vector species and warrants investigations into the symbiont’s diversity across sub-Saharan Africa.
Similar content being viewed by others
Introduction
Mosquitoes remain very important vectors of human disease, and several efforts are being made to reduce the burden they pose to human and animal health. As the progress of chemical-based vector control interventions is continually threatened, there is increased focus on alternative strategies including biotechnological ones for disease control1. Therefore, the search for natural mosquito-associated symbionts with the ability to reduce vector competence has been a growing interest.
It has recently been demonstrated that a vertically transmitted microsporidian prevalent in An. arabiensis in Kenya disrupts Plasmodium development2. Contrary to other mosquito-associated microsporidians, Microsporidia MB does not confer any significant negative effect on host’s fertility, fecundity, development, and longevity2,3,4. These characteristics make Microsporidia MB an appealing candidate for control of parasite transmission in Anopheles mosquitoes.
A recent discovery revealed the prevalence of Microsporidia MB only in An. arabiensis2. However, malaria is transmitted by a wide range of Anopheline species with different ecological ranges and varied efficiencies in transmitting Plasmodium5,6. In this study, we sought to extend the geographical range and identification of Microsporidia MB to other Anopheles species. We screened archived Anopheles mosquito DNA from field collections of several unrelated projects to detect the existence of Microsporidia MB among Anopheles populations in Ghana. To the best of our knowledge, this is the first study since the discovery of this microsporidian in An. arabiensis from Kenya that has investigated its incidence in different Anopheles mosquito species and from another geographically distant African population.
Results
Microsporidia MB detected in An. gambiae s.s and An. coluzzii
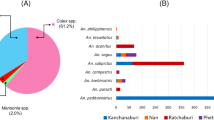
Microsporidia MB was detected in 133 out of 1158 DNA samples from mosquitoes collected across Ghana (Fig. 1). Accounting for the number of individual mosquitoes that went into pools for DNA extraction, a total of 7575 individual mosquitoes were involved in this analysis (Fig. S1). MB was found only in the DNA samples from single mosquitoes, giving an overall MB prevalence of 1.8% (133/7575) in the total number of mosquitoes analysed. Microsporidia MB was identified only in the DNA from An. gambiae s.s and An. coluzzii; the former constituting 87.2% of MB-positive mosquitoes (Fig. 2; Supplementary Table S1). The distribution of MB infection among An. gambiae s.s and An. coluzzii was 23.9% and 19.3%, respectively (Fig. 2) and this rate did not differ significantly between the two sibling species (χ2 = 0.66; p = 0.42).
Higher MB detection rate in field-caught larvae than adults
The DNA samples used in this study comprised those extracted from adults that emerged from field-collected larvae and those that were collected from the field as adults. We were interested in establishing whether infections were more prevalent in newly emerged adults (from field-caught larvae) or in the field-caught adults. This was also to assess which of the two life stages would increase the probability of finding MB in a field study aimed at MB discovery. For this analysis, An. arabiensis, An. melas and An. funestus were excluded since they did not show positive infection. Of the remaining 7534 An. gambiae s.l mosquitoes, ~ 90% were field-caught adults. However, among the remaining 10% of samples that were initially collected from the field as larvae, 17.3% recorded positive for MB. This was significantly higher than the prevalence (0.13%) observed in field-caught adults (χ2 = 1092; p < 0.0001).
Discussion
We make the first report of Microsporidia MB in An. gambiae s.s and An. coluzzii following identification of the symbiont in An. arabiensis. This does not only demonstrate the existence of the microsporidian in another predominant malaria vector species in Africa but also extends its incidence from East to West Africa. The prevalence of MB-positive mosquitoes was estimated to be 1.8%, which is within the rate of < 1–9% reported for An. arabiensis2. The present study took advantage of archived mosquito DNA samples which were either collected from the field as larvae or adults from different study sites over 5 years.
Anopheles gambiae s.s and An. coluzzii are the predominant malaria vectors in Ghana7,8,9. They are often found in sympatry with one species usually being more abundant9,10. Contrary to the study by Herren and colleagues2, a handful of An. arabiensis was analysed in the present study. Anopheles arabiensis is more commonly found in the arid north of Ghana where rainfall is observed within a few months in a year. In studies conducted in Ghana that are focused on Anopheles distribution, between 2–3% are An. arabiensis despite the collection of large numbers of mosquitoes7,11. However, we acknowledge that there were more collections from the south of the country, especially Greater Accra, which contributed 40% of the DNA samples used in this study and 89% of MB-positive mosquitoes. Further studies to investigate variations in mosquito species density and seasonal prevalence of Microsporidia MB will shed more light on the field dynamics of the symbiont in these mosquito populations.
DNA samples from mosquitoes initially collected as larvae from the field showed significantly higher MB-positivity than those collected as adults. Microsporidians can be transmitted both vertically and horizontally12. The efficiency with which MB is transmitted vertically depends on the intensity in the ovaries of the female parent. Horizontal transfer was initially speculated to occur in the larval habitat2 which would increase the spread of MB from few infected larvae to many in the breeding site, but this transmission route has recently been shown not to occur in larvae under laboratory conditions13. In effect, our detection of higher prevalence among newly emerged adult mosquitoes is more likely to be attributed to highly efficient vertical transmission from female parent to offspring2. In the laboratory, the intensity of MB in adult mosquitoes has been observed to increase with age2. However, in the wild where conditions are very dynamic, the effect of larval habitat conditions and adult age on MB intensity in mosquitoes may differ. For example, larval diet has been shown to affect the microbial composition of mosquitoes in their adult life stages14. It is also established that microbial diversity decreases in female adults mainly because of proliferation of certain bacteria species following sugar and/or blood feeding15. It, therefore, makes it challenging to explain variations in bacterial diversity among field-caught adult mosquitoes since their feeding histories are unknown16. Several environmental and physiological statuses could potentially affect the intensity of MB observed in adults and variations in infection prevalence could be governed by factors that are yet to be investigated. Given that collecting large numbers of adult mosquitoes in the field may prove more challenging than larval sampling, our results have also shown the increasing chances of finding MB infections in a population when larvae are collected in field studies2.
While the data presented here shows basic information about the ecological spread of Microsporidia MB, it has nonetheless demonstrated a potential widespread occurrence of Microsporidia MB among Anopheles mosquitoes across sub-Saharan Africa. It warrants further investigation of the diversity, environmental dynamics, and interactions with other mosquito symbionts for a clearer understanding of their possible use in malaria control.
Methods
Description of Anopheles samples
We retrieved Anopheles mosquito DNA samples from various studies that have been conducted at the Noguchi Memorial Institute for Medical Research, University of Ghana between 2014–2019. Mosquitoes had been collected from different sites across the country (Fig. 1) either as larvae or adults during the rainy season period. When collected as larvae, they were reared to adults for experimental assays before DNA was extracted. Most DNA samples were from single female mosquitoes while few were extracted from pools of 25 (Supplementary Table S1). The species information on the samples were retrieved from the different projects. Where the mosquitoes were only identified as An. gambiae s.l, these were further assessed for their sibling species identification using Restriction Fragment Length Polymorphism (RFLP)17 or SINE18 methods when they were positive for Microsporidium MB infection.
Quality check of archived samples
Different extraction methods, including CTAB, Trizol RNA/DNA and columns had been used by the various projects to obtain DNA. It was therefore expected that the DNA integrity would differ among samples. The samples would have also gone through some freeze–thaw cycles which would compromise the quality of the DNA and result in false negatives. To address this concern, we randomly selected samples for DNA quantification with Qubit Fluorometer (Thermofisher, UK) and purity using the BioDrop (Biochrom, UK). Average DNA concentration was 34.8 ng/μL (1.02—204 ng/μ) and average purity was 1.7 (1.14–2.17). To reduce potential contaminants that would limit PCR, DNA samples were diluted 1 in 10.
Detection of Microsporidia MB
A total of 1158 DNA samples were screened for Microsporidia MB using MB18SF/ MB18SR primers2 and with Microsporidia MB DNA obtained from Jeremy Herren’s lab as positive control in each set of reactions run. Diluted DNA samples were used in a first round of PCR reactions. The PCR included in the final reaction 1X One-Taq Master mix, 0.4 µM of each primer and 1µL of DNA template. The cycling conditions were as described in2. Products were loaded and run on a 2% agarose gel stained with SYBR Safe DNA stain (Invitrogen) and viewed under a blue-light transilluminator. The PCR reaction was repeated for samples that showed no band for Microsporidia MB using an increased volume of the diluted DNA and/or using 1µL of the stock DNA sample to confirm the initial results. The band size for Microsporidia MB detection was ~ 500 bp (Supplementary Fig. S2).
Statistical analyses
Contingency analyses to compare independence observed positivity between mosquito species was performed using a two-sided Chi-squared test with Yate’s correction. The distribution of An. gambiae s.s and An. coluzzii from the study sites was tested with a two-tailed unpaired Mann–Whitney non-parametric test. All test significance was accepted a p < 0.05.
References
Jones, R. T., Ant, T. H., Cameron, M. M. & Logan, J. G. Novel control strategies for mosquito-borne diseases. Philos. Trans. R. Soc. B Biol. Sci. 376, 20190802 (2021).
Herren, J. K. et al. A microsporidian impairs Plasmodium falciparum transmission in Anopheles arabiensis mosquitoes. Nat. Commun. 11, 2187 (2020).
Agnew, P., Bedhomme, S., Haussy, C. & Michalakis, Y. Age and size at maturity of the mosquito Culex pipiens infected by the microsporidian parasite Vavraia culicis. Proc. R. Soc. B Biol. Sci. 266, 947–952 (1999).
Undeen, A. H. & Alger, N. E. The effect of the microsporidan, Nosema algerae, on Anopheles stephensi. J. Invertebr. Pathol. 25, 19–24 (1975).
Wiebe, A. et al. Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar. J. 16, 85 (2017).
Sinka, M. E. et al. A global map of dominant malaria vectors. Parasit. Vectors 5, 69 (2012).
de Souza, D., Kelly-Hope, L., Lawson, B., Wilson, M. & Boakye, D. Environmental factors associated with the distribution of Anopheles gambiae s.s in Ghana; an important vector of lymphatic filariasis and malaria. PLoS ONE 5, e9927 (2010).
Mattah, P. A. D. et al. Diversity in breeding sites and distribution of Anopheles mosquitoes in selected urban areas of southern Ghana. Parasit. Vectors 10, 1–15 (2017).
Kudom, A. A. Larval ecology of Anopheles coluzzii in Cape Coast, Ghana: Water quality, nature of habitat and implication for larval control. Malar. J. 14, 447 (2015).
Akorli, J. et al. Seasonality and locality affect the diversity of Anopheles gambiae and Anopheles coluzzii midgut microbiota from Ghana. PLoS ONE 11, e0157529 (2016).
Hamid-Adiamoh, M. et al. Insecticide resistance in indoor and outdoor-resting Anopheles gambiae in Northern Ghana. Malar. J. 19, 314 (2020).
Andreadis, T. G. & Hall, D. W. Development, Ultrastructure, and Mode of Transmission of Amblyospora sp. (Microspora) in the Mosquito. J. Protozool. 26, 444–452 (1979).
Nattoh, G. et al. Horizontal transmission of the symbiont Microsporidia MB in Anopheles arabiensis. Front. Microbiol. 12, 1995 (2021).
Saab, S. A. et al. The environment and species affect gut bacteria composition in laboratory co-cultured Anopheles gambiae and Aedes albopictus mosquitoes. Sci. Rep. 10, 1–13 (2020).
Wang, Y., Gilbreath, T. M., Kukutla, P., Yan, G. & Xu, J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE 6, e24767 (2011).
Osei-Poku, J., Mbogo, C. M., Palmer, W. J. & Jiggins, F. M. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 21, 5138–5150 (2012).
Fanello, C., Santolamazza, F. & della Torre, A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med. Vet. Entomol. 16, 461–464 (2002).
Santolamazza, F. et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar. J. 7, (2008).
Acknowledgements
We thank Jeremy Herren, ICIPE, Kenya for sharing DNA sample of Microsporida MB which was used as positive control. This research was funded by the Wellcome Trust [220737/Z/20/Z]. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
J.A. conceived and co-ordinated the study. E.A.A., S.T., M.O. screened samples for microsporidian infection. G.A., R.P., M.A., D.A., S.P.-B., J.C., S.K.D. carried out or co-ordinated field collection and archiving of the mosquitoes used in the study. J.A. analysed the data and wrote the manuscript. All authors reviewed, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Akorli, J., Akorli, E.A., Tetteh, S.N.A. et al. Microsporidia MB is found predominantly associated with Anopheles gambiae s.s and Anopheles coluzzii in Ghana. Sci Rep 11, 18658 (2021). https://doi.org/10.1038/s41598-021-98268-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-98268-2
This article is cited by
-
High temperatures and low humidity promote the occurrence of microsporidians (Microsporidia) in mosquitoes (Culicidae)
Parasites & Vectors (2024)
-
First identification of Microsporidia MB in Anopheles coluzzii from Zinder City, Niger
Parasites & Vectors (2024)
-
First report of natural infection of Anopheles gambiae s.s. and Anopheles coluzzii by Wolbachia and Microsporidia in Benin: a cross-sectional study
Malaria Journal (2024)
-
Microsporidia MB in the primary malaria vector Anopheles gambiae sensu stricto is avirulent and undergoes maternal and horizontal transmission
Parasites & Vectors (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.