Abstract
Vertebrate photoreceptors are categorized into two broad classes, rods and cones, responsible for dim- and bright-light vision, respectively. While many molecular features that distinguish rods and cones are known, gene expression differences among cone subtypes remain poorly understood. Teleost fishes are renowned for the diversity of their photoreceptor systems. Here, we used single-cell RNA-seq to profile adult photoreceptors in zebrafish, a teleost. We found that in addition to the four canonical zebrafish cone types, there exist subpopulations of green and red cones (previously shown to be located in the ventral retina) that express red-shifted opsin paralogs (opn1mw4 or opn1lw1) as well as a unique combination of cone phototransduction genes. Furthermore, the expression of many paralogous phototransduction genes is partitioned among cone subtypes, analogous to the partitioning of the phototransduction paralogs between rods and cones seen across vertebrates. The partitioned cone-gene pairs arose via the teleost-specific whole-genome duplication or later clade-specific gene duplications. We also discovered that cone subtypes express distinct transcriptional regulators, including many factors not previously implicated in photoreceptor development or differentiation. Overall, our work suggests that partitioning of paralogous gene expression via the action of differentially expressed transcriptional regulators enables diversification of cone subtypes in teleosts.
Similar content being viewed by others
Introduction
In vertebrates, photoreceptor cells are categorized into two classes, rods and cones, which together are able to respond to a broad range of light intensities from dim starlight to bright sunshine1,2,3. Rods are primarily responsible for dim-light vision at night, whereas cones mediate bright-light vision and color discrimination4. Visual pigments, consisting of an opsin and a covalently bound chromophore, are the light-sensitive molecules of photoreceptors5,6. Absorption of a photon by a visual pigment activates the phototransduction cascade, which induces photoreceptor hyperpolarization and synaptic transmission to second-order neurons7 (Fig. 1A). Phototransduction pathways in rods and cones are composed of distinct opsins and signal transduction components8. Rod- and cone-specific phototransduction genes arose before or during two rounds of whole-genome duplication, which occurred in a chordate ancestor prior to the emergence of vertebrates ~ 600 million years ago (Mya)9,10. These gene duplications permitted subsequent partitioning of paralogous gene expression between rods and cones and fine-tuning of individual phototransduction components to meet the needs of dim- and bright-light vision. In this way, the duplex retina was established at an early stage of vertebrate evolution2,8.
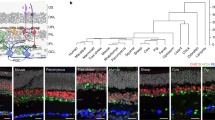
Transcriptome profiles of adult zebrafish photoreceptor subtypes. (A) Schematic representation of the major cell classes in the zebrafish retina based on a prior design7. Photoreceptor cell types and ON and OFF bipolar cells are highlighted in color, whereas other retinal cell types are in grey. The ON bipolar cell cluster in our single cell data expresses genes specific to both rod ON bipolar cells (prkcaa) and cone ON bipolar cells (gnao1b, gnb3a, trpm1a, rgs11, and isl1). See also Fig. S1. ONL outer nuclear layer, INL inner nuclear layer, GCL ganglion cell layer. (B) Isolation of rod and cone cells from transgenic adult zebrafish expressing green fluorescent protein (GFP). GFP-positive cells were collected from each line. A small percentage of GFP-negative cells was also included in the analysis. (C) Automatic clustering of single-cell expression profiles reveals six distinct photoreceptor populations. The plot shows a two-dimensional representation (UMAP) of global gene expression relationships among 2186 cells. (D) Heatmap showing top five differentially enriched genes for each cell population (rows). Columns correspond to single cells grouped by cell cluster. Each cell cluster is colored as in panel (C). Values are row-wise Z-scored gene-expression values. See also Fig. S1A. Full list of differentially enriched genes is provided in Supplementary Data S1.
Retinas detect color by comparing the relative activation of multiple cone subtypes, each maximally sensitive to distinct wavelengths. Maximal sensitivity (λmax) is primarily determined by the opsin subfamily a cone expresses and the chromophore it contains5,11. Prior to the two rounds of whole-genome duplication, four main cone opsin subfamilies emerged via local gene duplication and subsequent molecular diversification: ultraviolet (UV)- (opn1sw1; SWS1; range of λmax = 360–420 nm), blue- (opn1sw2; SWS2; λmax = 400–470 nm), green- (opn1mw; RH2; λmax = 460–510 nm) and red-sensitive opsins (opn1lw; LWS; λmax = 510–560 nm)12,13. Exclusive expression of one of these opsin subfamilies is the defining characteristic of UV, blue, green, and red cones in many vertebrate species.
Teleost fishes occupy a wide diversity of aquatic habitats and have expanded their opsin repertoires to adapt to these diverse photic niches. Zebrafish (Danio rerio) is widely used as a model system in photoreceptor research14,15. While the zebrafish genome encodes a single UV cone opsin gene (opn1sw1) and a single blue cone opsin gene (opn1sw2), it contains a syntenic array of four green cone opsins tuned to a range of wavelengths (opn1mw1, λmax = 467 nm; opn1mw2, λmax = 476 nm; opn1mw3, λmax = 488 nm and opn1mw4, λmax = 505 nm), as well as a tandem array of two red cone opsin genes (opn1lw1, λmax = 558 nm; opn1lw2, λmax = 548 nm)16. Prior studies showed differential expression of these green and red cone opsin paralogs across the retina and over developmental time17, but the physiological role unique to each individual opsin paralog remains largely unknown. More broadly, a teleost-specific whole-genome duplication occurred ~ 350 Mya at the origin of the teleost lineage18. Similar to the genome duplications that occurred earlier in vertebrate evolution9,10, this additional teleost whole-genome duplication produced numerous paralogous pairs of photoreceptor-expressed genes, but the expression pattern of these genes among zebrafish photoreceptor subtypes remains largely unknown.
Cellular identity is determined by the combinatorial expression of transcription factors and their cofactors. The multiplicity of cone photoreceptor subtypes in the zebrafish retina makes this species an ideal model for understanding how transcriptional regulators control the development and diversification of closely related, but distinct cell types. Previous studies have identified multiple transcription factors required for vertebrate photoreceptor development and function. In mammals, many transcriptional regulators play a role in both rod and cone development (OTX2, CRX, RAX, MEF2D, and NEUROD1)19,20,21. Whereas others are more specifically involved in rod (RORB, NRL, NR2E3, ESRRB, CASZ1, and SAMD7)22,23,24,25,26,27 or cone development and/or function (THRB, RXRG, RORA, COUP-TFI/COUP-TFII, and OC1/OC2)28,29. In zebrafish, studies have begun to identify additional transcription factors required for the development of specific cone subtypes: tbx2b in UV cones30, foxq2 in blue cones31, six6a, six6b, and six7 in blue and green cones32, and thrb in red cones33,34. Despite these advances, the architecture of the transcriptional regulatory networks that govern photoreceptor diversification in zebrafish remain largely unknown. Given the sophistication and complexity of their photoreceptor systems, it is likely that many additional transcriptional regulators remain to be discovered in fish.
Here, we used single-cell RNA-seq to profile adult zebrafish photoreceptors. We identified unique subpopulations of green and red cones in the ventral retina which express red-shifted opsin paralogs and share a specialized complement of phototransduction genes. In addition, we found that other cone subtypes differentially express phototransduction gene paralogs which arose either during the teleost-specific genome duplication or later in specific teleost sub-lineages. Lastly, we discovered numerous transcriptional regulators associated with differential gene expression across zebrafish photoreceptor subtypes; many of these factors were not previously known to be associated with photoreceptor gene regulation.
Results
Single-cell transcriptome profiling of adult zebrafish photoreceptors
To reveal the extent of gene expression diversity among adult zebrafish photoreceptor subtypes, we generated single-cell transcriptome data using a droplet-based approach (10 × Chromium single-cell RNA-seq). We obtained enriched populations of photoreceptor cells for our analysis by using two transgenic zebrafish, Tg(rho:EGFP)ja2Tg and Tg(gnat2:EGFP)ja23Tg, which express GFP in rods and all cone subtypes, respectively35,36. GFP-positive cells from each line and a small percentage of GFP-negative cells were isolated via fluorescence-activated cell sorting (FACS) and subjected to single-cell RNA-seq (scRNA-seq) analysis (Fig. 1B). To enhance our ability to detect photoreceptor-specific transcripts, we updated the existing transcript annotation with a publicly available transcriptome derived from adult zebrafish eye (see “Methods”). We used this updated annotation for all of our analyses. We then subjected the scRNA-seq data to multiple rounds of clustering, filtering, and selection to identify 2186 high-quality cells for subsequent bioinformatic analysis. In addition to photoreceptors, we included bipolar cells in our analysis to serve as an ‘outgroup’, since bipolar cells are the cell type most closely related to photoreceptors at the level of gene expression37,38.
Unsupervised clustering of the scRNA-seq data categorized cells into eight distinct populations, including five canonical photoreceptor subtypes (UV, blue, green, and red cones and rods) defined by their enriched expression of individual opsin genes: UV cone opsin (opn1sw1), blue cone opsin (opn1sw2), green cone opsins (opn1mw1, opn1mw2, and opn1mw3), red cone opsin (opn1lw2), and rod opsin (rho) (Fig. 1C,D and Fig. S1A). Unsupervised clustering also identified an additional cone population consisting of a mixture of opn1mw4-expressing green cones and opn1lw1-expressing red cones (Fig. 1D and Fig. S1A). We will discuss this unique opn1mw4+/opn1lw1+ population in greater detail below. In addition to these photoreceptor clusters, we identified two populations defined by the expression of the bipolar cell marker genes cabp5a and vsx139,40 (Fig. 1C,D). These two clusters express genes previously shown to be specific to either mouse ON cone bipolar cells (e.g., gnao1b, gnb3a, trpm1a, rgs11, and isl1) or rod bipolar cells (prkcaa), and OFF cone bipolar cells (e.g., fezf2, neto1, and zfhx4) (Fig. 1D, Fig. S1A, and S1B)41.
Analysis of differential gene expression among the eight clusters revealed ~ 1100 differentially expressed genes (Fig. 1D, Fig. S1, and Supplementary Data S1). These genes include subtype-defining opsin genes as well as previously identified subtype markers, such as rom1a (rod)42, foxq2 (blue)31, thrb (red)31, and si:busm1–57f23.1 (red)33. We also identified two pre-microRNAs (mir729 and mir726) expressed exclusively in UV and red cones, respectively, similar to what was previously described in medaka (Oryzias latipes)43 (Fig. S2). Hierarchical clustering of the top 15 differentially expressed genes from each cluster revealed that a considerable number of genes showed co-expression in multiple cone subtypes (e.g., red + green, UV + blue, etc.) (Fig. S1C). This analysis also showed that genes that are highly specific to single cone subtypes are quite rare (Fig. S1C and Supplementary Data S1) and include tbx2a (UV), grk7b (UV), tgfa (UV), mpzl2b (blue), fibcd1a (green), angptl4 (green), and glis3 (red). To validate these scRNA-seq results, we performed quantitative PCR analysis using reverse transcribed mRNA derived from GFP- or tdTomato-positive subpopulations of rods, cones, and bipolar cells isolated by FACS with lines of transgenic zebrafish (Fig. S3). This analysis confirmed the scRNA-seq results for 24 differentially expressed genes, underscoring the overall validity of our profiling data.
A unique subpopulation of double cones in the ventral retina
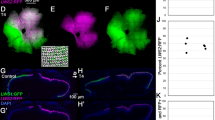
In addition to the four canonical cone subtypes, unsupervised clustering identified a unique subpopulation of cones expressing either opn1mw4 or opn1lw1 (referred to here as opn1mw4+/opn1lw1+). These cells occupied the region between canonical green and red cone clusters in the 2D plot produced by uniform manifold approximation and projection (UMAP, Fig. 1C) and were defined as members of a single cluster despite their expression of opsins from two different classes (opn1mw4 is a green cone opsin and opn1lw1 is a red cone opsin). In many teleosts including zebrafish, red and green cones form a closely apposed pair referred to as a ‘double cone’44. Given that opn1mw4 and opn1lw1 are expressed primarily in the ventral retina17, our data suggest that opn1mw4+ and opn1lw1+ cones together form a unique subtype of double cone with a transcriptional profile distinct from that of the green and red cones which comprise canonical double cones. Indeed, even when we subdivide the opn1mw4+/opn1lw1+ population into two sub-clusters (opn1mw4+ and opn1lw1+) based on opsin expression, we see that they share a distinctive combination of genes (Fig. 2A,B). Compared to the opn1mw1/2/3+/opn1lw2+ population, opn1mw4+/opn1lw1+ cones are enriched for three genes known to be expressed in the ventral retina: the phototransduction gene gngt2a45 and the transcription factors vax1 and vax246 (Fig. 2B). The expression of multiple ventrally expressed genes strongly suggests that opn1mw4+ and opn1lw1+ cones are localized to the ventral retina. The transcriptomes of opn1mw4+ and opn1lw1+ cones also show depletion of various genes, including multiple phototransduction components (gnb3b, pde6c, gngt2b, rcvrn2, and cnga3a), relative to canonical green and red cones (Fig. 2B). Lastly, opn1mw4 and opn1lw1 have the most red-shifted spectral sensitivity (λmax) of all green and red cone opsins encoded in the zebrafish genome (Fig. 2C)16. These opsins are well-adapted for detecting downwelling light which has a broader and more red-shifted spectral distribution than sidewelling or upwelling light (Fig. 2D,E). The potential functional significance of opn1mw4+ and opn1lw1+ cones is discussed in greater detail in the “Discussion”. Overall, these results show that photoreceptor subpopulations may be defined by region-specific gene expression signatures that supersede overly simplistic classifications based on opsin expression alone.
A distinctive subpopulation of red and green cones in the ventral retina. (A) Left: UMAP plot of cell clusters from Fig. 1C. Cell clusters except for green and red cones are colored gray. The opn1mw4+/opn1lw1+ cells were split into two sub-clusters (opn1mw4+ and opn1lw1+) based on the expression of opn1mw4+ and opn1lw1+. Right: Expression of green and red cone opsin genes within the cell populations enclosed by the dotted box in the UMAP plot. (B) Expression of the top 30 most differentially enriched genes (ranked by adjusted p-value) between ventral (opn1mw4+ and opn1lw1+) and dorsal/central (opn1mw1/2/3+ and opn1lw2+) green and red cones. Green and red cone clusters were identical to those in (A). Dot size reflects the percentage of cells within the cluster expressing the gene, and dot color indicates average expression level within the cluster. (C–E) Ventrally localized opn1mw4+ and opn1lw1+ cones are positioned to detect downwelling light. (C) Intensity/spectral distributions for two lines of sight (downwelling light and upwelling light, 20° and 150° from vertical, respectively). These spectra were measured at a depth of 3 m in the lagoon of Enewetok (formerly Eniwetok) Atoll in the Marshall Islands. Data are reproduced from a previous study62. The maximum sensitivity of green and red opsin genes are indicated as dotted lines overlying the intensity/spectral distributions16. (D) From an underwater vantage point, all light from above the water surface enters via a circular aperture known as Snell’s window, which subtends an angle of ~ 96° relative to the fish’s eye irrespective of depth. Scattering and absorption by water cause the dominant wavelengths of transmitted light to vary with the direction of the line of sight. (E) The approximate location of the opn1mw4+ and opn1lw1+ cones is based on a prior in situ hybridization study17
Partitioning of teleost-specific phototransduction gene paralogs among photoreceptor subtypes
Expression partitioning of phototransduction gene paralogs between rods and cones occurred early in vertebrate evolution8 and mediates key functional differences between these cell classes. The extent to which such partitioning occurs among cone subtypes is currently unknown. Yet, the unique combination of phototransduction genes expressed by opn1mw4+ and opn1lw1+ cones suggests that other cone subtypes might also differ with respect to the expression levels of phototransduction genes. We therefore examined the expression patterns and evolutionary origins of all phototransduction-related genes in the zebrafish genome. Guided by the published literature8,45,47,48, we identified a total of 63 phototransduction genes, including opsins (Fig. 3A), and found that 46 of these 63 genes arose either during the teleost-specific whole-genome duplication (3R) or during clade- or species-specific gene duplication events after 3R (Fig. 3B,C). The remaining 17 phototransduction genes arose during earlier vertebrate genome duplications (1R or 2R) or before.
Expression partitioning of paralogous phototransduction genes. (A) Schematic representation of vertebrate phototransduction cascade components. During phototransduction, light-activated opsin induces the detachment of the catalytic subunit Gα of the heterotrimeric G protein (transducin) from the inhibitory β/γ subunits. The activated Gα subunit then binds to the two inhibitory γ subunits of cGMP phosphodiesterase 6 (PDE6), thereby relieving inhibition on the catalytic subunits (α, β, and α′). The activated PDE subunits, in turn, catalyze the hydrolysis of the second messenger cGMP, leading to closure of cGMP-gated channels (CNG) on the plasma membrane and photoreceptor membrane hyperpolarization. Shut-off of the activated transducin is accelerated by a GTPase-activating protein complex (RGS9 and R9AP). The light-activated opsin is quenched via phosphorylation mediated by visual pigment kinases (GRK) and by the subsequent binding of arrestins. The activity of GRKs is regulated by binding of recoverin in a calcium-dependent manner. In the recovery/adaptation process, guanylyl cyclase activating protein (GCAP) enhances the synthesis of the second messenger cGMP through guanylyl cyclase (GC) in a calcium-dependent manner. Na+/Ca2+, K+ exchanger (NCKX) is involved in maintaining the dynamic equilibrium of calcium ions in the outer segment. In cones, the ion channel (CNG) and exchanger (NCKX) are located in the plasma membrane, whereas in rods they are located in the disc membrane. Figure design is adapted from Larhammar et al., 200980. (B) Phylogenetic tree showing the approximate time points at which various genome duplications occurred. (C) Evolutionary scenario for gene duplications of vertebrate phototransduction cascade genes (Left panels) and heatmap showing their expression levels in each cell population (Right panels). Left: The four dotted vertical lines mark the events, ‘1R’, ‘2R’, ‘II’, ‘III’, described in (B). The horizontal axis is not to scale. White circles indicate putative ancestral genes. Black circles indicate genes encoded in the zebrafish genome. Evolutionary branching patterns for each gene family are described according to the described previous studies8,45,47,48 and our BLAST searching results. Figure design is adapted from Lamb, 20208. Right: heatmap showing average expression levels of phototransduction genes in each cluster. Values are row-wise Z-scored gene-expression values. rcvrnb expression is not detected.
Inspection of gene expression patterns reveals that 59 out of 63 phototransduction genes are differentially expressed among photoreceptor and bipolar populations (Fig. 3C, Fig. S4, and Supplementary Data S1). As expected, we found differentially expressed gene ‘pairs’ between rods and cones (gnat1 vs. gnat2, gnb1a/gnb1b vs. gnb3b, pde6a/pde6b vs. pde6c, pde6ga/pde6gb vs. pde6ha/pde6hb, cnga1a/cnga1b vs. cnga3a/cnga3b, cngb1a vs. cngb3.1/cngb3.2, and saga/sagb vs. arr3a/arr3b). All of these gene pairs arose during early vertebrate whole-genome duplications, and their differential expression between rods and cones is conserved between fish and amniotes (e.g., mice and chickens)37,49.
We also identified extensive expression partitioning among cone subtypes. All differentially partitioned genes except for opn1sw1 and opn1sw2 arose during the teleost-specific whole-genome duplication (arr3a/arr3b, cnga3a/cnga3b, grk7a/grk7b, gngt2a/gngt2b, and rcvrn2/rcvrn3) or later (opn1mw1/2/3/4, opn1lw1/2, guca1e/guca1e.2, and cngb3.1/cngb3.2) (Fig. 3C). We noted above the suite of phototransduction genes (cnga3a/cnga3b, gngt2a/gngt2b, and rcvrn2/rcvrn3) differentially expressed between ventral (opn1mw4 and opn1lw1) and dorsal/central (opn1mw1/2/3 and opn1lw2) green and red cones (Fig. 2B). Additionally, we found multiple pairs of paralogous genes that were differentially enriched between UV cones (grk7b, cngb3.2, and guca1e) and other cone types (grk7a, cngb3.1, and guca1e.2). We also detected partitioning of cone arrestin paralogs between UV and blue cones (arr3b) and green and red cones (arr3a), as previously described50. On the other hand, four pairs of paralogous rod phototransduction genes (cnga1a/cnga1b, gnb1a/gnb1b, pde6ga/pde6gb, and saga/sagb), which arose during the teleost-specific whole-genome duplication, are both expressed in rods. The expression of rgs9a/rgs9b, another pair of genes that arose during the teleost-specific duplication, is partitioned between rods (rgs9b) and cones (rgs9a). Finally, we found that two teleost-specific cone-type transducin β genes, gnb3a and gnb3b, were partitioned between cones (gnb3b) and ON bipolar cells (gnb3a). In contrast, the single Gnb3 ortholog in mouse and chicken is expressed in both cones and ON bipolar cells37,49. The expression patterns of several phototransduction gene pairs were confirmed by RT-qPCR analysis of FACS-isolated photoreceptor subtypes and bipolar cells (Fig. S3). In summary, single-cell transcriptome profiling and phylogenetic analysis demonstrate extensive expression partitioning of cone-expressed genes that arose during the teleost-specific whole-genome duplication or later. These differences may mediate differential tuning of the light response in these individual cone subtypes.
Transcriptional regulatory networks in zebrafish photoreceptors
Transcription factors, cofactors, and chromatin regulators play crucial roles in controlling cell fate and regulating gene expression. To elucidate the relationship between transcriptional regulators and their target genes in zebrafish photoreceptors, we employed a machine learning-based approach, GEne Network Inference with Ensemble of trees (GENIE3) in SCENIC51,52. GENIE3 calculates weight scores for each transcriptional regulator (out of a total of 1932), measuring its respective relevance for predicting the expression of each of 59 differentially expressed phototransduction genes (Fig. 4 and Fig. S5). In the following paragraphs, we highlight those transcriptional regulators most strongly implicated in control of phototransduction gene expression by this approach (see “Methods”).
Candidate transcriptional regulators responsible for expression of phototransduction genes. Heatmap showing positive (red) and negative (blue) associations between transcriptional regulators (transcription factors and cofactors) and differentially expressed phototransduction genes (target genes) calculated by GENIE3 algorithm in SCENIC. Rows and columns are arranged according to divisive hierarchical clustering (dividing clusters in a top-down manner). The (dis)similarity of observations was calculated using Euclidean distances. Cell type expression patterns of the transcriptional regulators are presented in Fig. S5A. Gene#1: zgc:114046; Gene#2: zgc:110269; Gene#3: si:ch211–288g17.3.
We identified a total of 61 different transcriptional regulators associated with expression of phototransduction genes (Fig. 4). These regulators can be roughly categorized by cell class based on the expression pattern of genes they control: bipolar cells (7 regulators), rods (14), all cones (18), and cone subtypes (22). Our analysis ‘rediscovered’ most rod and cone transcriptional regulators known from previous studies in zebrafish and other vertebrates. In addition, it nominated multiple regulators not previously implicated in photoreceptor gene regulation. Among regulators of rod genes, casz1, nr2e3, rorb, and nrl showed the strongest positive association with rod-specific target genes. Two of these genes (nr2e3 and nrl) are known to be essential for rod development in both mice23,24 and zebrafish53,54, and mouse orthologs of casz1 and rorb are important in photoreceptor gene regulation22,26. Other known photoreceptor transcriptional regulators positively associated with rod gene targets include neurod1, samd11, and esrrd (related to mouse Esrrb)25,55,56. Regulators not previously associated with rod gene expression include hmgb2a, mafba, pbx3b, tead3b, tp53inp1, ybx1, and znf536.
We also identified numerous transcriptional regulators broadly associated with gene expression across cone subtypes or within specific subtypes. Known regulators active in multiple cone subtypes include: six6b, six7, roraa, rx1 (rax2a), rx2 (rax2b), and rxrga. The first two genes (six6b and six7) were previously shown to be required for development of blue and green cones in zebrafish32,36, and orthologs of the latter four have all been implicated in photoreceptor development in birds or mammals19,57,58. Novel potential cone regulators include: crema, foxo1a, foxo3b, hif1ab, hipk2, hsf4-like, lbh-like, lrrfip1a, mef2cb, sall1a, tfe3a, and zfand5a. Our analysis also implicated 22 transcriptional regulators in the control of cone subtype-specific gene expression. Known regulators such as tbx2b, foxq2, and thrb show the strongest positive associations with UV, blue, and red cone-specific marker genes, respectively30,31,34. We also discovered novel candidate regulators of UV cone genes (tbx2a), UV and blue cone genes (skor1a), and red cone genes (sox6 and glis3). Additionally, we identified vax1 and vax2 as putative regulators in opn1mw4+ and opn1lw1+ cones; these two factors showed strong positive associations with the target genes: opn1mw4, opn1lw1, and gngt2a. The tandemly arrayed cone opsin genes (opn1mw1/2/3/4 and opn1lw1/2) were positively, but weakly, associated with several regulators (cxxc4, fosab, mier1b, pbx1a, ripply1, tie2b, thrap3a, zic2a, and zic6).
We next used SCENIC to perform gene regulatory network analysis with a set of differentially expressed non-phototransduction target genes (Fig. S5 and S6, see “Methods” for further detail). We identified many of the same transcriptional regulators that we found for phototransduction gene targets. In addition, we found additional known regulators of rod and cone gene targets, including six6a and otx5 (both positively associated with cone gene expression). CRX, an ortholog of otx5, plays a critical role in photoreceptor gene regulation in mammals21, and six6a (along with six6b and six7) was previously shown to be required for blue and green cone gene expression in zebrafish32. This new analysis also highlights multiple additional regulators that are weakly associated with cone gene expression: cbx4, hlfa, hmga1b, hsf1, meis1b, mlf1, mn1a, nr2f1b, phf19, rxrba, si:ch73-386h18.1, ss18l2, top1l, tsc22d3, tshz3a, and xbp1. Collectively, these analyses highlight many transcriptional regulators of photoreceptor gene expression known from prior studies in zebrafish and other vertebrates and reveal a wide range of novel factors potentially involved in the regulation of rod-, pan-cone-, or cone-subtype-specific gene expression.
Discussion
In the present study, we used scRNA-seq to profile adult zebrafish photoreceptors. We identified a distinctive subpopulation of green and red cones concentrated in the ventral region of the retina, which expresses red-shifted opsin paralogs and a unique complement of phototransduction genes. We also found that canonical UV, blue, green, and red cone subtypes differentially express paralogous phototransduction genes, which arose either during the teleost-specific genome duplication or later. Lastly, we discovered numerous transcriptional regulators associated with differential gene expression across zebrafish photoreceptor subtypes. This work lays a foundation for future studies aimed at understanding how molecular differences among cone subtypes affect photoreceptor function.
Early vertebrate whole-genome duplications (1R and 2R) provided the raw genetic material for the subsequent evolution of distinct rod- and cone-specific opsins and phototransduction components. Similarly, we find that zebrafish cone subtypes show differential expression of many phototransduction genes, but in this case, nearly all differentially expressed paralog pairs appear to have arisen during the teleost-specific whole-genome duplication (3R) ~ 350 Mya or later. This finding suggests that partitioning of paralogous gene expression may be a common mechanism of cell type diversification that permits physiological fine-tuning of serially ‘paralogous’ cell types. Interestingly, additional whole-genome duplication events have occurred in teleosts: in the common ancestor of salmonids (~ 80 Mya)59 and in the common ancestor of goldfish (Carassius auratus) and the common carp (Cyprinus carpio) (~ 14 Mya)60. Future analyses of differential gene expression among photoreceptor subtypes in salmon or goldfish may reveal whether expression partitioning of paralogous phototransduction components invariably follows genome duplication and how rapidly it occurs in evolution.
We observed distinct patterns of opsin and phototransduction gene expression between dorsal/central double cones (opn1mw1/2/3 and opn1lw2) and ventral double cones (opn1mw4 and opn1lw1) (Fig. 2). The partitioning of gene expression between these two populations suggests that opn1mw4+ and opn1lw1+ cones may be specialized for the detection of light with special qualities. In the wild, zebrafish are known to inhabit shallow, slow-moving streams and pools61. A classic study of light in a shallow, tropical marine environment showed that downwelling light along the solar axis is more red-shifted, of far greater intensity, and noisier (due to surface turbulence) than light along all other lines of sight62 (Fig. 2C–E). This downwelling light is expected to impinge upon the retina in the distribution of the opn1mw4+ and opn1lw1+ cones when the fish is in a horizontal position (Fig. 2D,E), suggesting that the expression of red-shifted opsin paralogs in these cells may serve to enhance detection of red-shifted downwelling light. Conversely, it has been proposed that reduction of gnb3b expression in the ventral zebrafish retina may have evolved to protect photoreceptors from high-intensity downwelling light by decreasing the gain of the phototransduction cascade45. In either case, our findings reveal a complex suite of changes in the expression of phototransduction genes in ventral cones, suggesting the existence of functional adaptations at multiple levels of the phototransduction cascade.
We also found expression partitioning of 3R-duplicated phototransduction genes among canonical cone subtypes, most notably between UV cones and non-UV cones (Fig. 3). The functional role of this distinctive gene expression signature in adult UV cones is currently unknown, but a prior study of larval zebrafish retina found distinctive gene expression in UV cones within the ‘strike zone’, a region of the ventral-temporal retina specialized for the detection of UV-reflective prey63. The adult UV cone-enriched paralogs are all involved in calcium-mediated feedback regulation of the phototransduction shut-off cascade and recovery/adaptation in cones (Fig. 3)8,14,50,64. Future functional studies will be required to determine the precise functional role of this adult UV cone-enriched gene expression program.
Our analysis of transcriptional regulators in zebrafish photoreceptors implicated dozens of factors in the control of rod, pan-cone, and cone subtype genes (Fig. 4 and Fig. S6). A role for many of these factors in photoreceptor development had been previously demonstrated in other vertebrates, but not in zebrafish. Thus, our findings underscore a striking degree of evolutionary conservation within vertebrate photoreceptor transcriptional networks, extending from fish to mammals. Zebrafish retain the full set of four canonical cone subtypes inferred to have been present in the common ancestor of fish and mammals, while mammals lost two of those cell types (opn1sw2-expressing blue cones and opn1mw-expressing green cones) in the course of evolution65. So-called ‘green’ cones in mice and humans express orthologs of fish opn1lw (not opn1mw) and therefore arose from ancestral red cones12. The retention of the four ancestral cone types in zebrafish makes this species an excellent system for discovering features of vertebrate photoreceptor transcription networks that may have been lost in mammals. In addition, the present study suggests an even greater degree of complexity within the photoreceptor transcription network than previously suspected, revealing many transcriptional regulators not previously implicated in photoreceptor development or differentiation. Some of these novel factors likely play determinative roles in photoreceptor cell fate, whereas others may fine-tune gene expression in more subtle ways.
Methods
Zebrafish husbandry
Zebrafish were raised and maintained according to established protocols66. All experiments were designed according to the ARRIVE guidelines, carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and approved by the Washington University in St. Louis Institutional Animal Care and Use Committee (protocol# 19-1110). Adult fish were raised in a 14-h light/10-h dark cycle and fed with dry food once per day and with rotifers twice per day. Tg(rho:EGFP)ja2Tg35, Tg(gnat2:EGFP)ja23Tg36, Tg(-5.5opn1sw1:EGFP)kj9Tg67, Tg(-3.5opn1sw2:EGFP)kj11Tg68, and Tg(opn1mw2:EGFP)kj4Tg69 fish were obtained from the Zebrafish International Resource Center and National BioResource Project Zebrafish. Tg(thrb:Tomato)q22Tg34 was obtained from Dr. Rachel Wong at the University of Washington. TgBAC(vsx1:GFP)nns5Tg70 was obtained from Dr. Ryan B. MacDonald at University College London.
Isolation of rod and cone photoreceptors by fluorescence-activated cell sorting (FACS)
For isolation of rod and cone photoreceptors, we used the transgenic zebrafish lines, Tg(rho:EGFP)ja2Tg35 and Tg(gnat2:EGFP)ja23Tg36, which express GFP in rods and all cone subtypes, respectively. Five-month-old adult zebrafish were euthanized by submersion in ice water and their retinas were harvested at around zeitgeber time 3 (ZT 3). The dissected retinas were washed twice with calcium- and magnesium-free Hanks’ balanced salt solution (HBSS). Two retinas were incubated for 10 min at 37 °C in 400 µl of an activated papain dissociation solution (50 mM HEPES, 2.5 mM l-Cysteine, 0.5 mM EDTA, 23 U/ml Papain Suspension [LS0003126, Worthington] in calcium- and magnesium-free HBSS), that had been pre-activated by incubation for 10 min at 37 °C. After papain incubation, the retinas were centrifuged at 1500×g for 30 s. The supernatant was removed, and the retinas were further incubated for 5 min at 37 °C in 600 µl of 10% fetal bovine serum (FBS) in Dulbecco modified Eagle medium (DMEM) with 5 mM magnesium and 5 U DNaseI (Cat. No. 04716728001, Roche). The incubated samples were then gently triturated five times with a P1000 pipette to generate a single-cell suspension. The dissociated retinas were then centrifuged at 300×g for 5 min. After drawing off the supernatant, the samples were gently triturated again five times with a P1000 pipette in 300 µl of sorting buffer (20 mM HEPES and 0.04% bovine serum albumin in Calcium- and magnesium-free HBSS, pH 7.4). Four retinas from two individuals were combined for Tg(rho:EGFP) fish, while six retinas from three individuals were combined for Tg(gnat2:EGFP) fish. The combined samples were each passed through a 35 µm nylon mesh filter into a polypropylene FACS tube. Cell viability was evaluated by incubating the cells in a solution of propidium iodide (10 µg/ml) on ice for 5 min before cell sorting. The filtered samples were also incubated in a solution of Hoechst 33342 (5 µg/ml) to label nuclei. GFP-positive cells were isolated with a fluorescence activating cell sorter (FACSAria, BD Biosciences). Cells were initially filtered by forward- and side-scatter signals. Dead cells were then removed based on propidium iodide positivity. Intact rods and cones were then selected based on the presence of both blue (Hoechst 33342) and green fluorescence (GFP). About 35,000 viable, intact GFP-positive cells (PI-, GFP+, Hoechst+) and 1500 GFP-negative cells (PI-, GFP-, Hoechst+) were collected from Tg(gnat2:EGFP) fish, while 10,000 viable, intact GFP-positive cells and 1500 GFP-negative cells were collected from Tg(rho:EGFP) fish. These isolated cells were collected in 600 µl of the sorting buffer in 1.5 ml microtubes. The collected cells were then centrifuged at 300×g for 5 min and the supernatant was removed. Cell density was quantified on a hemocytometer, and ~ 6000 cells were used for sequencing library preparation.
Assembling an adult zebrafish eye transcriptome
We retrieved publicly available strand-specific RNA-seq data for the adult zebrafish eye (European Nucleotide Archive, ERR4029230)71. StringTie (v2.1.4)72 was used to assemble a genome-guided transcriptome with an improved annotation file (v4.3.2.gtf)73 as an initial guide. RNA-seq reads were mapped onto the reference transcripts in a strand-specific manner using the STAR aligner74 with the command-line options --outSAMattrIHstart 0 --outFilterIntronMotifs RemoveNoncanonical --outSAMstrandField intronMotif. Next, StringTie was used with the command-line options (--rf -t -G) to assemble new transcripts based on the RNA-seq reads with the reference annotation file (v4.3.2.gtf) guiding the assembly process. StringTie was then rerun with the command line option (merge) to obtain an updated transcript annotation, which contained both reference transcripts and non-redundant assembled transcripts predicted by the sequencing reads. The novel transcripts were named according to StringTie’s naming convention (e.g., MSTRG.19429). Some of the novel, de novo loci may correspond to non-coding RNAs or enhancer RNAs. We included these ‘genes’ in our analysis to enhance cell clustering. The StringTie merge mode concatenates transcript IDs of multiple genes when those transcripts overlap with each other, and the expression levels of these concatenated genes are counted as a single gene in the 10X Genomics Cell Ranger pipeline. To determine which of the concatenated genes is actually differentially expressed among cell clusters, we manually inspected pseudo-bulk RNA seq reads described in the following section. The gene symbol of the differentially expressed gene was then used to replace the corresponding concatenated name.
Single-cell RNA-seq
Sample preparation and sequencing
Single-cell libraries were prepared using the Chromium v3 platform (10X Genomics, Pleasanton, CA) according to the manufacturer's instructions. Both GFP-positive and GFP-negative cells were collected from adult zebrafish Tg(rho:EGFP)ja2Tg and Tg(gnat2:EGFP)ja23Tg as described in the previous section, and approximately 6000 single cells were used for library preparation. Single cells were partitioned into Gel beads in EMulsion (GEMs) using the GemCode instrument, followed by cell lysis and reverse transcription of RNA, amplification, shearing, adaptor ligation, and sample index attachment. Libraries were sequenced on an Illumina NovaSeq machine (540 million paired-end reads: Read 1: 28 bp, Read 2: 98 bp). Sample demultiplexing, alignment to the genomic reference (GRCz11), quantification, and initial quality control was performed using Cell Ranger software (version 6.0.0, 10X Genomics). The eye-specific transcript reference assembly described above was used for the alignment of reads. The GFP transcript sequence was added manually to the reference assembly as an extra chromosome. We initially obtained a matrix consisting of 27,931 genes × 12,833 cells. The greater number of the recovered cells (~ 13,000 cells) than expected (~ 6000 cells) suggested that a sizeable fraction of GEMs contain only ambient RNA or organelles such as mitochondria. These ‘cells’ were removed during subsequent data processing.
Data processing
Data were analyzed using the Seurat R package (v4.0.0)75. We retained all cells that expressed > 500 genes, and we required all genes to be expressed in at least five cells. Cells with greater than 30% mitochondrial gene content (likely representing dead cells) or > 40,000 unique molecular identifiers (likely representing doublets/multiplets) were removed from the analysis. For the remaining cells (8793 cells), a gene expression matrix was normalized to total cellular read counts using the negative binomial regression method implemented in the Seurat SCTransform function with the method set to glmGamPoi. The 3000 most variable genes, identified by the SCTransform function, were used for Principal Component Analysis (PCA). The top 30 principal components were selected for subsequent analysis according to the elbow plot. Graph-based clustering was performed to obtain a set of transcriptionally distinct clusters. At this point in the analysis, we deliberately set parameters to "over cluster" the data, to avoid combining distinct cell types and to identify sub-populations of low-quality cells for removal. In addition to photoreceptor cells, we initially identified several classes of retinal cells such as bipolar cell, horizontal cell, retinal pigment epithelium, and Müller glia. Of these non-photoreceptor cell types, we only retained bipolar cells which were used as an outgroup in our subsequent analyses.
Cell clustering and filtering
To retain high-quality photoreceptors and bipolar cells only, we subjected our data to multiple rounds of clustering, filtering, and selection. In the first round, we retained those clusters characterized either by the presence of one or more of the following opsin genes or phototransduction genes (rho, opn1sw1, opn1sw2, opn1mw1, opn1mw2, opn1mw3, opn1mw4, opn1lw1, opn1lw2, gnat1, or gnat2) or bipolar-specific genes (e.g., gnao1b, vsx1, cabp2a, cabp5a, and cabp5b) among the top 20 most differentially expressed genes as identified by the FindAllMarkers function in Seurat with the Wilcoxon rank sum test. We also removed clusters consisting of low-quality cells with low total gene counts (500–1000 genes/cell) compared with high-quality photoreceptor clusters (1000–3000 genes/cell for rods and 1000–4000 genes/cell for cones). A total of 2602 cells were retained after the first round of filtering. In the second round of clustering and selection, we removed clusters that showed co-expression of photoreceptor genes and Müller glial genes (icn, fxyd6l, mt2, rlbp1a, and glula). Previous single-cell studies showed that zebrafish Müller glia often show aberrant photoreceptor gene expression, likely due to adherence of fragments of photoreceptor cytoplasm to the cells76. We also removed one cluster showing co-expression of photoreceptor and bipolar genes and with low total gene counts. In the final round of clustering and selection, we removed a rod subpopulation with low total gene counts. A final set of 2186 high-quality cells was used for subsequent bioinformatic analyses.
Differential expression test
Genes differentially expressed among clusters were identified using the FindAllMarkers function in Seurat with arguments “test.use = ‘wilcox’, min.pct = 0.25, logfc.threshold = log2(1.5)”. This list of differentially expressed genes is available in Supplementary Data S1. Genes differentially expressed between ON and OFF bipolar cells were identified using the FindMarkers function in Seurat with arguments “test.use = ‘wilcox’, min.pct = 0.1”. Genes differentially expressed between dorsal/central and ventral green/red cones were identified using the same approach used for bipolar cells.
Pseudo-bulk average expression profile
The mapped sequence reads (BAM file) were subsetted using cellranger-dna bamslice to generate pseudo-bulk read counts for each cell cluster. The subsetted reads were each counted using subread featureCount v2.0.077 with an improved annotation file (v4.3.2.gtf)73. Normalized RNA sequencing reads (transcripts per million, TPM) of differentially expressed genes for each cluster are included in the list of differentially expressed genes (Supplementary Data S1).
Gene nomenclature
Phototransduction genes were manually curated from the Ensembl database [http://www.ensembl.org/; (Release 104)] according to the literature8,45,47,48. Some of these gene names were revised according to NCBI Gene database [www.ncbi.nlm.nih.gov/gene; (cited 2021 May)] as well as on the basis of manual BLAST searches. The revised gene names and their accession numbers are listed in Supplementary Table S1.
Transcriptional regulatory network analysis
We used the SCENIC R package51 (v1.2.4) to identify associations between transcriptional regulators and target genes in our datasets. One hundred cells were randomly chosen from each of the eight clusters, and standardized gene expression scores (scale.data in Seurat) derived from the total set of 800 cells were used for the analysis. A list of transcription factors, transcription cofactors, and chromatin regulators was retrieved with ZebrafishMine using the following gene ontology terms: “negative regulation of transcription, DNA-templated”, “regulation of transcription, DNA-templated”, “positive regulation of transcription, DNA-templated”, “DNA-binding transcription factor activity, RNA polymerase II-specific”, “RNA polymerase II cis-regulatory region sequence-specific DNA binding”, and “DNA binding”. We retained transcription factor/cofactor/chromatin regulators that were detected in at least 1% of the cells and which were represented by at least eight transcripts (normalized for each cell by the total expression and multiplied by a scale factor, 10,000) in total across all samples, yielding a total of 1932 genes. Phototransduction genes (target genes) were manually curated from Ensembl genome and/or NCBI databases according to the literature8,45,47,48. We only retained 59 phototransduction genes, which are included in the list of differentially expressed genes among clusters (Supplementary Data S1).
For the analysis of non-phototransduction-related target genes, we retained the top 10 differentially expressed genes for each photoreceptor cluster (Supplementary Data S1) after excluding both phototransduction genes and transcriptional regulatory genes. The GENIE3 algorithm52 was implemented in SCENIC to generate random forest weights of transcriptional regulators for each target gene. Weights reflect the predictive power of each regulator in determining the expression level of each target gene. In parallel, Spearman correlation coefficients between regulators and target genes were calculated using the runCorrelation function. To indicate whether a transcriptional regulator had an activating or repressive effect on a target gene’s expression, we preserved the sign of the Spearman correlation coefficient (i.e., a positive coefficient indicates an activating effect and a negative coefficient indicates a repressive effect). We used the “top5” cutoff in SCENIC to only display the strongest regulatory linkages identified by the algorithm. The selected transcriptional regulators and target genes were clustered using the Heatmap function in ComplexHeatmap78 with hierarchical clustering of the weights for visualization. We used customized versions of some SCENIC functions, regarding geneFiltering and runGenie3 to allow use of our gene lists. The custom scripts were made referring to the previous study79.
RT-qPCR validation of single-cell profiling results
Ages and genomic features for each transgenic fish used for RT-qPCR are described in Supplemental Table S2. Dissociated retinal cells were prepared for each transgenic fish as described in the section above, but without propidium iodide and Hoechst 33342 staining. Cells were filtered by forward- and side-scatter signals, and then 10,000 GFP- or tdTomato-positive were collected into 300 µl of lysis buffer (Buffer RL, Norgen Biotek Corporation) in 1.5 ml microtubes. Total RNA was extracted with a Single Cell RNA purification kit (Norgen Biotek Corporation). The extracted RNA was reverse-transcribed with SuperScript IV (Invitrogen) and oligo(dT) primers according to manufacturer’s instructions. The reverse-transcribed cDNA was subjected to quantitative PCR using Power SYBR Green Master Mix (Thermofisher Scientific) and the QuantStudio 3 Real-time PCR system (Thermofisher Scientific) according to manufacturer’s instructions. Expression levels were calculated by the relative standard curve method. The standard curve was prepared with serial dilutions of cDNA samples reverse-transcribed from total RNA of zebrafish eye. The transcript levels were normalized to ribosomal protein L13a (rpl13a) transcript levels in all analyses. Primers used for quantitative PCR are listed in Supplemental Table S3.
Statistical analysis
Sample sizes were determined based on prior literature and best practices in the field. The Tukey–Kramer HSD (honestly significant difference) test was used to determine the statistical significance among multiple datasets (the ‘multcomp’ package v1.4-16 in R, version 4.0.0).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). The datasets generated in the current study are available in Gene Expression Omnibus (GSE175929).
References
Lamb, T. D., Collin, S. P. & Pugh, E. N. Evolution of the vertebrate eye: Opsins, photoreceptors, retina and eye cup. Nat. Rev. Neurosci. 8, 960–976 (2007).
Morshedian, A. & Fain, G. L. The evolution of rod photoreceptors. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160074 (2017).
Ebrey, T. & Koutalos, Y. Vertebrate photoreceptors. Prog. Retin. Eye Res. 20, 49–94 (2001).
Kawamura, S. & Tachibanaki, S. Rod and cone photoreceptors: Molecular basis of the difference in their physiology. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 150, 369–377 (2008).
Corbo, J. C. Vitamin A1/A2 chromophore exchange: Its role in spectral tuning and visual plasticity. Dev. Biol. 475, 145–155 (2021).
Palczewski, K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 (2006).
Baden, T., Euler, T. & Berens, P. Understanding the retinal basis of vision across species. Nat. Rev. Neurosci. 21, 5–20 (2020).
Lamb, T. D. Evolution of the genes mediating phototransduction in rod and cone photoreceptors. Prog. Retin. Eye Res. 76, 100823 (2020).
Lagman, D. et al. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol. Biol. 13, 238 (2013).
Hisatomi, O. & Tokunaga, F. Molecular evolution of proteins involved in vertebrate phototransduction. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133, 509–522 (2002).
Yokoyama, S. Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9, 259–282 (2008).
Bowmaker, J. K. Evolution of vertebrate visual pigments. Vis. Res. 48, 2022–2041 (2008).
Okano, T., Kojima, D., Fukada, Y., Shichida, Y. & Yoshizawa, T. Primary structures of chicken cone visual pigments: Vertebrate rhodopsins have evolved out of cone visual pigments. Proc. Natl. Acad. Sci. U.S.A. 89, 5932–5936 (1992).
Zang, J. & Neuhauss, S. C. F. Biochemistry and physiology of zebrafish photoreceptors. Pflugers Arch. https://doi.org/10.1007/s00424-021-02528-z (2021).
Stenkamp, D. L. Development of the vertebrate eye and retina. Prog. Mol. Biol. Transl. Sci. 134, 397–414 (2015).
Chinen, A., Hamaoka, T., Yamada, Y. & Kawamura, S. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163, 663–675 (2003).
Takechi, M. & Kawamura, S. Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J. Exp. Biol. 208, 1337–1345 (2005).
Meyer, A. & Van De Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). BioEssays 27, 937–945 (2005).
Andzelm, M. M. et al. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron 86, 247–263 (2015).
Swaroop, A., Kim, D. & Forrest, D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11, 563–576 (2010).
Furukawa, T., Morrow, E. M. & Cepko, C. L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531–541 (1997).
Jia, L. et al. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc. Natl. Acad. Sci. U.S.A. 106, 17534–17539 (2009).
Mears, A. J. et al. Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447–452 (2001).
Corbo, J. C. & Cepko, C. L. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 1, 0140–0153 (2005).
Onishi, A. et al. The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc. Natl. Acad. Sci. U.S.A. 107, 11579–11584 (2010).
Mattar, P., Stevanovic, M., Nad, I. & Cayouette, M. Casz1 controls higher-order nuclear organization in rod photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 115, E7987–E7996 (2018).
Omori, Y. et al. Samd7 is a cell type-specific PRC1 component essential for establishing retinal rod photoreceptor identity. Proc. Natl. Acad. Sci. U.S.A. 114, E8264–E8273 (2017).
Ng, L. et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98 (2001).
Brzezinski, J. A. & Reh, T. A. Photoreceptor cell fate specification in vertebrates. Development 142, 3263–3273 (2015).
Alvarez-Delfin, K. et al. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc. Natl. Acad. Sci. U.S.A. 106, 2023–2028 (2009).
Ogawa, Y., Shiraki, T., Fukada, Y. & Kojima, D. Foxq2 determines blue cone identity in zebrafish. bioRxiv. https://doi.org/10.1101/2021.03.14.435350 (2021).
Ogawa, Y. et al. Six6 and Six7 coordinately regulate expression of middle-wavelength opsins in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 116, 4651–4660 (2019).
Volkov, L. I. et al. Thyroid hormone receptors mediate two distinct mechanisms of long-wavelength vision. Proc. Natl. Acad. Sci. U.S.A. 117, 15262–15269 (2020).
Suzuki, S. C. et al. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. U.S.A. 110, 15109–15114 (2013).
Asaoka, Y., Mano, H., Kojima, D. & Fukada, Y. Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 99, 15456–15461 (2002).
Ogawa, Y., Shiraki, T., Kojima, D. & Fukada, Y. Homeobox transcription factor Six7 governs expression of green opsin genes in zebrafish. Proc. Biol. Sci. 282, 20150659 (2015).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Murphy, D. P., Hughes, A. E. O., Lawrence, K. A., Myers, C. A. & Corbo, J. C. Cis-regulatory basis of sister cell type divergence in the vertebrate retina. Elife 8, 648824 (2019).
Vitorino, M. et al. Vsx2 in the zebrafish retina: Restricted lineages through derepression. Neural Dev. 4, 14 (2009).
Glasauer, S. & Neuhauss, S. Expression of CaBP transcripts in retinal bipolar cells of developing and adult zebrafish. Matters. https://doi.org/10.19185/matters.201604000009 (2016).
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308-1323.e30 (2016).
Sun, C., Galicia, C. & Stenkamp, D. L. Transcripts within rod photoreceptors of the zebrafish retina. BMC Genomics 19, 127 (2018).
Daido, Y., Hamanishi, S. & Kusakabe, T. G. Transcriptional co-regulation of evolutionarily conserved microRNA/cone opsin gene pairs: Implications for photoreceptor subtype specification. Dev. Biol. 392, 117–129 (2014).
Larison, K. D. & Bremiller, R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development 109, 567–576 (1990).
Lagman, D., Callado-Pérez, A., Franzén, I. E., Larhammar, D. & Abalo, X. M. Transducin duplicates in the zebrafish retina and pineal complex: Differential specialisation after the teleost tetraploidisation. PLoS One 10, e0121330 (2015).
Lupo, G. et al. Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc. Natl. Acad. Sci. U.S.A. 108, 8698–8703 (2011).
Imanishi, Y. et al. Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: Characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1-8) in pufferfish (Fugu rubripes). J. Mol. Evol. 59, 204–217 (2004).
Lagman, D., Sundström, G., Daza, D. O., Abalo, X. M. & Larhammar, D. Expansion of transducin subunit gene families in early vertebrate tetraploidizations. Genomics 100, 203–211 (2012).
Yamagata, M., Yan, W. & Sanes, J. R. A cell atlas of the chick retina based on single cell transcriptomics. Elife 10, 1–39 (2021).
Renninger, S. L., Gesemann, M. & Neuhauss, S. C. F. Cone arrestin confers cone vision of high temporal resolution in zebrafish larvae. Eur. J. Neurosci. 33, 658–667 (2011).
Aibar, S. et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS One 5, 1–10 (2010).
Oel, A. P. et al. Nrl is dispensable for specification of rod photoreceptors in adult zebrafish despite its deeply conserved requirement earlier in ontogeny. iScience 23, 101805 (2020).
Xie, S. et al. Knockout of Nr2e3 prevents rod photoreceptor differentiation and leads to selective L-/M-cone photoreceptor degeneration in zebrafish. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1273–1283 (2019).
Kubo, S. et al. Functional analysis of Samd11, a retinal photoreceptor PRC1 component, in establishing rod photoreceptor identity. Sci. Rep. 11, 1–11 (2021).
Morrow, E. M., Furukawa, T., Lee, J. E. & Cepko, C. L. NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126, 23–36 (1999).
Kon, T. & Furukawa, T. Origin and evolution of the Rax homeobox gene by comprehensive evolutionary analysis. FEBS Open Bio 10, 657–673 (2020).
Forrest, D. & Swaroop, A. Minireview: The role of nuclear receptors in photoreceptor differentiation and disease. Mol. Endocrinol. 26, 905–915 (2012).
Lien, S. et al. The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200–205 (2016).
Chen, Z. et al. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci. Adv. 5, eaav0547 (2019).
Parichy, D. M. Advancing biology through a deeper understanding of zebrafish ecology and evolution. Elife 4, 1–11 (2015).
McFarland, W. N. & Munz, F. W. Part II: The photic environment of clear tropical seas during the day. Vis. Res. 15, 1063–1070 (1975).
Yoshimatsu, T., Schröder, C., Nevala, N. E., Berens, P. & Baden, T. Fovea-like photoreceptor specializations underlie single UV cone driven prey-capture behavior in zebrafish. Neuron 107, 320-337.e6 (2020).
Zang, J., Keim, J., Kastenhuber, E., Gesemann, M. & Neuhauss, S. C. F. Recoverin depletion accelerates cone photoresponse recovery. Open Biol. 5, 150086 (2015).
Jacobs, G. H. Evolution of colour vision in mammals. Philos. Trans. R. Soc. B Biol. Sci. 364, 2957–2967 (2009).
Monte, W. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th edn. (University of Oregon Press, 2000).
Takechi, M., Hamaoka, T. & Kawamura, S. Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett. 553, 90–94 (2003).
Takechi, M., Seno, S. & Kawamura, S. Identification of cis-acting elements repressing blue opsin expression in zebrafish UV cones and pineal cells. J. Biol. Chem. 283, 31625–31632 (2008).
Tsujimura, T., Chinen, A. & Kawamura, S. Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 12813–12818 (2007).
Kimura, Y., Satou, C. & Higashijima, S. I. V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development 135, 3001–3005 (2008).
Gillard, G. B. et al. Comparative regulomics supports pervasive selection on gene dosage following whole genome duplication. Genome Biol. 22, 103 (2021).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Lawson, N. D. et al. An improved zebrafish transcriptome annotation for sensitive and comprehensive detection of cell type-specific genes. Elife 9, 1–76 (2020).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888-1902.e21 (2019).
Goldman, D. Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 15, 431–442 (2014).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Shafer, M. E. R. et al. Gene family evolution underlies cell type diversification in the hypothalamus of teleosts. bioRxiv. https://doi.org/10.1101/2020.12.13.414557 (2020).
Larhammar, D., Nordstrom, K. & Larsson, T. A. Evolution of vertebrate rod and cone phototransduction genes. Philos. Trans. R. Soc. B Biol. Sci. 364, 2867–2880 (2009).
Acknowledgements
We thank L. Volkov, D. Murphy, and M. Toomey for their comments on the manuscript. Tg(rho:EGFP)ja2Tg and Tg(gnat2:EGFP)ja23Tg fish lines were a generous gift from Dr. Yoshitaka Fukada (University of Tokyo). Tg(-5.5opn1sw1:EGFP)kj9Tg, Tg(-3.5opn1sw2:EGFP)kj11Tg, and Tg(opn1mw2:EGFP)kj4Tg fish lines were kindly provided by Dr. Shoji Kawamura (University of Tokyo). Tg(thrb:Tomato)q22Tg fish were kindly provided by Dr. Rachel Wong (University of Washington, Seattle). TgBAC(vsx1:GFP)nns5Tg were kindly provided by Dr. Ryan B. MacDonald (University College London). We also thank the Genome Technology Access Core (GTAC) in the Department of Genetics at Washington University in St. Louis for 10× library preparation and next generation sequencing, and the Flow Cytometry Core in the Department of Pathology and Immunology at Washington University in St. Louis for FACS services. This work was supported by the Japan Society for the Promotion of Science Overseas Research Fellowship (202060618 to YO) and the National Institutes of Health (EY030075 to JCC). Funding for this project was also provided by the Children's Discovery Institute of Washington University and St. Louis Children's Hospital.
Author information
Authors and Affiliations
Contributions
Y.O. and J.C.C. conceived of the study and designed experiments. Y.O. performed the experiments and analyzed the data. Y.O. and J.C.C. prepared the manuscript and figures. Both authors reviewed and edited the manuscript. J.C.C. oversaw the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ogawa, Y., Corbo, J.C. Partitioning of gene expression among zebrafish photoreceptor subtypes. Sci Rep 11, 17340 (2021). https://doi.org/10.1038/s41598-021-96837-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-96837-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.