Abstract
The aim of this study was to assess the effects of irrigation frequency and nitrogen fertilization rate on the abundance, diversity, and composition of soil bacteria in winter wheat. Irrigation, but not nitrogen fertilization, significantly affected the bacterial alpha diversity index. Among the 50 phyla obtained in these treatments, Proteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, Gemmatimonadetes, and Firmicutes were the predominant phyla. The LEfSe analysis of different treatments indicated that irrigation had a stronger effect on soil bacteria community composition than nitrogen fertilization. Moreover, the soil pH, moisture, available phosphorus (AP), and available potassium (AK) significantly correlated with the relative abundance of dominant bacteria at the phylum, genus, and operational taxonomic unit (OTU) levels. Overall, after three years of irrigation and fertilization treatments, the effect of irrigation on soil bacteria abundance, diversity, and composition of winter wheat was stronger than that of nitrogen fertilization, highlighting the importance of water availability for bacteria communities in semi-arid ecosystems. Inorganic and organic fertilizers should be applied in rotation.
Similar content being viewed by others
Introduction
During the growing season of winter wheat, a large amount of fertilizer and groundwater irrigation are needed, resulting in groundwater pollution and a continuous decline in groundwater level, which causes the North China Plain to face a severe water resource crisis1. The North China Plain is an important grain production base in China, which has a long history of wheat–maize rotation agricultural system2. Water shortage is clearly a limiting factor for sustainable agricultural development in the North China Plain and other semi-arid areas. Wheat is one of the most important crops globally3. Therefore, it is important to study the effects of nitrogen fertilizer combined with irrigation on the quantity, diversity, and community structure of soil microorganisms.
The microbial community is an essential component of the soil and plays an important role in maintaining the ecological functions of the soil. It is directly involved in nutrient cycling, energy flow, and degradation of organic matter4. The microbial community is an important index for evaluating the health and quality of the soil. Soil microbial communities are very sensitive to soil changes, such as disturbances due to land tillage and irrigation water5. Maintaining the complexity and diversity of soil microbial communities is critical to the sustenance of soil fertility because soil microbes mediate the biogeochemical cycles of carbon and nitrogen, and serve as important reservoirs for plant nutrients6. Fertilization and irrigation are the main agricultural management measures in agricultural production that can promote crop growth and increase yield, and it can also affect soil microbial community7. Fertilization does not only provide the necessary nutrients for plant growth but also improves the characteristics of the soil due to the improvement in the nutrient utilization rate8. He et al. reported that long-term fertilization changed soil pH, total carbon, total nitrogen, and available phosphorus, which may lead to significant differences in microorganisms8. However, Yang et al. found that fertilization did not significantly change the composition and structure of the bacterial community, and the effect on bacterial diversity was not significant, because fertilization only increased the content of organic carbon and did not affect the pH value9. In this study, nitrogen fertilizer was used to clarify the mechanism of the effect on microorganisms.
Nitrogen is an essential element that plays a crucial role in natural and managed ecosystems, including agro-ecosystems, where it is often a common limiting factor in plant productivity and yield10. Nitrogen fertilizer plays an irreplaceable role in fertilization, therefore, rational and efficient application is particularly important. Nitrogen fertilization has been shown to alleviate the negative effects of plant-soil interaction11. Nitrogen availability largely impacts microbial communities12. Zeng et al. observed a change in the soil community structure and a decrease in bacteria diversity13. Fierer et al. found that nitrogen fertilization may directly or indirectly lead to changes in major microbial life-story strategies, which is conducive to a more active and copiotrophic microbial community14. This pattern is similar to the frequently observed K-selected plants being replaced by r-selected plants, while the nitrogen value of r-selected plants increased.
Yuan et al. found that the effect of irrigation water-type on soil microbial community structure was greater than that of nitrogen fertilizer treatment15. Soil irrigation management also affects the quantity, community structure, and diversity of soil microorganisms16. Soil water content changes the composition and activity of soil microbial communities through its effect on the transport of soil nutrients, substrate availability, and other soil properties17. Scarcity of water reduces soil organic carbon, thereby altering the soil microbial community structure18. Bai et al. found that moderate water supplementation, especially at the tasseling, postulation, and maturity stages, promotes maize growth and the diversity and composition of related rhizosphere microorganisms19. Moreover, the richness and evenness of bacteria increased with an increase in the soil relative water content. Soil microorganisms are important for the availability of underground nitrogen, and many processes of soil organic matter decomposition, mineralization, and biological nitrogen fixation driven by soil microorganisms are affected by nitrogen and water availability20. Hence, water and nitrogen availability interactively affect the soil microbial community and agricultural ecosystem functioning. The distribution of soil bacteria in natural and agricultural ecosystems has been extensively studied21. Bacterial community diversity and activity are considered more sensitive than other biota in response to changes in soil conditions such as pH, organic carbon, and nitrogen status22. Therefore, we will consider bacteria as the object of this study to evaluate their responses to different treatments.
In recent years, with the rapid development in DNA sequencing technology, next-generation sequencing (high-throughput sequencing) has been widely applied in the study of microbial communities. Next-generation sequencing technology has the advantages of a deep sequencing degree and large amount of data, which can reveal the complexity and diversity of the microbial community more accurately, and accelerate the research of non-culturable and trace microorganisms in the environment23. Previous studies have focused on the effects of single practices, such as nitrogen fertilization or irrigation, on soil bacterial communities. There have been few systematic studies on the effects of the combined management of nitrogen fertilization and irrigation on soil bacteria communities24. In this study, IonS5 sequencing was used to determine the soil bacteria community structure, diversity, and abundance in the Xinji Experimental Station, following three years of nitrogen fertilization and irrigation. Based on the data analysis, we can explore the influence of nitrogen fertilizer and irrigation on the bacteria community of wheat soil and provide a basis for optimizing the system of nitrogen fertilizer and irrigation, thereby improving soil fertility, maintaining soil microbial ecosystems, and achieving sustainable agricultural development in the North China Plain. Based on the results of previous studies, we hypothesized that nitrogen fertilizer and irrigation would affect the soil bacteria community composition and diversity by increasing or decreasing the relative abundance of specific bacteria classifications. In addition, we hypothesized that nitrogen fertilizer and irrigation would improve soil fertility by affecting soil chemical properties.
Results
Soil chemical characteristics and grain yields
After three years of conducting the experiment, the irrigation frequency was observed to have had significant effects on pH (p < 0.01), moisture (p < 0.001), total nitrogen (TN) (p < 0.001), C/N ratio (p < 0.001), and grain yield of winter wheat (p < 0.01), but not on available phosphorus (AP), available potassium (AK), or bulk density (BD). With increasing irrigation frequency, the pH decreased significantly, but moisture significantly increased (Table 1). TN was significantly higher under W0 (no irrigation) than under W1 (irrigation once at jointing) and W2 (irrigation twice at jointing and anthesis). The C/N ratio was significantly higher under W1 than under W0. Grain yield was significantly higher under W1 and W2 than under W0. Nitrogen fertilization rate had significant effects on TN (p < 0.001), SOC (p < 0.001), C/N ratio (p < 0.001), and grain yield (p < 0.001), but not on pH, moisture, AP, AK, or BD. With the increase in nitrogen fertilization rate, TN increased significantly, but the C/N ratio significantly decreased. The SOC and grain yield of winter wheat were significantly higher under N1 (nitrogen fertilization rate: 120 kg ha−1) and N2 (nitrogen fertilization rate: 240 kg ha−1) than under N0 (nitrogen fertilization rate: 0 kg ha−1). The interaction between irrigation frequency and nitrogen fertilization rate had no significant effect on any of the soil variables.
Sequencing results and diversity indices
In total, we obtained 2,875,721 high-quality sequences from all 27 samples, and 102,258–110,990 sequences were obtained per sample with an average of 106,508. The read lengths ranged from 220 to 260 bp (mean = 252 bp). The values of the Good's coverage estimators were in the range of 0.982 and 0.985 at a 97% similarity cutoff, which suggested that the current numbers of the sequence reads were sufficient to capture the diversity of the soil bacteria community. When grouped at the 97% similarity level, there were a total of 11,360 OTUs in the complete data set (Table 2).
As shown in Table 3, irrigation frequency had a significant effect on richness (Sobs and chao1) (p < 0.05) and diversity (Shannon and PD) (p < 0.05), but not on evenness (Shannon even). Richness and diversity were significantly higher under W1 and W2 than under W0. Nitrogen fertilization rate and the interaction between irrigation frequency and nitrogen fertilization rate had no significant effect on the alpha diversity of bacteria.
Phylogenetic diversity of bacteria communities
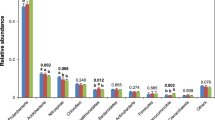
The relative abundance of dominant bacteria phyla across all the soil samples is shown on the right side of Fig. 1. The dominant bacteria phyla were the same across all treatments, but their relative abundance in soil samples varied: Proteobacteria (28.46–37.78%), Bacteroidetes (11.36–19.65%), Actinobacteria (8.01–17.27%), Acidobacteria (10.76–15.30%), Gemmatimonadetes (3.84–6.74%), Firmicutes (3.13–6.6%), Planctomycetes (3.41–4.96%), Chloroflexi (2.90–4.34%), Verrucomicrobia (2.24–3.81%), and Nitrospirae (0.64–1.68%). On the left side of Fig. 1, cluster analyses showed the relationships among the different treatments.
PCoA analysis of soil samples explained 37.02% of the variation in bacterial community composition along the two axes (Fig. 2). The three replicates of irrigation and nitrogen fertilization treatments usually clustered closely, indicating the reproducibility of the profiles of the bacteria community. The different irrigation frequency treatments were significantly different from each other. Irrigation significantly affected the bacteria community composition of soils (PERMANOVA: F = 4.1107, p < 0.001), whereas no significant effects in nitrogen fertilization were observed (PERMANOVA: F = 0.6528, p = 0.758).
Taxonomic composition
We conducted LEfSe analysis to identify the statistical significance of differentially abundant bacteria taxa and the biological relevance of the species in different treatments. Supplementary Figure S1 and Supplementary Fig. S2, and Supplementary Fig. S3 show the taxa abundance at different irrigation frequencies under the same N fertilization rate. When the nitrogen fertilization rate was 0, the phylum Gemmatimonadetes was more abundant in the soils of W2 than in the other treatments (Supplementary Fig. S1). Irrigation also significantly increased the abundance of the bacterial orders Xanthomonadales, Gemmatimonadales, Deltaproteobacteria, Gemmatimonadetes, OM190, and family Gemmatimonadaceae (Supplementary Fig. S1). At a nitrogen fertilization rate of 120 kg ha−1, the phylum Proteobacteria, class Gammaproteobacteria, and order Xanthomonadales were more abundant in the soils of W2 than in the other treatments (Supplementary Fig. S2). The order Pseudonocardiales was more abundant in the soils of W0 than in the other treatments (Supplementary Fig. S2). At nitrogen fertilization rate of 240 kg ha−1, the phyla Saccharibacteria, class Bacteroidia, order Bacteroidales, family Porphyromonadaceae, and genus Adhaeribacter were more abundant in the soils of W0 than in other treatments (Supplementary Fig. S3). The class Deltaproteobacteria was more abundant in the soils of W1 than in the other treatments (Supplementary Fig. S3). The phylum Nitrospirae, class Nitrospira, and genus Nitrospira were more abundant in the soils of W2 than in the other treatments (Supplementary Fig. S3).
Supplementary Figure S4 Supplementary Fig. S5, and Supplementary Fig. S6 show the taxa abundance at different nitrogen fertilization rates under the same irrigation frequency. At the irrigation frequency of 0, the classes Clostridia, Clostridiales, Porphyromonadaceae, Bacteroidales-S24-7-group, genera Parabacteroides, and Schlesneria were more abundant in the N0 soil than in the other treatments (Supplementary Fig. S4). In contrast, at the irrigation frequency of 1, the order Xanthomonadales, family Xanthomonadaceae, and genus Lysobacter were more abundant in the soil of N2 than in the other treatments (Supplementary Fig. S5). The family CHAB-XI-27 was more abundant in the soils of N1 than in the other treatments (Supplementary Fig. S5). However, at the irrigation frequency of 2, the abundance of bacteria in different treatments of nitrogen fertilization rate was not significantly different (Supplementary Fig. S6).
The taxa abundance resulting from different irrigation frequencies (irrespective of nitrogen fertilization rate) and nitrogen fertilization rates (irrespective of irrigation frequency) are shown in Figs. 3 and 4. The phyla Proteobacteria, Gemmatimonadetes, and Nitrospirae were more abundant in the soils of W2 than in the other treatments, whereas Bacteroidetes, Actinobacteria, and Firmicutes were more abundant in the soils of W0 (Fig. 3). Only phylum Planctomycetes was found to be more abundant in W1 than in the other treatments (Fig. 3). The class Betaproteobacteria and family Xanthomonadaceae were more abundant in the soils of N2 (Fig. 4). In general, irrigation significantly influenced the abundance of many bacteria, but nitrogen fertilization had no significant effect on bacteria (Fig. 4).
A linear discriminant analysis effect size (LEfSe) method identifies the significantly different abundant taxa of bacteria in different treatments of irrigation frequency (irrespective of nitrogen fertilization rate). The taxa with the absolute LDA > 3.5 and p < 0.05 are shown. This figure is drawn by LEfSe (Version 1.0, http://huttenhower.sph.harvard.edu/galaxy/root?tool_id=lefse_upload) software.
A linear discriminant analysis effect size (LEfSe) method identifies the significantly different abundant taxa of bacteria in different treatments of nitrogen fertilization rate (irrespective of irrigation frequency). The taxa with the absolute LDA > 3.5 and p < 0.05 are shown. This figure is drawn by LEfSe (Version 1.0, http://huttenhower.sph.harvard.edu/galaxy/root?tool_id=lefse_upload) software.
At the OTU level, 11,360 OTUs were identified in all the soils (Supplementary Table S1). Among the 10 most abundant OTUs, six OTUs were identified as dominantly enriched with a relative abundance > 0.5% in at least one sample. The six OTUs included OTU82 (norank-c-Acidobacteria), OTU95 (Thermomonas), OTU1428 (RB41), OTU1740 (norank-f-Gemmatimonadaceae), OTU2822 (norank-f-Nitrosomonadaceae), and OTU2894 (norank-c-Gemmatimonadetes). Irrigation frequency had significant effects on eight OTUs, but not on OTU82 (norank-c-Acidobacteria) or OTU6495 (norank-o-Subgroup-7). With the increase in irrigation frequency, OTU1428 (RB41) decreased significantly, whereas the other OTUs significantly increased. Nitrogen fertilization had no significant effect on the 10 OTUs (p > 0.05). The interaction between irrigation frequency and nitrogen fertilization rate had significant effects on OTU95 (Thermomonas) (p < 0.01), OTU1428 (RB41) (p < 0.05), OTU2822 (norank-f-Nitrosomonadaceae) (p < 0.01), and OTU6387 (norank-o-Xanthomonadales) (p < 0.001), but not on other OTUs.
Correlations between alpha diversity, relative abundance of dominant bacteria, and soil properties
There was a significant negative correlation between pH and moisture content (Supplementary Table S2). The total nitrogen (TN) and soil organic carbon (SOC) were significantly and negatively correlated with the C/N ratio. There was a significant positive correlation between TN and SOC. SOC was significantly and positively correlated with the yield of winter wheat, whereas the C/N ratio and yield were negatively correlated.
The Pearson correlations between alpha diversity and soil properties showed that AP and AK were significantly and positively correlated with alpha diversity, whereas the correlation between pH and alpha diversity was significant and negative (Supplementary Table S2). Moisture was positively correlated with alpha diversity, but it was significantly correlated only with chao1 and PD. The correlations between the other indices and alpha diversity were not significant.
The Pearson correlations between the relative abundance of dominant bacteria and soil properties confirmed that pH, moisture, AP, and AK were the dominant factors correlating with bacterial community structures (Supplementary Table S2). The correlations between several soil properties and the relative abundance of many dominant bacteria were significant. At the phylum level, Firmicutes and Chloroflexi were significantly and positively correlated with the yield of winter wheat. Additionally, at the OTU level, OTU6387 (norank-o-Xanthomonadales) was significantly and positively correlated with yield.
Discussion
Soil pH is the main driving force shaping soil microbial community25. Soil moisture content is also the main factor affecting soil microbial composition and activity26. The pH and moisture of the soil were mainly affected by irrigation, but not by nitrogen fertilizer (Table 1). Similar results were observed by Ding et al., who thought that the significant decrease in pH value in paddy soil might result from the increase in proton concentration in the soil solution due to the accumulation of organic matter under irrigation27. However, the correlation between pH and soil organic carbon (SOC) was not significant in this study, whereas the correlation between pH and moisture was negative and significant (Supplementary Table S2). This is probably because irrigation causes more nitrates and other negative ions to dissolve in the soil, which reduces soil pH28. Zhong et al. observed that the soil pH showed no significant differences among different nitrogen fertilization treatments, which is similar to our study24. Inconsistencies have also been reported in other studies that soil pH decreased with increased nitrogen fertilization29. It is likely that three years of nitrogen fertilization may still be too short to affect soil pH. Soil total nitrogen (TN) increased with increasing nitrogen fertilization and decreased with increasing irrigation. Higher soil moisture can foster microbial growth and promote metabolic activities30, which increases the efficiency of nitrogen utilization, thus lowering TN. Compared with W0N0, irrigation and nitrogen fertilization significantly increased the yield of winter wheat. However, the difference between W2 and W1 was not significant, as was the case with N1 and N2, which was consistent with the results of Wang et al.31 who found no significant relationship between crop yield and bacterial community, but a greater relationship with soil organic carbon, which is fully illustrated in this study (Supplementary Table S2).
Microbial diversity plays a key role in ecosystem stability and soil productivity by regulating soil chemical processes32, which it is essential for the sustainable development of agriculture33. Two-way ANOVA analysis of alpha diversity revealed the effects of irrigation and nitrogen fertilization (Table 3). Irrigation had a significant effect on alpha diversity, but nitrogen fertilization did not. As mentioned above, it is likely that irrigation significantly reduced the pH of the soil, but nitrogen fertilization did not. Peralta et al. reported that moderate wetting could improve the diversity of soil bacterial community structure, while excessive drying and wetting could do the opposite34. Some studies have found that water quantity is an important factor affecting microbial community diversity, and it was more important than land management or type of fertilizer35. The correlation between pH and alpha diversity was significant and negative (Supplementary Table S2). When the soil pH was close to neutral, the alpha diversity of the bacterial community was the highest36. These results indicate that a neutral environment is suitable for the survival of microorganisms. There may be two major effects of pH on bacteria. First, PH directly restricts the physiological process of soil bacteria, and if the soil pH value exceeds a certain range, it will reduce the growth of individual groups that cannot survive. Second, soil pH may not directly change bacterial diversity and community structure, but as a composite variable, the composite index providing soil conditions has many soil characteristics that are often directly or indirectly related to soil pH and may drive observed changes in diversity and community composition. In recent years, pH has been extensively studied in various soils as a major factor in determining the diversity and composition of soil bacteria36. Long-term nitrogen fertilization has been shown to increase microbial diversity37; however, no significant change may be observed in the short term over several years38, mainly because there was no change in pH, which was consistent with our study. Moreover, abundant nutrients can promote the growth of some bacteria39, which may explain why available phosphorus (AP) and available potassium (AK) were significantly and positively correlated with alpha diversity (Supplementary Table S2).
Soil microorganisms are the core component of terrestrial ecosystems. Their composition and structure can reflect, not only the efficiency of biological transformation, but also the state of soil fertility40. In this study, the Ion S5™ XL sequencing platform was used to study the structure of soil bacteria communities in nine treatments of nitrogen fertilization and irrigation. Fifty phyla, 137 classes, and 1242 genera were identified (Table 2). The results show that the dominant phyla in the different treatments were similar, but the relative abundances of the dominant phyla in the different treatments were different (Fig. 1). In agreement with previous studies41, the dominant phyla in each treatment were Proteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, and Gemmatimonadetes. Different nitrogen fertilization and irrigation treatments may alter the physicochemical properties of soil, which are often closely related to soil bacterial communities. Our results show that the soil chemical characteristics had different effects on the relative abundance of bacteria at different taxonomic levels. For example, the phylum, Nitrospirae, was not significantly correlated with soil AP, but the genus, Norank-f-Nitrosomonadaceae, was significantly and positively correlated with soil AP. Consistent with our study, Shen et al. observed that nitrogen fertilization and irrigation could alter soil bacterial community structure, but the effects of nitrogen fertilization and irrigation on bacteria community structure were different (Figs. 1 and 2)42. A similar community structure was found in the treatments with the same irrigation frequency, which indicated that the effects of irrigation on soil bacterial community structure were more significant than those of nitrogen fertilization.
The results of the different treatments indicated that irrigation had a stronger effect on soil bacteria community composition than nitrogen fertilization in this study (Supplementary Table S1, Fig. 3, and Fig. 4). The relative abundance of phyla Proteobacteria, Gemmatimonadetes, and Nitrospirae were higher in the W2 treatment, whereas the relative abundances of Bacteroidetes, Actinobacteria, and Firmicutes were higher in the W0 treatment (Fig. 3). Although other studies have shown similar results, this is still remarkable because all bacteria phyla are phylogenetic, and there could still be wide metabolic differences among them, making some bacteria phyla dominant in the soil, while others are always quite rare. Proteobacteria is a gram-negative bacterium that is highly susceptible to environmental disturbance and water-limiting stress43. Consistent with the results of our study, Dennis et al. found that the relative abundance of Proteobacteria increased with an increase in the irrigation amount44. This is because increasing soil moisture is beneficial for crop growth and metabolism, which promotes the secretion of some rhizomatous sediments. Proteobacterium can synthesize carbohydrates and proteins from the secretions of crops. Gemmatimonadetes were associated with phosphorus metabolism45, which showed a significant positive correlation with AP (Supplementary Table S2). Nitrospirae contain nitrite-oxidizing bacteria that are capable of nitrification and are part of the biogeochemical nitrogen cycle46. The correlation between the relative abundance of Bacteroidetes and Actinobacteria and soil pH was positive and has been observed in a previous study47, which is consistent with our study (Supplementary Table S2). Bacteroidetes play an important role in degrading cellulose in soils48. The relative abundance of Actinobacteria was strongly associated with long-term drought. Barnard et al. found that the relative abundance of Actinobacteria in grasslands decreased with increased irrigation49. Ammonifying and nitrifying bacteria play important roles in soil nitrogen metabolism. The common ammonifying bacteria in soils are Pseudomonas, Bacillus, Clostridium, Arthrobacter, and Proteus. In this study, there were significant differences in the abundance of some bacteria among the different treatments, but not in the ammonifying bacteria (Fig. 3 and Fig. 4). We found that short-term nitrogen application did not affect soil ammoniation, in contrast to previous research results50. Ammonifying bacteria are not particularly sensitive to environmental changes, and small changes in temperature, water content, and pH are do not affect their activity. Compared with ammonifying bacteria, nitrifying bacteria are more sensitive to changes in the soil environment. There were significant differences in the presence of nitrifying bacteria among the different irrigation treatments (Fig. 3 and Fig. 4). The abundance of Nitrospira in W2 was significantly higher than that in W0. Nitrospira can oxidize ammonia to nitrite, which is key to controlling the nitrification rate. Fan et al. found that the abundance of Nitrospira would increase with an increase in nitrogen application rate, which is a more accepted view at present51. However, irrigation had a significant effect on the abundance of Nitrospira in this study, and there were no significant differences between the different nitrogen fertilization treatments. As nitrate easily dissolves in water, excessive irrigation will cause nitrate loss, and nitrate leaching caused by irrigation is more serious than nitrogen application52. We speculated that nitrate leaching led to the reduction of nitrifying bacteria reaction products, which promoted the increase of Nitrospira by feedback to ensure the absorption of nitrogen by wheat roots. Due to the ability of wheat to absorb nitrogen and water, the structure and composition of soil microbial communities are affected by intensive agricultural practices.
In summary, our results suggest that most changes in bacteria community parameters were influenced by soil water and pH, which were significantly correlated with irrigation levels, while nitrogen fertilizer had no significant effect on bacteria. Long-term application of inorganic fertilizer shows that nitrogen fertilizer could effectively increase crop yield, but also significantly reduce bacterial diversity. However, some studies have indicated that the application of nitrogen fertilizer has little or no effect on bacteria communities, especially in a short time53. This may be due to the fact that although some harmful local conditions appeared after urea application, the short-term effect of fertilization on soil microbial community was minimal. The application of urea and other nitrogen fertilizers resulted in a high concentration of ammonia nitrogen in the local crop soil, which may have a strong inhibition or killing effect on microorganisms. However, it should be noted that the harsh environment caused by nitrogenous fertilizers is generally limited in space. Although studies have shown that concentrations of ammonium ions in long-term fertilized soil may be higher than those in unfertilized soil. Concentrations at most times and places, in the short term, may be well below levels that are toxic to microorganisms54. Under laboratory culture conditions, approximately 200 kg N was applied to urea as a nitrogen fertilizer. After 10 days, the microbial community composition changed significantly; however, after 90 days, the microbial community returned to its original level55. The above results show that the effect of nitrogen fertilizer would be more significant after long-term application. The effect of irrigation on bacteria communities may be more pronounced than that of nitrogen fertilization. Some studies have emphasized that drought inhibits microbial activity, which in turn affects soil fertility-related microorganisms and impedes their mediated biogeochemical processes56. Genomic analysis showed that under the condition of reduced irrigation, the decomposition rate of organic matter slowed down, which was related to the change in community structure57. The influence of relative soil water concentration on the microbial community composition was significant. We speculate that reducing irrigation will reduce soil enzyme activity and have a long-term impact on soil nutrient availability and plant supply, and thus have a significant impact on crop yield. Based on the results from this and previous studies, it can be concluded that increased water availability due to irrigation treatment has important implications for microbial communities, soil function, and crop production in semi-arid ecosystems.
Water and nitrogen interacted with each other, and irrigation and nitrogen fertilization at higher levels had a significant inhibitory effect on other management measures. Water dissolved all kinds of nutrients, including nitrogen, and sufficient soil water indicate efficient nutrient supply. High irrigation diluted the high concentration of ammonia nitrogen caused by local nitrogen fertilization, so that nitrogen was evenly distributed in the crops. However, too much nitrogen had irreversible effects, permanently affecting the bacteria and even causing death. This feedback between irrigation and nitrogen fertilization is shown in Supplementary Fig. S4, Supplementary Fig. S5, and Supplementary Fig. S6. In the absence of irrigation, nitrogen fertilizer did not dissolve sufficiently and was likely to accumulate and cause bacterial damage, and this effect was more significant with the increase in nitrogen fertilization rate (Supplementary Fig. S4). Irrigation significantly altered this toxic effect (Supplementary Fig. S6). When irrigated in large quantities, the effect of water on bacteria was much greater than that of nitrogen fertilizer, resulting in no significant difference between different nitrogen treatments (Supplementary Fig. S6).
Conclusion
This study shows that water is the key factor affecting the abundance, diversity, and composition of soil bacteria. Our results suggest that most changes in bacteria community parameters were influenced by soil pH, which is significantly correlates with irrigation levels. Soil pH and moisture were significantly correlated with the relative abundance of dominant bacteria at the phylum, genus, and OTU levels. Irrigation affects the composition of bacteria communities by changing the abundance of certain bacteria, such as Proteobacteria, Gemmatimonadetes, Nitrospirae, Bacteroidetes, Actinobacteria, and Firmicutes. This could serve as basis for optimizing the system of nitrogen fertilizer and irrigation, reducing environmental pollution, maintaining soil microbial ecosystems, and achieving sustainable agricultural development.
Materials and methods
Experimental site
The study was conducted over three growing seasons for winter wheat from 2015–2017 at the Xinji Experimental Station (37°48′18'' N, 115°18′40'' E) of Hebei Agricultural University (Fig. 5). The typical cropping system is a winter wheat and summer maize double-cropping system. The area is semi-arid with a monsoon climate and an average annual temperature of 13 °C and average annual precipitation of 466.4 mm. More than 70% of the precipitation occurred from July to September. Monthly distributions of precipitation for 2015–2018 is shown in Table 4. According to the U.S. soil taxonomy, the soil was classified as an aquic inceptisol with a sandy loam texture. The chemical parameters of the soil at the start of the experiment were as follows: pH 7.86, bulk density (BD) 1.20 g cm−3, 18.70 g kg−1 soil organic carbon (SOC), 0.60 g kg−1 total nitrogen (TN), 37.05 mg kg−1 available phosphorus (AP), and 165.74 mg kg−1 available potassium (AK).
Experimental design
The experiments were designed as a split plot arrangement, with irrigation frequency (no irrigation, irrigation once at jointing, and irrigation twice at jointing and anthesis, expressed by W0, W1, and W2, respectively) as the main plots, and nitrogen fertilization rate (0, 120, and 240 kg ha−1, expressed by N0, N1, and N2, respectively) as the sub-plots. The amount of irrigation was 600 m3 ha−1 per time. The main plot size was 45 m × 17 m and the sub-plot was 15 m × 17 m, and each main plot was separated from each other by a 1 m zone. Each treatment combination was replicated three times. A local winter wheat (Triticum aestivum L.) cultivar, Jimai 585, was planted in the 2015–2016, 2016–2017, and 2017–2018 growth seasons.
Every year, comminuted maize straw was plowed as a base fertilizer, and the same amount of chemical fertilizer (135 kg ha−1 P2O5, and 150 kg ha−1 K2O) was applied before planting. Half of the nitrogen fertilizer (CH4N2O) in W1 and W2 was applied as a base fertilizer before planting, and half was applied at the jointing stage. All of the nitrogen fertilizers in W0 were applied as base before planting. The management practices for controlling pests, diseases, and weeds complied with local practices for agricultural production.
Soil sampling and chemical analyses
In the 2018 harvest stage, which represented the 3rd year of the experiment, five soil samples were collected from the 0–10 cm soil layer in each treatment plot using a soil-drilling sampler. All five soil samples were composited and sieved through a 2-mm mesh to thoroughly homogenize and remove the roots, plant residues, and stones. A portion of each soil sample was collected in a 50-mL centrifuge tube, placed in an ice-box, and transferred to the laboratory. The tubes were archived at − 80 °C until DNA extraction. The physicochemical properties of the other soil samples were determined.
All analyses of soil properties were performed using routine methods58. Soil moisture content was measured by oven-drying the soil to a constant weight at 105 °C for 24 h. Soil pH values were determined by dilution (5:1) with a 0.01 M CaCl2 solution. Soil organic carbon (SOC) was measured using the K2Cr2O7 oxidation method. Total nitrogen (TN) was analyzed using the semi-Kjeldahl method. The available phosphorus (AP) content was first extracted from the soil in a 0.5 M NaHCO3 solution and then determined using the vanadium molybdate yellow colorimetric method59. The available potassium (AK) was extracted from the soil by ammonium acetate and ascertained by flame photometry (FP-6410; Xinyi Instruments Co. Ltd., Shanghai, China) as described by Bao59. The top soil bulk density (BD) was measured during field sampling with a 100 cm3 steel cylinder.
DNA extraction and PCR amplification
We used the CTAB/SDS method to extract total genomic DNA from all the soil samples. DNA concentration and purity were measured using a 1% agarose gel. Depending on the concentration, DNA was diluted to 1 ng μL−1 using sterile water. The V4 regions of the 16S rRNA genes were amplified using specific primers: 515F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). All PCR reactions were performed in 30 μL mixture with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of forward and reverse primers, and about 10 ng of template DNA. The reactions were carried out in a thermal cycler (Bio-Rad Laboratories, Hercules, CA) using the following program: initial denaturation at 98 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s; and finally, 72 °C for 5 min.
The obtained PCR products were mixed with the same volume of 1 × loading buffer (containing SYBR green) and were checked on 2% agarose gel to ascertain the specificity of 16 s rRNA gene amplification. The PCR products were mixed in equal density ratios and then purified using the GeneJETTM Gel Extraction Kit (Thermo Scientific). According to the manufacturer's instructions, the Ion Plus Fragment Library Kit (48 reactions, Thermo Scientific) was used to generate sequencing libraries. The library quality was assessed on a Qubit 2.0 Fluorometer (Thermo Scientific). Finally, the libraries were sequenced on the Ion S5™ XL platform, and 400 bp/600 bp single-end reads were generated.
Bioinformatics and statistical analysis
According to the unique barcode, single-end reads were assigned to samples and truncated by cutting off the barcodes and primer sequences. With the Cut adapt (Version 1.9.1, http://cutadapt.readthedocs.io/en/stable/) quality-controlled process, the raw reads were filtered under specific filtering conditions to obtain high-quality clean reads. The reads were compared with the Sliva database (https://www.arb-silva.de/). The chimera sequences were detected using the UCHIME algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html), and then removed60. Finally, high-quality clean reads were obtained. Sequence analysis was performed using the Uparse software (Version 7.0.1001, http://drive5.com/uparse/)61. High-quality sequences with ≥ 97% similarity were assigned to the same operational taxonomic units (OTUs). Representative OTU sequences were selected for further annotation. The Silva Database (https://www.arb-silva.de/) was used to annotate taxonomic information using the Mothur algorithm for each representative sequence62. To ascertain the differences in the dominant species in different samples and the phylogenetic relationship of different OTUs, the MUSCLE software (Version 3.8.31, http://www.drive5.com/muscle/) was used to conduct multiple sequence alignments63. The OTU abundance information of each sample was normalized to that of the sample with the least sequences. The original sequence obtained by sequencing was submitted to the NCBI database, and the sequence number was obtained: SRR13767216.
The complexity of species diversity for samples was analyzed using alpha diversity, including observed richness (Sobs), Chao1, Shannon, Shannon, and phylogenetic diversity (PD) indices. All the indices were calculated using the free online platform, Majorbio I-Sanger Cloud Platform (www.i-sanger.com). Unweighted pair-group method, while the arithmetic means (UPGMA) clustering was performed as a type of hierarchical clustering method to interpret the distance matrix using average linkage and was conducted using QIIME software (Version 1.7.0). The differences between groups in the beta diversity index were evaluated using principal coordination analysis (PCoA) based on the composition of the bacteria using the Bary-Curtis metrics by R software (Version 2.15.3). SPSS (Version 20.0) was used to perform ANOVA to calculate significant differences in the dominant microbial taxon composition, alpha diversity, and soil variables. The interactions among alpha diversity, relative abundance of bacteria (phylum, genus, and OTUs), and soil properties were confirmed through Pearson's correlation analysis using SPSS (Version 20.0).
The bacteria community features in the different treatments of irrigation and nitrogen fertilization were characterized using the linear discriminant analysis effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/). LEFSe is an algorithm for high-dimensional biomarker discovery that emphasizes both statistical significance and biological relevance64. The non-parametric factorial Kruskal–Wallis sum rank test was used to identify the bacteria taxa with different treatments using LEfSe. Linear discriminant analysis (LDA) was used to estimate the effect of size on each differentially abundant feature. LDA > 3.5, and p < 0.05, were used for the biomarkers evaluated.
Ethical statement
Wheat collection was approved by the local government and scientific research institutions.
The authors declare that the methods of our study are in full compliance with the relevant institutional, national, and international guidelines and legislation.
References
Dikgwatlhe, S. B., Chen, Z. D., Lal, R., Zhang, H. L. & Chen, F. Changes in soil organic carbon and nitrogen as affected by tillage and residue management under wheat-maize cropping system in the North China Plain. Soil Till. Res. 144, 110–118 (2014).
Liu, Y., Wang, E., Yang, X. & Wang, J. Contributions of climatic and crop varietal changes to crop production in the North China Plain, since 1980s. Glob. Chang. Biol. 16, 2287–2299 (2010).
Fischer, R. A. T. & Edmeades, G. O. Breeding and cereal yield progress. Crop Sci. 50, S-85 (2010).
Chen, Z. M. et al. Changes in soil microbial community and organic carbon fractions under short-term straw return in a rice-wheat cropping system. Soil Till. Res. 165, 121–127 (2017).
Frenk, S., Hadar, Y. & Minz, D. Resilience of soil bacterial community to irrigation with water of different qualities under Mediterranean climate. Environ. Microbiol. 16, 559–569 (2014).
Trivedi, P. et al. Soil aggregation and associated microbial communities modify the impact of agricultural management on carbon content. Environ. Microbiol. 19, 3070–3086 (2017).
Feng, Y. Z. et al. Bacillus asahii comes to the fore in organic manure fertilized alkaline soils. Soil Biol. Biochem. 81, 186–194 (2015).
He, J. Z., Zheng, Y., Chen, C. R., He, Y. Q. & Zhang, L. M. Microbial composition and diversity of an upland red soil under long-term fertilization treatments as revealed by culture-dependent and culture-independent approaches. J. Soils Sediments 8, 349–358 (2008).
Yang, Y. D., Wang, Z. M. & Zeng, Z. H. Effects of long-term different fertilization and irrigation managements on soil bacterial abundance, diversity and composition. Sci. Agric. Sin. 51, 290–301 (2018) ((in Chinese)).
Duc, L., Noll, M., Meier, B. E., Bürgmann, H. & Zeyer, J. High diversity of diazotrophs in the forefield of a receding alpine glacier. Microb. Ecol. 57, 179–190 (2009).
Guo, Q. et al. Influence of soil qualities on intra- and interspecific competition dynamics of Larix kaempferi and L. olgensis. Environ. Exp. Bot. 135, 96–105 (2017).
Treseder, K. K. et al. Arbuscular mycorrhizal fungi as mediators of ecosystem responses to nitrogen deposition: a trait-based predictive framework. J. Ecol. 106, 480–489 (2018).
Zeng, J. et al. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 92, 41–49 (2016).
Fierer, N. et al. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017 (2012).
Yuan, X. et al. Plant community and soil chemistry responses to long-term nitrogen inputs drive changes in alpine bacterial communities. Ecology 97, 1543–1554 (2016).
Morugán-Coronado, A., García-Orenes, F., Mcmillan, M. & Pereg, L. The effect of moisture on soil microbial properties and nitrogen cyclers in Mediterranean sweet orange orchards under organic and inorganic fertilization. Sci. Total Environ. 655, 158–167 (2019).
Han, X. M. et al. Effects of vegetation type on soil microbial community structure and catabolic diversity assessed by polyphasic methods in North China. J. Environ. Sci. 19, 1228–1234 (2007).
Bastida, F., Torres, I. F., Hernández, T. & García, C. The impacts of organic amendments: do they confer stability against drought on the soil microbial community?. Soil Biol. Biochem. 113, 173–183 (2017).
Bai, H. et al. Influences of irrigation amount on the rhizospheric microorganism composition and carbon dioxide flux of maize crops. Geoderma 343, 1–9 (2019).
Inglett, P. W., Rivera-Monroy, V. H. & Wozniak, J. R. Biogeochemistry of nitrogen across the Everglades landscape. Crit. Rev. Environ. Sci. Technol. 41, 187–216 (2011).
Gumiere, T., Durrer, A., Bohannan, B. J. M. & Andreote, F. D. Biogeographical patterns in fungal communities from soils cultivated with sugarcane. J. Biogeogr. 43, 2016–2026 (2016).
Griffiths, B. S., Bonkowski, M., Roy, J. & Ritz, K. Functional stability, substrate utilisation and biological indicators of soils following environmental impacts. Appl. Soil Ecol. 16, 49–61 (2001).
Avershina, E., Frisli, T. & Rudi, K. D. novo semi-alignment of 16s rRNA gene sequences for deep phylogenetic characterization of next generation sequencing data. Microbes Environ. 28, 211–216 (2013).
Zhong, W. et al. Nitrogen fertilization induced changes in ammonia oxidation are attributable mostly to bacteria rather than archaea in greenhouse-based high n input vegetable soil. Soil Biol. Biochem. 93, 150–159 (2016).
Ochoa-Hueso, R. et al. Soil fungal abundance and plant functional traits drive fertile island formation in global drylands. J. Ecol. 106, 242–253 (2018).
Guenet, B. et al. The impact of long-term CO2 enrichment and moisture levels on soil microbial community structure and enzyme activities. Geoderma 170, 331–336 (2012).
Ding, L. J., Su, J. Q., Li, H., Zhu, Y. G. & Cao, Z. H. Bacterial succession along a long-term chronosequence of paddy soil in the Yangtze River Delta, China. Soil Biol. Biochem. 104, 59–67 (2017).
Wang, J. et al. Effects of nitrogen and water on soil enzyme activity and soil microbial biomass in Stipa baicalensis steppe, Inner Mongolia of north China. J. Agric. Resour. Environ. 31, 237–245 (2014).
Carrara, J. E. et al. Interactions among plants, bacteria, and fungi reduce extracellular enzyme activities under long-term N fertilization. Glob. Chang. Biol. 24, 2721–2734 (2018).
Liu, W. X., Zhang, Z. & Wan, S. Q. Predominant role of water in regulating soil and microbial respiration and their responses to climate change in a semiarid grassland. Glob. Chang. Biol. 15, 184–195 (2009).
Wang, C. Y. et al. Effects of different irrigation and nitrogen regimes on root growth and its correlation with above-ground plant parts in high-yielding wheat under field conditions. Field Crops Res. 165, 138–149 (2014).
Chen, J. H. et al. Consistent increase in abundance and diversity but variable change in community composition of bacteria in topsoil of rice paddy under short term biochar treatment across three sites from south china. Appl. Soil Ecol. 91, 68–79 (2015).
Ferris, H. & Tuomisto, H. Unearthing the role of biological diversity in soil health. Soil Biol. Biochem. 85, 101–109 (2015).
Peralta, A. L., Ludmer, S., Matthews, J. W. & Kent, A. D. Bacterial community response to changes in soil redox potential along a moisture gradient in restored wetlands. Ecol. Eng. 73, 246–253 (2014).
Colombo, F., Macdonald, C. A., Jeffries, T. C., Powell, J. R. & Singh, B. K. Impact of forest management practices on soil bacterial diversity and consequences for soil processes. Soil Biol. Biochem. 94, 200–210 (2016).
Lauber, C. L., Hamady, M., Knight, R. & Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Ma, G. et al. Bacterial community structure and predicted function in wheat soil from the north china plain are closely linked with soil and plant characteristics after seven years of irrigation and nitrogen application. Front. Microbiol. 11, 506 (2020).
Zhao, J. et al. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS ONE 9, e85301 (2014).
Jiao, S. et al. Bacterial communities in oil contaminated soils: biogeography and co-occurrence patterns. Soil Biol. Biochem. 98, 64–73 (2016).
Mäder, P. et al. Soil fertility and biodiversity in organic farming. Science 296, 1694–1697 (2002).
Bei, S. K. et al. Response of the soil microbial community to different fertilizer inputs in a wheat-maize rotation on a calcareous soil. Agric. Ecosys. Environ. 260, 58–69 (2018).
Shen, J. P., Zhang, L. M., Guo, J. F., Ray, J. L. & He, J. Z. Impact of long-term fertilization practices on the abundance and composition of soil bacterial communities in northeast China. Appl. Soil Ecol. 46, 119–124 (2010).
Schimel, J., Balser, T. C. & Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394 (2007).
Dennis, P. G., Miller, A. J. & Hirsch, P. R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities?. FEMS Microbiol. Ecol. 72, 313–327 (2010).
Takaichi, S., Maoka, T., Takasaki, K. & Hanada, S. Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2′-dirhamnoside. Microbiology (Reading) 156, 757–763 (2010).
Altmann, D., Stief, P., Amann, R., De Beer, D. & Schramm, A. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol. 5, 798–803 (2003).
Chu, H. Y. et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006 (2010).
Fernández-Gómez, B. et al. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037 (2013).
Barnard, R. L., Osborne, C. A. & Firestone, M. K. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J. 9, 946–957 (2015).
Zhong, Y. Q. W., Yan, W. M. & Shangguan, Z. P. Impact of long-term N additions upon coupling between soil microbial community structure and activity, and nutrient-use efficiencies. Soil Biol. Biochem. 91, 151–159 (2015).
Fan, M. C., Li, J. J., Yan, W. M., Shi, H. & Shangguan, Z. P. Shifts in the structure and function of wheat root-associated bacterial communities in response to long-term nitrogen addition in an agricultural ecosystem. Appl. Soil Ecol. 159, 103852 (2021).
Guan, D. H. et al. Tillage practices effect on root distribution and water use efficiency of winter wheat under rain-fed condition in the North China Plain. Soil Till. Res. 146, 286–295 (2015).
Börjesson, G., Menichetti, L., Kirchmann, H. & Kätterer, T. Soil microbial community structure affected by 53 years of nitrogen fertilization and different organic amendments. Biol. Fertil. Soils 48, 245–257 (2012).
Ai, C. et al. Different roles of rhizosphere effect and long-term fertilization in the activity and community structure of ammonia oxidizers in a calcareous fluvo-aquic soil. Soil Biol. Biochem. 57, 30–42 (2013).
Stark, C., Condron, L. M., Stewart, A., Di, H. J. & O’Callaghan, M. Influence of organic and mineral amendments on microbial soil properties and processes. Appl. Soil Ecol. 35, 79–93 (2007).
Hueso, S., García, C. & Hernández, T. Severe drought conditions modify the microbial community structure, size and activity in amended and unamended soils. Soil Biol. Biochem. 50, 167–173 (2012).
Bastida, F. et al. Combined effects of reduced irrigation and water quality on the soil microbial community of a citrus orchard under semi-arid conditions. Soil Biol. Biochem. 104, 226–237 (2017).
Lu, R. K. Methods of Soil and Agro-Chemical Analysis (China Agricultural Science and Technology Press, 2000) ((in Chinese)).
Bao, S. D. Analytical Methods of Soil Agrochemistry 3rd edn. (China Agricultural Press, Beijing, 1999).
Haas, B. J. et al. Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011).
Edgar, R. C. Uparse: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Quast, C. et al. The silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Acknowledgements
This work was supported by the National Key Research and Development Plan of China (2017YFD0300909), and Technology System in Modern Wheat Industry, Chinese Ministry of Agriculture (CARS-03-05), and Science and Technology Research Project of University, Hebei province (QN2018219), and Science and Technology Program of Baoding, China (1911ZN010).
Author information
Authors and Affiliations
Contributions
R.Q.L. conceived the project and set the scientific objectives; H.R.L., H.G.W., B.J., D.X.L., and Q.F. conducted the experiments and analyzed the data; H.R.L. wrote the manuscript; H.R.L. and R.Q.L. revised the manuscript. All authors discussed the results and read and approved the final manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, H., Wang, H., Jia, B. et al. Irrigation has a higher impact on soil bacterial abundance, diversity and composition than nitrogen fertilization. Sci Rep 11, 16901 (2021). https://doi.org/10.1038/s41598-021-96234-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-96234-6
This article is cited by
-
Dynamic of active microbial diversity in rhizosphere sediments of halophytes used for bioremediation of earthen shrimp ponds
Environmental Microbiome (2023)
-
Soil bacterial and fungal communities respond differently to Bombax ceiba (Malvaceae) during reproductive stages of rice in a traditional agroforestry system
Plant and Soil (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.