Abstract
The result of improper treatment has led to the rise of Multidrug-resistant (MDR) strains. This concern still exists in Pakistan. In order to save energy, time and resources an early detection of resistant cases is imperative. Thus, a treated group of 100 isolates and a control group of 56 untreated isolates were studied. PCR and gene sequencing showed mutations at codon 531 and 513 in the rpoB gene. 12% of cases showed a double mutation in the rpoB gene. katG gene showed mutations at codon 315 and 299. 28.6% of the control group cases were positive for MDR whereas 100% of the treated group were positive for MDR. This study explores the significantly increasing ratio of MDR-TB among Pakistani population. This study provides prevalent MDR mutations among Pakistanis and suggests developing such molecular assays that are time and cost effective. Importance: Pakistan is a developing country and has fourth highest incidence rate of MDR-TB. The treatment of MDR-TB is the use of second line drugs that has severe side effects as well as it requires long time span. One of the strategies to control the spread of MDR-TB is to decipher the aberrations at molecular level in order to formulate potent drugs that can treat the patients within short span of time. Determining the mutation profile of MDR in Pakistani populations will open new horizons for the improvement of drug treatment regimens to make it more effective or for the development of novel potent drugs and vaccines to better treat the drug-resistant TB. Moreover, this study will be help in disease control program.
Similar content being viewed by others
Introduction
Pakistan is a developing country with a crowded population of 210 million people, out of which 1.5 million suffer from TB. Pakistan ranks sixth highest in the world for estimated number of tuberculosis cases. With the passage of time, the emergence of strains with drug-resistance and multiple drug resistance (MDR) has lethaly increased. MDR has become the leading health concern in Pakistan which currently ranks fourth among 27 countries highly burdened by MDR-TB1. According to a WHO report, 18% of MDR cases reported in the year 2018 were previously on treatment and 3.5% of cases did not receive prior treatment2. MDR is associated with the combined resistance to first line drugs, Rifampicin and Isoniazid. Rifampicin resistance (RIFr) is of particular epidemiological importance. Isoniazid resistance coexists with RIFr in more than 90% of isolates3,4. Mutations in rpoB that confer resistance to Rifampicin hinder the formation of mRNA transcripts and RNA elongation, which thereby prevent binding to the beta subunit of RNA Polymerase5,6. Published data related to Rifampicin resistance illustrates 87 unique point mutations or short indels positioned in an 81-bp rifampin resistance-determining region (RRDR) that encodes 27 amino acids (codons 507 to 533). The katG gene encodes Catalase Peroxidase enzymes (~ 740-bp) that are indispensable for the conversion of Isoniazid, the first line drug, into its active form7,8 in Mycobacterium tuberculosis. Several genetic alterations in katG correspond to codons 315, 316, and 3099. A study based on a cohort of 163 patients in Belarus identified four types of mutations in codon 315: AGC→ACC (present in 85% of patients), AGC→AGG (2.3%), AGC→ACC (4.7%), and AGC→GGC (2.3%). One type of mutation was identified at codon 316 which was GGC→AGC (41.4%). Four types of mutations were found in codon 309: GGT→GGT (16.1%), GGT→GCT (9.2%), GGT→GTC (6.9%), GGT→GGG (2.7%)9. Moreover, a mutation at codon 315 was observed in 64% of cases around the globe10. Although isoniazid resistance due to inhA mutation has been observed but this resistance is of low level11. Various molecular techniques depend upon these genetic factors to diagnose MDR. Identification of MDR by detection of known mutations in rpoB and katG is as efficient as drug susceptibility testing (DST)11. MDR and extensive drug-resistant TB is not just limited to a few countries. It is now a world-wide phenomenon12. The most effective patient treatment and efficient disease control rely absolutely on the early detection of the drug-resistant M.tuberculosis strains.
Pakistan is the fourth highly vulnerable country in the world for MDR-TB1. It is, therefore, imperative that Pakistan should adopt such strategies as to control the disease at an early stage.
The type and severity of drug resistance in different populations lead to varied mutations. Therefore, extensive research deciphering changes at molecular level of mycobacterial genes will form the foundation for new treatment regimens. To identify mutations, automated DNA sequencing was adopted as it is a universally practiced method with high sensitivity and selectivity to characterize mutations13,14,15. This study was conducted to investigate the type and rate of occurrence of drug resistance found among M. tuberculosis clinical isolates that demonstrated an MDR phenotype. The objective of this study in Pakistan is to develop low-cost molecular assays depending upon already know mutations. This molecular test is time efficient and as accurate as drug susceptibility test (DST).
Our approach was to pick and select number of mutation sites that could be used for swift screening of isolates to detect MDR-TB in Pakistan.
Results
Amplification and nucleotide analysis
rpoB and katG genes’ selected regions were amplified by using the designated primers and were sequenced to unravel the mutations. The sequences were analyzed for aberrations against the reference sequence, GI: 57,116,681. The strain sequences were submitted to GenBank and accession numbers of the sequences are: JN315348.1, JQ394887-JQ394890, JN626460, JN626461, JN626462, JN626463, JN626464, JN626465, JN315348, JN315349, JN315350, JN315351, JN315352, JN315353, JN315354, JN315355, JN315356, and JN626466.
Analysis of rpoB gene mutations
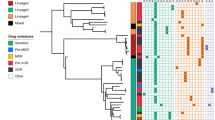
DNA sequence analysis of 156 M. tuberculosis samples taken from the people coming from different regions of Pakistan showed MDR. No polymorphism was observed in the control strain (H37Rv strain). 67% of the treated group had a mutation at codon 531 in rpoB. 21% of the treated group had an insertional mutation at codon 513 in rpoB. However, 12% of the treated group had mutation at both codons 531 and 513. Most of the mutations in MDR-TB strains were single nucleotide mutations in rpoB. 28.6% of the control group showed mutation at 531 in rpoB. No mutation was observed in the region outside the hot spot of rpoB gene (350 bp upstream of codons 507 to 533). The total number of cases found resistant to Rifampicin are provided in (Table 1). Table 2 shows the nucleotide and amino acid changes in the specific regions of rpoB. Table 3 compares the nucleotide sequence of rpoB gene with the control strain.
Analysis of katG gene mutations
MDR strains were analyzed via DNA sequencing of a 1120-bp katG fragment. In the katG gene, a mutation at codon 315 was found in 88% of treated cases where the wild type codon AGC was mutated to ACC (Ser→Thr). 12% of the treated group demonstrated double mutations, S315T and G299S. The latter mutation was found to be a novel mutation based on publicly available data. 28.6% of the control group showed S315T mutation. The H37Rv strain was used as a control strain and no mutations were found. Single nucleotide mutation was observed in the isolates. Table 1 shows the number of Isoniazid resistant cases. katG amino acid changes are shown in the Table 2.
Statistical analysis
Depending upon the identification of different mutations in rpoB and katG during MDR-TB, the treated and the control groups were divided into 3 categories. Category A has individuals with mutations at rpoB [S531L] and katG [S315T], category B has individuals with mutations at rpoB [513Ins (Arg)], katG [S315T] and category C has individuals with double mutations at rpoB [513Ins (Arg), S531L], katG [S315T, G299S]. Mann Whitney U test was applied to this data to compare the two groups. The p-value was 0.03 that is significant.
Discussion
Pakistan is one such country as lacks latest information and a vast majority of the masses lives under or close to the poverty line. These factors still exist even in the twenty-first century. This makes it easier to understand why mutated strains spring up. The standard procedure of observing MDR-TB is by finding resistance in rpoB and katG against rifampicin and isoniazid. Studies revealed that katG and rpoB are the commonly known genes that show mutations due to inconsistent medical treatment. For the study 156 samples were collected. 100 were of the patients already treated inconsistently and 56 of the patients who had not gone through any treatment. All the patients went through the standard operational procedure before the initiation of the treatment and conclusion.
Current research detected MDR-TB strains by culturing smear positive respiratory samples followed by drug sensitivity tests. It has now been established that mutations in rpoB gene and katG gene of M. tuberculosis are associated with resistance to Rifampicin and Isoniazid respectively. These mutations, along with novel ones, can be determined very easily and in a timely manner through sequence analysis. Previous studies from other countries affirm our findings in a Pakistani cohort that 95% of mutations appear in the 81-bp RRDR of rpoB. The most frequent mutations in RRDR are at codons 531, 526, and 516 which are listed in descending order of their occurrence in the population6,16,17,18.
Present study data is in accordance with the data presented by Thailand that the mutation in rpoB531 (58%) and katG Ser315 (100% of 38 strains) was commonly found in the study group19. An Australian study reported rpoB S531L (40.6%) and katG Ser315 (84.1%) as the most frequent mutations17 that has also been reported by a global survey conducted in 201518. An Iranian study revealed the rpoB mutation at codon 531 (55.68%) and was among highly prevalent mutations in northeast Iran19. Another study showed katG gene mutation at codon 315 (70%) is frequently found in Iran20. High prevalence of mutations at codon 531 of rpoB10,21,22,23,24 and at 315 of katG10,21,22,23,,23,24 have been reported from China, Vietnam, India and Nepal.
The region of gene that is 350-bp upstream of the RRDR has been reported to have mutations at codon 145, 170, 173, 174, 176, 180, 181 and 1844,25,26,27,28. A study conducted in China showed that the frequency of mutation at codon 531 was the highest and our study found similar results in rpoB29. The current study revealed that 67% of the treated group had mutation at codon 531 whereas 21% of the cases had mutation at codon 513 in rpoB gene. Therefore, the S531L substitution mutation is found to be common in RRDR. These results confirm the most prevalent mutation is found at the rpoB S531L and katG S315T codons in the Pakistani population sampled. Moreover, a novel mutation was also identified in this research where an extra Arginine was inserted at codon 513. Sequence analysis did not show any polymorphism in control strains. Studies conducted in Pakistan and China showed mutations in codon 315 of katG17,18,29 that is in agreement with our results.
Alarmingly, among 56 control group patients (unmedicated tuberculosis), 16 patients showed mutation at codon 531 in rpoB gene and 315 in katG. This fact reveals that drug resistance that was previously thought to be due to improper treatment and medication has now infected untreated patients. This striking fact may reflect the direct transfer of mutated TB strains from already infected patients. Moreover, statistical analysis showed a significant increase of MDR-TB cases among Pakistani population. So, the issue should be redressed.
It can be concluded from the results of the current study that previously reported mutations as well as novel mutations exist in the Pakistani M. tuberculosis strains. The current study further reveals that the most frequent mutation in rpoB is at the 531 codon and in katG at the codon 315. The double mutations in rpoB at positions 513 and 531 and in katG at 315 and 299 are notable. The presence of mutated strains in un-medicated patients is a concerning fact. Determining the mutation profile of MDR in Pakistani populations will open new horizons for the improvement of drug treatment regimens to make it more effective or for the development of novel potent drugs and vaccines to treat the drug resistance in TB. The results and molecular assessment of isoniazid and rifampicin resistance in this study were remarkable. It suggests that low-cost molecular assays may effectively be used in Pakistan for the detection of MDR-TB in clinical settings with reasonable sensitivity.
Material and methods
Sample collection
M. tuberculosis samples were collected from patients at Pakistan Medical Research Council (PMRC) Centre for Tuberculosis and Lung Diseases Mayo Hospital, Lahore. Informed consent was obtained from all subjects. Ethical Committee of Centre of Excellence in Molecular Biology, University of the Punjab approved the study. All methods were performed in accordance with the relevant guidelines and regulations.
Most specimens received by the laboratory were sputum samples. Samples were received from a total of 156 TB patients. The stringency criteria for the selection of patients was that they should have no comorbidity and should have been receiving inconsistent treatment for 2 to 5 years. 56 samples included cases of TB that had not undergone any medical treatment before and were labeled as the control group. A total of 100 samples were selected based on the long duration of the disease. These patients had received Rifampicin and Isoniazid treatment for 2–5 years inconsistently due to limited resources and poverty and were referred as the treated group. When the treatment was initiated, the treated group was not MDR. All the patients were carefully studied before and after treatment concluded. Smear slides were prepared to identify Acid Fast Bacteria to confirm the presence of M. tuberculosis. The selected samples were then grown on Lowenstein-Jensen (LJ) media and tested for drug susceptibility (Rifampicin and Isoniazid). The H37Rv strain (GenBank Id: AP018036.1) (a susceptible strain) was also used as a control.
DNA extraction and PCR amplification
M. tuberculosis DNA was extracted from samples by following the Gentra Puregene Blood Kit (Qiagen, USA) instructions. Extracted DNA was then amplified by using the desired primers. Primer 3 was used to design the primers30. This study was designed to identify mutations in the two regions of rpoB, the RRDR and another region 350 bps upstream of the RRDR region, and in the katG gene of M. tuberculosis in Pakistani isolates. A set of primers, POB-F 5′-CGAGCTGATCCAAAACCAGA-3′ and POB-R GCTCCAGGAAGGGAATCATC, were designed to amplify a 650-bp region of rpoB gene and another set of primers, RRB-F 5′-CTTCTCCGGGTCGATGTCGTTG-3′ and RRB-R 5′-CGCGCTTGTCGACGTCAAACTC-3′, were designed to amplify a 365 bp region of rpoB gene31. Similarly, a region of 1120-bp of katG gene was amplified by Kat-F: 5′-ACTACGGGCCGCTGTTTATC-3′ and Kat-R:5′-TGAGACAGTCAATCCCGATG-3′.
The amplification conditions were the same for both POB and Kat primers: initial denaturation at 94 °C for 2 min followed by 35 cycles consisting of denaturation at 94 °C for 45 s, primer annealing at 50 °C for 45 s, primer extension at 72 °C for 3 min, followed by a final extension of 10 min. The annealing temperature used for the RRB set of primers was 64 °C for 1 min. The amplified products were visualized on a 1.5% agarose gel stained with ethidium bromide under UV transilluminator after electrophoresis.
DNA sequencing
For DNA purification from the agarose gel, the Vivantis GF-1 Nucleic Acid Extraction Kit was used and the manufacturer’s protocol was followed. DNA samples were sequenced by using Big Dye chemistry (Applied Bioscience Inc., USA). The samples were sequenced with the related forward and reverse primers using the Applied Biosystems Prism Dye Termination method according to the manufacturer’s instructions (Big Dye Deoxy Terminators; Applied Biosystems, Weiterstadt, Germany). Sequencing was performed on an automated sequencer (Applied Biosystems; 3100 DNA Analyzer). Chromatograms of sequences were obtained and submitted to GenBank. The accession numbers of the sequences are: JN315348.1, JQ394887-JQ394890, JN626460, JN626461, JN626462, JN626463, JN626464, JN626465, JN315348, JN315349, JN315350, JN315351, JN315352, JN315353, JN315354, JN315355, JN315356, and JN626466.
Statistical analysis
GraphPad Prism v. 8.032 was used to apply Mann Whitney U test to find the probability (p-value is significant at p < 0.05) of the experiment done.
Ethical approval
Ethical Committee of Centre of Excellence in Molecular Biology, University of the Punjab approved the study. All methods were performed in accordance with the relevant guidelines and regulations.
References
http://www.emro.who.int/pak/programmes/stop-tuberculosis.html
https://www.who.int/tb/areas-of-work/drug-resistant-tb/MDR-RR_TB_factsheet_2018_Apr2019.pdf?ua=1
Drobniewski, F. & Wilson, S. The rapid diagnosis of isoniazid and rifampicin resistance in mycobacterium tuberculosis—A molecular story. J. Med. Microbiol. 47, 189–196 (1998).
Mani, C., Selvakumar, N., Narayanan, S. & Narayanan, P. Mutations in the rpo b gene of multidrug resistant mycobacterium tuberculosis clinical isolates from India. J. Clin. Microbiol. 39, 2987–2990 (2001).
Lin, W. et al. Structural basis of mycobacterium tuberculosis transcription and transcription inhibition. Mol. Cell. 66(2), 169–179 (2017).
Yue, J. et al. Mutations in the rpob gene of multidrug-resistant mycobacterium tuberculosis isolates from china. JCM. 41(5), 2209–2212 (2003).
Ando, H. et al. Identification of katg mutations associated with high-level isoniazid resistance in mycobacterium tuberculosis. AAC. 54(5), 1793–1799 (2010).
Purkan, P. et al. Molecular analysis of katg encoding catalase-peroxidase from clinical isolate of isoniazid-resistant mycobacterium tuberculosis. J. Med. Life. 11(2), 160 (2018).
Bostanabad, S., Titov, L., Bahrmand, A. & Nojoumi, S. Detection of mutation in isoniazid resistant mycobacterium tuberculosis isolates from tuberculosis patients in belarus. Indian J. Med. Microbiol. 26, 143–147 (2008).
Seifert, M., Catanzaro, D., Catanzaro, A. & Rodwell, T. C. Genetic mutations associated with isoniazid resistance in mycobacterium tuberculosis: A systematic review. PLoS ONE 10(3), e0119628 (2015).
Rosales-Klintz, S. et al. Drug resistance-related mutations in multidrug-resistant mycobacterium tuberculosis isolates from diverse geographical regions. Int. J. Mycobacteriol. 1(3), 124–130 (2012).
Fox, G. J. et al. Preventing the spread of multidrug-resistant tuberculosis and protecting contacts of infectious cases. CMI. 23(3), 147–153 (2017).
Telenti, A. et al. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. JCM. 31(2), 175–178 (1993).
Lee, H., Park, H., Cho, S., Bai, G. & Kim, S. Species identification of mycobacteria by pcr-restriction fragment length polymorphism of the gene. J. Clin. Microbiol. 38, 2966–2971 (2000).
King, M. D. et al. Assays and enumeration of bioaerosols-traditional approaches to modern practices. Aerosol. Sci. Tech. 54(5), 611–633 (2020).
Patra, S., Jain, A., Sherwal, B. & Khanna, A. Rapid detection of mutation in rrdr of rpo b gene for rifampicin resistance in mdr-pulmonary tuberculosis by dna sequencing. Ind. J. Clin. Biochem. 25, 315–318 (2010).
Ali, A. et al. Characterization of mutations conferring extensive drug resistance to mycobacterium tuberculosis isolates in pakistan. AAC. 55(12), 5654–5659 (2011).
Li, Q. et al. Characterisation of drug resistance-associated mutations among clinical multidrug-resistant mycobacterium tuberculosis isolates from hebei province, china. J. Glob. Antimicrob. Resist. 18, 168–176 (2019).
Prammananan, T. et al. Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin. Microbiol. Infect. 14(5), 446–453 (2008).
Lam, C. et al. Value of routine whole genome sequencing for mycobacterium tuberculosis drug resistance detection. IJID. (2021).
Tajbakhsh, A. et al. Investigation of the rpob mutations causing rifampin resistance by rapid screening in mycobacterium tuberculosis in northeast of iran. Iran. J. Pathol. 13(4), 429–437 (2018).
Motavaf, B. et al. Detection of genomic mutations in katg and rpob genes among multidrug-resistant mycobacterium tuberculosis isolates from Tehran, Iran. NMNI 41, 1079 (2021).
Liu, Y. et al. Evaluation of the frequency of mutation genes in multidrug-resistant tuberculosis (mdr-tb) strains in Beijing, China. Epidemiol. Infect. 149 (2021).
Singhal, R. et al. Early detection of multi-drug resistance and common mutations in mycobacterium tuberculosis isolates from delhi using genotype mtbdrplus assay. Indian J. Med. Microbiol. 33 (2015).
Yao, C. et al. Detection of rpob, katg and inha gene mutations in mycobacterium tuberculosis clinical isolates from chongqing as determined by microarray. CMI. 16(11), 1639–1643 (2010).
Siu, G. K. et al. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in mycobacterium tuberculosis. J. Antimicrob. Chemother. 66(4), 730–733 (2011).
Chen, L. et al. rpob gene mutation profile in rifampicin-resistant mycobacterium tuberculosis clinical isolates from guizhou, one of the highest incidence rate regions in China. J. Antimicrob. Chemother. 65(6), 1299–1301 (2010).
Lingala, M., Srikantam, A., Jain, S., Rao, K. & Rao, P. Clinical and geographical profiles of gene mutations in mycobacterium tuberculosis isolates from Hyderabad and Koraput in India. J. Microbiol. Antimicrob. 2, 13–18 (2010).
Khan, S.N. et al. 2013. Molecular characterization of multidrug-resistant isolates of mycobacterium tuberculosis from patients in Punjab, Pakistan. Pak. J. Zool. 45(1) (2013).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucl. Acids Res. 40(15), e115–e115 (2012).
Heep, M. et al. Frequency of rpob mutations inside and outside the cluster I region in rifampin-resistant clinical mycobacterium tuberculosis isolates. JCM. 39(1), 107–110 (2001).
GraphPad Prism Mann Whitney U test was performed using GraphPad Prism version 8.0.0 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com.
Acknowledgements
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. The authors thank to Pakistan Medical Research Council (PMRC) TB Research Centre and Lung Diseases Mayo Hospital, Lahore for donating M. tuberculosis strains. The technical help of Dr. Rizwan Iqbal, Mr. Zia, Professor Javeed Naeem is also highly appreciated.
Author information
Authors and Affiliations
Contributions
M.I. conceived the idea of study. A.A. performed experimental work. A.A. and S.A. drafted the manuscript. Statistical analysis was done by Z.Q. All authors have reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aftab, A., Afzal, S., Qamar, Z. et al. Early detection of MDR Mycobacterium tuberculosis mutations in Pakistan. Sci Rep 11, 16736 (2021). https://doi.org/10.1038/s41598-021-96116-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-96116-x
This article is cited by
-
Health-Related Quality of Life (HRQoL) in Tuberculosis Patients: a Cross-Sectional Study Using the EuroQoL EQ-5D-3L Scale
SN Comprehensive Clinical Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.