Abstract
Staphylococcus aureus is a major etiological agent of clinical and subclinical bovine mastitis. Owing to the mostly backyard dairy practices, we hypothesized that genetic diversity among mastitis-associated S. aureus from India would be high, and investigated 166 isolates obtained mostly from the Southern State of Karnataka, but also from a few other states. The results revealed (a) 8 to 13 fragments in pulsed-field gel electrophoresis (PFGE), forming 31 distinct patterns, and (b) 34 spa types, of which three (t17680, t18314, and t18320) were newly identified. Multi-locus sequencing typing (MLST) identified 39 sequence types (STs), with ST2454 (34.4%) and ST2459 (24%) being the most commonly represented, which clustered to clonal complexes (CC) CC9 and CC97, respectively; 12 STs were newly identified. Thirty-four (20.5%) of the 166 isolates displayed oxacillin resistance. On the other hand, whereas none were mecC+, 44 (26.5%) isolates were mecA+, with a predominance of SCCmecIVb (26/32 isolates, others being untypeable); 24 isolates (14.46%) were oxacillin-susceptible methicillin-resistant S. aureus (OS-MRSA; mecA+ but OS). Integrated analysis revealed that CC9-ST2454- and CC97-ST2459-SCCmecIVb were the predominant MRSA, although the distribution of CC9 and CC97 was similar between methicillin-resistant and -susceptible isolates. By PCR, 56.25%, 28.75% and 47.5% of the 166 isolates were positive for hlg, tsst and pvl genes, respectively. Our results, for the first time describe the application of a combination of various molecular methods to bovine mastitis-associated S. aureus isolates from India, corroborate the worldwide distribution of CC97 and CC9, and suggest pathogenic potential of the isolates.
Similar content being viewed by others
Introduction
Infections and dynamics of disease caused by the same pathogen differ from host to host and from one geographical location to another. Some clonal lineages of a particular pathogen tend to be specific to a host, and others appear to transmit easily between hosts. To gain knowledge about similarities among strains, sources of infection, modes of transmission and carriage of virulence and resistance genes, one can employ molecular typing methods which can reveal genetic diversity of the isolates of a particular pathogen.
Staphylococcus aureus is found associated with a variety of human infections, with some evidence for overlap in the distribution of certain clonal populations between humans and animals1,2,3,4. The organism is known to produce a number of virulence factors such as toxins and antibiotic resistance mediators which facilitate its survival in the host as well as in the environment5,6. In addition, methicillin-resistant S. aureus (MRSA), which encode multiple antimicrobial resistance genes, are a major problem in human medicine7. Bovine mastitis is a very common livestock production-related disease, and S. aureus is the major contributor, resulting in a range of manifestations, including a large proportion of subclinical and chronic cases8,9. Isolates from bovine mastitis have been shown to produce various types of hemolysins and enterotoxins, as well as Panton-Valentine leucocidin (PVL) and toxic shock syndrome toxin (TSST), although the proportions of the isolates expressing each type of these toxins vary widely10,11,12. Although early studies suggested much lower prevalence of MRSA among bovine mastitis-associated S. aureus compared to those isolated from human infections, recent studies have shown trends towards higher prevalence13,14,15,16,17,18,19,20,21,22,23,24,25,26. This has led to focus towards the longitudinal spread and sharing of particular MRSA clones and their related strains among and between animals and humans, in order to understand the consequence of animal-human interface in infections and diseases caused by MRSA.
Multiple genetic types of bovine mastitis-associated S. aureus exist worldwide. Studying the origin, clonal populations, genetic variants and virulence determinants of S. aureus is important for the understanding of epidemiology, ecology and host adaptation, zoonotic potential, association with disease, and possibly to predict clinical outcome and control of disease. Typing methods for S. aureus include pulsed-field gel electrophoresis (PFGE), staphylococcal protein A (spa) typing and multi-locus sequence typing (MLST)7. PFGE provides reliable epidemiological comparisons among isolates, and is based on macro-restriction fragment polymorphism of genomic DNA. Spa typing analyses sequence repeats within the X region of the spa gene27. MLST analyses the sequence polymorphism of seven housekeeping genes of each isolate; MLST facilitates comparisons of population structures with high levels of discrimination and reproducibility28. These typing methods provide tools to examine the epidemiological picture in terms of diversity and clonality of S. aureus.
Most of the studies investigating genotypes of mastitis-associated S. aureus have been from large dairy herds, where it may not be surprising to see high level of clonality. Indian dairy industry is majorly based on pooling practices where individual holders keeping a few animals contribute to pooling of milk through dairy cooperatives. Because of the scattering of animals, we hypothesized that mastitis-associated S. aureus strains from India would be quite diverse. Very few studies have examined genotypes of bovine mastitis-associated S. aureus in India. In one study, where 39 MRSA isolates from three different states (Andhra Pradesh, Karnataka, Telangana) were investigated, spa types t037 and t267 and sequence types (ST) 72, ST245 and ST239 were reported to be predominating despite some diversity29. In another study with 100 isolates, spa types t359, t7867 and t3841 were found to be predominant in Karnataka30. In a third study, where 16 isolates were identified as MRSA, 50% of the isoltes belonged to ST168731. Another recent report from West Bengal described the existence of t304 and t6267 types among nine MRSA isolates32. In this study, we analysed 166 isolates of S. aureus obtained from bovine mastitis cases from six different states of India using spa, MLST and PFGE, and found the existence of limited CCs with diversity at genotype level.
Results
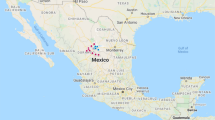
In this study, 166 bovine mastitis-associated S. aureus isolates from six different states of India were studied. The isolates included 98 from Karnataka, 19 each from Gujarat and Telangana, 12 from Uttar Pradesh and 5 from Maharashtra (Fig. 1).
Map of India showing location of the six states from where bovine S. aureus isolates were obtained. For each state, the predominant spa type, sequence type and clonal complex are provided. GJ = Gujarat, KA = Karnataka, MH = Maharashtra, MG = Meghalaya, TG = Telangana, UP = Uttar Pradesh. The map was modified from one downloaded from the URL https://d-maps.com/carte.php?&num_car=24867&lang=en; from where up to 10 maps can be used per publication for free, as per their terms and conditions of use.
By spa typing, only 145 isolates were typeable and belonged to 34 different spa types. The predominant types among isolates from Karnataka and Gujarat were t359 (31/98, 30.3%) and t1965 (8/19, 42.1%), respectively. On the other hand, t7867 was the most frequent type in Uttar Pradesh (6/12, 50%), Telangana (2/5, 40%) and Meghalaya (6/19, 31.5%) (Fig. 2). Three new types (t17680, t18314 and t18320) were identified in this study.
Minimum spanning trees showing spa types in each of the states. Each circle with different color represents spa type and size of the circle indicates the number of isolates for a particular spa type. Major spa type in each state is highlighted by a grey halo around the circle. The branch thickness (dotted lines, dashed lines and increasing thickness of solid lines in that order) indicates inverse of the distance between the types.
By MLST, 39 sequence types (STs) were identified, with ST2454 (57/166, 34.4%) and ST2459 (40/166, 24%) being the most commonly represented. The most frequent STs from Karnataka and Gujarat were ST2459 (38/98, 38.7%) and ST5408 (4/19, 21%), respectively, whereas ST2454 predominated in Uttar Pradesh (12/12, 100%), Telangana (9/13, 69.2%), Meghalaya (13/19, 68.4%), and Maharashtra (4/9, 44.4%). In this study, 12 STs (ST5407, ST5408, ST5410, ST5411, ST5413, ST5414, ST5415, ST5416, ST5418, ST5419, ST5420 and ST5689) were newly identified. The allelic profiles for all the isolates are shown in Supplementary Table S2 and the minimum spanning tree showing the relatedness among the different STs is shown in Fig. 3.
Minimum-spanning tree of MLST data from 166 S. aureus isolates in each state. Each circle with different color represents a different sequence type (ST) and the size of the circle is directly proportional to the number of isolates for a particular ST. Major clonal complex (CC) in each state is highlighted by a grey halo around the circle. The branch thickness (dotted lines, dashed lines and increasing thickness of solid lines in that order) indicates inverse of the distance between the STs.
The 39 STs could be grouped into seven CCs: CC1, CC5, CC8, CC9, CC30 and CC97, with CC97 and CC9 being predominant (Fig. 3). The dominant cluster from Gujarat (11/19, 57.8%) and Karnataka (53/98, 54%) was CC97, whereas CC9 dominated in Uttar Pradesh (12/12, 100%), Maharashtra (5/5, 100%), Meghalaya (17/19, 89.4%) and Telangana (11/13, 84.6%). Isolates belonging to CC1, CC5, CC8 and CC30 (20/166, 12%) were only identified from the states of Gujrat (6/20, 30%) and Karnataka (14/20, 70%).
By PFGE, only 144 of the 166 isolates produced bands upon SmaI digestion (Supplementary Fig. S1). The number of fragments ranged from 8 to 13, with 31 distinct patterns. Nine pulsotypes were identified at an 80% similarity level (Fig. 4). The majority of the isolates, 71.6% (101/141), clustered into two major pulsotypes C and E, followed by A and D (14% of the isolates). Majority of the isolates from Gujarat (10/12, 83.3%), Karnataka (34/90, 37.7%) and Meghalaya (7/13, 53.8%) clustered into pulsotype C. For pulsotype E, the proportions were 100% (12/12) for Telangana, 26.6% (24/90) for Karnataka, 70% (7/10) for Uttar Pradesh and 46.1% (6/13) for Meghalaya isolates. All four isolates from Maharashtra belonged to pulsotype A (Table 1).
To examine virulence determinants with the potential for pathogenicity, we subjected the isolates to disk diffusion test, broth microdilution for MIC for determining susceptibility to oxacillin and to PCR for mecA and mecC genes. Out of the 166 isolates, 34 (20.4%) showed resistance to oxacillin, with MICs of 8 to 64 µg/mL, whereas mecA amplification was observed with 44 (26.5%) isolates (Supplementary Table S3); none of the isolates contained mecC (data not shown). Out of the 44 mecA-positive isolates, 20 showed oxacillin MIC of > 4 µg/mL and 14 isolates showed oxacillin resistance in the absence of mecA or mecC (Table 1). By the definition of oxacillin susceptibility and presence of mecA, 24 of the 166 isolates (14.46%) could be categorised as OS-MRSA (Supplementary Table S4). Among 44 mecA+ isolates, 32 were typeable for SCCmec I to V, and most of the isolates (26/32) belonged to SCCmec IVb, whereas 12 isolates did not amplify any bands for these types (Table 1). The predominant MRSA clones were CC9-ST2454-SCCmecIVb-t7867 and CC97-ST2459-SCCmecIVb-t359 (Tables 1, 2).
PCR was also carried out to test the presence of the toxin genes hlg, tsst and pvl, which encode for the most common virulence determinants. The results showed that 56.25%, 28.75% and 47.5% of the isolates harboured hlg, tsst and pvl genes, respectively (Table 1). Notably, a high proportion of Karnataka isolates harboured hlg (87.7%) and pvl (82.2%) genes. Among these, isolates belonging to pulsotypes C2 and C3 had both hlg and pvl genes, and pulsotypes E5, E6 and E7 were positive for all the three genes (Table 1). All of the B, G, H and I pulsotypes carried the pvl gene, although there were only one to six isolates in these pulsotypes, and among pulsotypes C and E, which each contained more than 50 isolates, pvl carriage ranged between 39.62% and 44.23% (Table 1).
Discussion
Mastitis is a major concern for the dairy industry, as it not only affects animal’s health, but also results in direct and indirect losses33. Despite producing highest amount of milk, Indian dairy industry is crippled by mastitis34, with an estimated annual loss of INR 71.6551 billion (USD 1.6 billion) as of two decades ago35. Loss per incidence of INR 400 to 2086 (about USD 6 to 31) and annual loss per animal of INR 1500 to 1700 (about USD 22 to 25) have been reported36,37,38,39.
Although bovine mastitis is a multi-factorial, multi-etiological disease, S. aureus is one of the most common pathogens associated with it worldwide, including India40,41,42. Infection with S. aureus can lead to a wide range of manifestations including clinical as well as subclinical, chronic, persistent and recurrent mastitis. Transmission of S. aureus usually occurs from one animal to another through milking, but can also occur through contagion. Complete information on the population structure, virulence and resistance characteristics of mastitis-associated S. aureus is desired to monitor its dissemination among animal populations and for risk assessment, so as to improve interventions. This is the first study to report the genotypes of bovine mastitis-associated S. aureus from different locations in India, through the simultaneous application of spa typing, MLST and PFGE.
Among several genotypes of S. aureus, only a few appear to be associated with bovine mastitis43. As far as spa types are concerned, from bovine milk, t002, t008, t267, t359 and t6877 have been reported from Brazil, Canada and Japan44,45,46,47; t127 and t2279 were reported to be common in China48. The spa types t267 and t359 were also observed to be predominant among our isolates from Karnataka, and have been previously reported from this state29; other types such as t7867 and t3841 are also common in Karnataka30. On the other hand, t7867 was observed in all the states, suggesting that t267, t359 and t7867 may be commonly associated with bovine mastitis in India.
We observed a predominance of ST2454 and ST2459 among our isolates, although there were also 12 newly identified STs. Other types, ST72, ST245, ST239 and ST1687 have been previously reported from Andhra Pradesh, Telangana and Karnataka29, implying the existence of diverse genetic populations. Integrated data of our isolates showed that CC97-ST5414-t1965 and CC97-ST2459-t359 were predominant in the states of Gujarat and Karnataka, respectively, whereas CC9-ST2454-t7867 was frequent among isolates of the remaining four states studied.
Early phenotypic studies from Europe and the US showed low prevalence of MRSA among livestock13,14,15,18. By specific culturing or PCR for mecA or mecC, other studies have reported a relatively lower prevalence of 0% to 9.7% from Europe, US, Japan and South Korea17,19,20,25,26,45, while higher prevalence ranging from 16.7 to 49.6% has been reported from Germany, Brazil, Japan, Sweden and China16,21,22,23,24. We observed that 12.04% of our 166 isolates were methicillin-resistant both phenotypically and genotypically, whereas 14.46% were OS-MRSA. In total, 58 isolates (34.94%) were either oxacillin-resistant or mecA+. Earlier studies from India, which have performed either phenotypic or genotypic tests (not both), have reported a range from 5 to 27%49,50,51,52. It is possible that the high levels (34.94%) of MRSA within our isolates could just be a consequence of the application of both genopytic and phenotypic tests, but is similar to higher proportions observed in several parts of the world as stated above. It may, however, be noted that defining methicillin resistance has been problematic since a variety of conditions such as modifications in native penicillin-binding proteins, hyper-production of β-lactamase, or production of methicillinase can lead to resistance53. These studies together suggest that classifying an isolate as MRSA is complex, and in the context of a clinical setting, the phenotypic assay may be more important, although it is possible that genotypically positive, phenotypically negative isolates may still gain resistance in response to environmental stimuli such as encounter with salts or antibiotics54,55,56.
A large majority of our isolates, irrespective of methicillin resistance/susceptibility, clustered among CC97 and CC9. Historically, livestock-associated MRSA (LA-MRSA) were first reported from pig production systems, and found to belong to ST39857. Later, ST398 was also reported from bovine mastitis cases58,59, as well as other samples from various other livestock57. Majority of bovine mastitis-associated MRSA reported worldwide belong to CC39860,61,62,63, whilst less represented clones include those belonging to CC1, CC5, CC9, CC97, CC130 and a few others64,65,66,67. It is now apparent that strains belonging to CC9 are the predominant LA-MRSA in Asian countries whereas ST398 is predominant in Europe and North America2,60,61,62,63. In addition, CC97 MRSA have also been demonstrated to be predominant in dairy cattle and pigs in Italy68. On the other hand, a large proportion of MSSA from a number of countries have been reported to belong to CC398 and CC9768,69,70,71,72,73,74,75,76,77. ST398, an ST which clusters under CC398, is also predominant among MSSA from Japan and China78,79. In addition, MSSA from China also belonged to ST97 and ST9-SCCmecXII-t899 whilst MRSA belonged to ST1-SCCmecI-t193962,80. Whereas CC97 and CC9 were widely represented in our MRSA as well as MSSA isolates, we did not find any isolate belonging to ST398, suggesting commonality as well as variance with studies worldwide.
The exact origin of the predominant strains in our study is not discernible since neither this is a longitudinal study, nor is there sufficient data from within India to compare and analyse. One of the major cluster of strains from our study was CC9-ST2454. This particular clone has been previously reported from pigs in China and from water buffalos in the Philippines81,82. It may be argued that our CC9-ST2454 strains originated from pigs. However, most of the places from where isolates were obtained for our study were from semi-urban or village areas from states which have very less pig population. In India, the majority of pig rearing occurs in the Northeast region and the state of Kerala. In fact, a little more than 70% of the pig population of India is in the Northeast region, and only the state of Meghalaya belonged to this geographical location within the isolates that we obtained. In addition, most of the other locations from where isolates were obtained were free from domesticated or feral pigs. Hence, we consider it highly unlikely that ST2454-SCCmecIVb MRSA in our study may have originated directly from pigs, although we cannot rule out transmission events which predate isolation of the organisms used in our study.
Majority (26/32) of our mecA-containing isolates were typed as SCCmecIV, which has also been reported in other studies elsewhere83,84. On the other hand, several recent studies have reported on oxacillin-susceptible mecA+ S. aureus (OS-MRSA)22,85,86,87,88,89. In our study, 14.46% of the isolates were OS-MRSA. In Karnataka, OS-MRSA belonged to CC97-ST2459-SCCmecIVb with diverse spa types. In other states, OS-MRSA belonged to CC9-ST2454-SCCmecIVb-t7867 or t4522. A report from China identified high incidence (35.9%) of OS-MRSA, and with SCCmecV-t267 whereas OS-MRSA isolates from Shanghai belonged to different spa and SCCmec types22. One study from India reported 48.7% OS-MRSA, with diverse STs and spa types87. Only two of our OS-MRSA isolates belonged t267 and the remaining belonged to diverse types indicating clonal diversity among OS-MRSA.
By PFGE, we observed 31 pulsotypes among 144 isolates. Comparatively, narrower as well as wider ranges (from 10 among 146 isolates to 33 among 66 strains to 39 among 245 strains10,90) have been reported by others. Although PFGE has been projected to be the gold standard for non-sequence-based typing of bacterial strains, there are very few PFGE typing studies for bovine mastitis-associated S. aureus isolates, possibly owing to the cost of the equipment and the requirement for technical expertise. In addition, the results of PFGE typing for strains from different laboratories is not easily comparable without an extensive cataloguing of the banding patterns. It may, however, be noted that the majority of ST2454 and ST2459 strains clustered to pulsotypes E and C (30/57 and 21/40), respectively, suggesting corroboration between the two typing methods.
A large proportion of the isolates from Karnataka showed the presence of both hlg and pvl genes, suggesting pathogenic potential of these isolates. It has been hypothesized that different clones might display differential patterns of antimicrobial resistance and virulence factors48. We observed that isolates expressing the toxin genes hlg, tsst and pvl belonged to particular pulsotypes (C2, C3, E5, E6 and E7), lending credence to this hypothesis. It should be noted that C and E pulsotypes encompassed the largest groups with 52 and 53 strains, respectively, but only five sub-pulsotypes among these contained the majority of the isolates carrying the pvl gene, suggesting potential association between certain pulsotypes and virulence.
In conclusion, the majority of the isolates that we studied belonged to two major clonal complexes CC97 and CC9, similar to what has been observed globally. And yet, genetic diversity was evident as supported by the existence of more than 30 different genetic types (by any of the three typing methods). While heterogeneity at the micro-level would appear to support our hypothesis that mastitis-associated S. aureus strains from India show diversity, the diversity appeared to be within the two CCs. Among the oxacillin-resistant strains, the majority of the isolates belonged to CC97-ST2454-SCCmecIVb-t359 in Karnataka and CC9-ST2454-SCCmecIVb-t7867 in the other states except Maharashtra. These strains may therefore represent the predominant clones in these regions. Importantly, ST2454- and ST2459-SCCmecIVb appear to be the major MRSA clones in the country. However, a wider inference of the study is limited by the higher number of isolates from some states than others. Hence, more extensive, structured studies involving more number of states and isolates are required to better understand the genetic diversity of S. aureus isolates causing mastitis in India. This could help in elucidating strain variation, studying movement and transmission events, and in designing better control and prevention strategies for bovine mastitis.
Materials and methods
Isolates and genomic DNA isolation
For this study, 166 isolates of S. aureus curated at the Department of Microbiology, Veterinary College, Bangalore, Department of Microbiology, Veterinary College, Shivamogga, Karnataka State, and the National Institute of Animal Biotechnology, Hyderabad, were used. The study was therefore retrospective, and focussed on obtaining complete set of typing data for as many isolates as possible. The isolates had been obtained at various times from clinical and subclinical mastitis cases from six different states: Gujarat (Anand), Karnataka (North and South), Maharashtra (Mumbai), Meghalaya (unknown locations), Telangana (Hyderabad) and Uttar Pradesh (unknown locations) in India. Whereas details of the locations and dates are available for most of the isolates from Karnataka (Supplementary Table S1), no details are available for other locations as the isolates were obtained from collaborators. The isolates had been identified earlier as S. aureus by biochemical tests as well as by polymerase chain reaction (PCR), and were re-confirmed before beginning this study by PCR for the nuc gene91.
The isolates were cultured for 24 h at 35 °C on tryptic soy agar plates, and a single colony was further propagated in tryptic soy broth for 24 h. The cells were pelleted and genomic DNA was extracted using the HiYield Genomic DNA Mini Kit (Bacteria) (Real Biotech Corporation, Taiwan).
Spa typing
The spa repeat region was amplified using primers and conditions as per RidomSpa server [spa-1113f. (5′-TAAAGACGATCCTTCGGTGAGC-3′) and spa-1514r (5′-CAGCAGTAGTGCCGTTTGCTT-3′)]92. PCR was performed with genomic DNA (500 ng), dNTPs (200 µM), each primer (10 pmol), and Taq DNA polymerase (1.25 U; New England Biolabs). Thermal cycling included initial denaturation (5 min at 80 °C) followed by 35 cycles of denaturation (45 s at 94 °C), annealing (45 s at 60 °C), and extension (90 s at 72 °C), with a single final extension (10 min at 72 °C). A portion of the PCR product was run on 1.5% agarose gel, and the remaining was purified using a kit (Real Biotech Corporation) and sequenced (Bioserve Biotechnologies, Hyderabad, India). Both forward and reverse reads were analyzed using Ridom SpaServer (https://spa.ridom.de/index.shtml) and spaTyper (http://spatyper.fortinbras.us/).
Multi-locus sequence typing and analysis
The designated seven housekeeping genes of S. aureus were amplified by PCR with the specific primers as prescribed by Enright et al.93 (Staph MLST database). PCRs was performed with genomic DNA (1 μg of), primers (500 ng each), and Master Mix (Takara Bio). The thermal cycling included an initial denaturation at 95 °C for 5 min, then 30 cycles of annealing at 55 °C for 1 min, extension at 72 °C for 1 min, and denaturation at 95 °C for 1 min, followed by a final extension of 72 °C for 5 min. The amplified fragments were purified and sequenced. Consensus sequences were obtained by aligning sequences from both orientations and analyzed using the PubMLST database to determine the ST. Minimum spanning tree was constructed based on STs and the advanced cluster analysis was performed to define the clonal complexes (CCs) by using the BioNumerics software version 7 (Applied Maths NB, Belgium). A CC was defined by similarity in at least four or more loci.
Pulsed-field gel electrophoresis
The bacterial isolates were grown for 24 h at 37 °C on tryptic soy agar plates. 1.8% SeaKem gold agarose was boiled in TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA), aliquoted into micro-centrifuge tubes, and equilibrated to 60 °C in a heat block. Three to five colonies of each bacterial isolate were suspended in TEN buffer (0.1 M Tris–HCl, 0.15 M NaCl, 0.1 M EDTA; 125 µL) and adjusted to approx. 3.0 McFarland. Lysostaphin (5 µL of 1 mg/Ml; Sigma-Aldrich, India) was added to the cell suspension followed by the addition of 1.8% SeaKem gold agarose (125 µL) and transferred to the plug mould. Solidified plugs were placed in EC buffer (6 mM Tris–HCl, 1.0 M NaCl, 0.1 M EDTA, 0.5% Brij 58, 0.2% deoxycholate, 0.5% sarkosyl; 1 mL) and incubated in a 37 °C water bath for 4 h. The EC buffer was replaced with ESP buffer (10 mM Tris–HCl, 1.0 mM EDTA, 1% SDS, 1 mg/mL of proteinase K, pH 8.0; 1 mL) and incubated overnight at 55 °C in a water bath. The ESP buffer was removed and the plugs were washed three times for 30 min each in TE buffer (10 mL). Plugs were digested with SmaI (30 U; New England Biolabs) for 2 to 3 h at room temperature, and then sealed with 1% of Seakem GTG agarose in 0.5X TBE buffer (0.045 M Tris base, 0.045 M boric acid, pH 8.3, 1 mM EDTA). Electrophoresis was performed in CHEF-DRIII PFGE system (Bio-Rad Laboratories) with 5 s initial switch time and 40 s final switch time for 20 h at 120° angle, with 6-V/cm gradient, and chiller set at 14 °C. After electrophoresis, the gel was stained with ethidium bromide solution (1 mg/mL) and de-stained in water. The gel image was captured and subjected to analysis using BioNumerics software version 7. The percent similarity was attained on DICE coefficients derived from the unweighted pair group method with arithmetic averages (UPGMA). A coefficient similarity of 80% was fixed to assemble PFGE clusters. Band position tolerance was set at 1.0%.
Evaluation of methicillin susceptibility and the presence of mec genes
For these studies, methicillin-sensitive ATCC-29213 and methicillin-resistant ATCC-335913 strains of S. aureus were used as control strains. The antibiotic susceptibility of the isolates was evaluated by the disk diffusion test according to the Kirby-Bauer method94. Each isolate was subjected to testing three times along with a methicillin-sensitive and a –resistant reference strain (ATCC 29,213 and 335,913, respectively). Briefly, the isolates were inoculated into brain heart infusion broth (1 Ml; HiMedia) and incubated overnight at 37 °C. A suspension (0.5 mL) of the resultant culture was adjusted to 0.5 on McFarland scale using sterile phosphate-buffered saline (PBS, pH 7.2), and then spread plated onto Mueller–Hinton agar (HiMedia). Discs containing oxacillin or cefoxitin (HiMedia), as well as a sterile disc serving as a negative control, were gently placed on the dried inoculum, and the plates were incubated at 37 °C for 24 h. The zone of inhibition, inclusive of borders, was measured in mm and the breakpoints were applied as recommended for veterinary pathogens by the Clinical and Laboratory Science Institute95.
All the isolates were subjected to amplification of mecA and mecC genes using primers and conditions described previously96. All the isolates which showed mecA amplification were subjected to SCCmec typing using multiplex PCR for SCCmec types I to V as described previously97.
Further, minimum inhibitory concentration (MIC) of oxacillin was determined for all the isolates which showed either mecA amplification or resistance in disc diffusion, following the broth micro-dilution method recommended by CLSI95.
Detection of toxin genes
Presence of toxin genes (hlg, tsst and pvl) was confirmed by individual PCRs using DNA template (100 ng) and gene-specific primer pairs (0.5 μM of each) added to a PCR Master Mix (Takara Bio, Japan), and amplification was performed with the following cycle: 15 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 1 min at 53 °C, and 1 min at 72 °C, with a final 10 min elongation step at 72 °C. PCR products were analysed by 2% agarose gel electrophoresis and UV transillumination. The primers were F: GCCAATCCGTTATTAGAAAATGC and R: CCATAGACGTAGCCACGGAT for hlg, F: ACCCCTGTTCCCTTATCATC and R: TTTTCAGTATTTGTAACGCC for tsst, and F: GGCGCTGAGGTAGTCAAAAG and R: TCGGAATCTGATGTTGCAGT for pvl98,99.
Data availability
All data generated or analysed during this study are included in this article and its Supplementary Information files.
References
Alba, P. et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE 10, e0137143. https://doi.org/10.1371/journal.pone.0137143 (2015).
Fang, H. W., Chiang, P. H. & Huang, Y. C. Livestock-associated methicillin-resistant Staphylococcus aureus ST9 in pigs and related personnel in Taiwan. PLoS ONE 9, e88826. https://doi.org/10.1371/journal.pone.0088826 (2014).
Loncaric, I. et al. Carriage of meticillin-resistant staphylococci between humans and animals on a small farm. Vet. Dermatol. 27, 191-e148. https://doi.org/10.1111/vde.12321 (2016).
Momoh, A. H., Kwaga, J. K. P., Bello, M., Sackey, A. K. B. & Larsen, A. R. Antibiotic resistance and molecular characteristics of Staphylococcus aureus isolated from backyard-raised pigs and pig workers. Trop. Anim. Health Prod. 50, 1565–1571. https://doi.org/10.1007/s11250-018-1596-5 (2018).
Barkema, H. W., Schukken, Y. H. & Zadoks, R. N. Invited review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J. Dairy Sci. 89, 1877–1895. https://doi.org/10.3168/jds.S0022-0302(06)72256-1 (2006).
Gordon, R. J. & Lowy, F. D. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46(Suppl 5), S350-359. https://doi.org/10.1086/533591 (2008).
Turner, N. A. et al. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218. https://doi.org/10.1038/s41579-018-0147-4 (2019).
Krishnamoorthy, P., Goudar, A. L., Suresh, K. P. & Roy, P. Global and countrywide prevalence of subclinical and clinical mastitis in dairy cattle and buffaloes by systematic review and meta-analysis. Res. Vet. Sci. 136, 561–586. https://doi.org/10.1016/j.rvsc.2021.04.021 (2021).
Krishnamoorthy, P. et al. An understanding of the global status of major bacterial pathogens of milk concerning bovine mastitis: A systematic review and meta-analysis (scientometrics). Pathogens 10, 545. https://doi.org/10.3390/pathogens10050545 (2021).
Schmidt, T., Kock, M. M. & Ehlers, M. M. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 8, 511. https://doi.org/10.3389/fmicb.2017.00511 (2017).
Vrieling, M. et al. LukMF’ is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci. Rep. 6, 37759 (2016).
Wang, X. et al. Antimicrobial resistance and toxin gene profiles of Staphylococcus aureus strains from Hostein milk. Lett. Appl. Microbiol. 58, 527–534 (2014).
Cortimiglia, C. et al. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy Region (Northern Italy). Epidemiol. Infect. 144, 3046–3051. https://doi.org/10.1017/S0950268816001576 (2016).
De Oliveira, A. P., Watts, J. L., Salmon, S. A. & Aarestrup, F. M. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and the United States. J. Dairy Sci. 83, 855–862. https://doi.org/10.3168/jds.S0022-0302(00)74949-6 (2000).
Erskine, R. J., Walker, R. D., Bolin, C. A., Bartlett, P. C. & White, D. G. Trends in antibacterial susceptibility of mastitis pathogens during a seven-year period. J. Dairy Sci. 85, 1111–1118. https://doi.org/10.3168/jds.S0022-0302(02)74172-6 (2002).
Hata, E. Bovine mastitis outbreak in Japan caused by methicillin-resistant Staphylococcus aureus New York/Japan clone. J. Vet. Diagn. Invest. 28, 291–298. https://doi.org/10.1177/1040638716643126 (2016).
Huber, H., Koller, S., Giezendanner, N., Stephan, R. & Zweifel, C. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 15, 19542 (2010).
Makovec, J. A. & Ruegg, P. L. Antimicrobial resistance of bacteria isolated from dairy cow milk samples submitted for bacterial culture: 8,905 samples (1994–2001). J. Am. Vet. Med. Assoc. 222, 1582–1589. https://doi.org/10.2460/javma.2003.222.1582 (2003).
Matyi, S. A. et al. Isolation and characterization of Staphylococcus aureus strains from a Paso del Norte dairy. J. Dairy Sci. 96, 3535–3542. https://doi.org/10.3168/jds.2013-6590 (2013).
Moon, J. S. et al. Phenotypic and genetic antibiogram of methicillin-resistant staphylococci isolated from bovine mastitis in Korea. J. Dairy Sci. 90, 1176–1185. https://doi.org/10.3168/jds.S0022-0302(07)71604-1 (2007).
Oliveira, C. J. et al. Methicillin-resistant Staphylococcus aureus from Brazilian dairy farms and identification of novel sequence types. Zoonoses Public Health 63, 97–105. https://doi.org/10.1111/zph.12209 (2016).
Pu, W. et al. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS ONE 9, e88134. https://doi.org/10.1371/journal.pone.0088134 (2014).
Schlotter, K. et al. Multiple cases of methicillin-resistant CC130 Staphylococcus aureus harboring mecC in milk and swab samples from a Bavarian dairy herd. J. Dairy Sci. 97, 2782–2788. https://doi.org/10.3168/jds.2013-7378 (2014).
Spohr, M. et al. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 58, 252–261. https://doi.org/10.1111/j.1863-2378.2010.01344.x (2011).
Tenhagen, B. A. et al. Short communication: Methicillin-resistant Staphylococcus aureus in conventional and organic dairy herds in Germany. J. Dairy Sci. 101, 3380–3386. https://doi.org/10.3168/jds.2017-12939 (2018).
Virgin, J. E., Van Slyke, T. M., Lombard, J. E. & Zadoks, R. N. Short communication: Methicillin-resistant Staphylococcus aureus detection in US bulk tank milk. J. Dairy Sci. 92, 4988–4991. https://doi.org/10.3168/jds.2009-2290 (2009).
Yi, Y. et al. Analysis of the genetic diversity in methicillin-resistant Staphylococcus aureus isolates from bovine subclinical mastitis case in Xinjiang, China. Foodborne Pathog. Dis. 15, 568–575. https://doi.org/10.1089/fpd.2018.2424 (2018).
Smith, E. M., Green, L. E., Medley, G. F., Bird, H. E. & Dowson, C. G. Multilocus sequence typing of Staphylococcus aureus isolated from high-somatic-cell-count cows and the environment of an organic dairy farm in the United Kingdom. J. Clin. Microbiol. 43, 4731–4736. https://doi.org/10.1128/JCM.43.9.4731-4736.2005 (2005).
Mitra, S. D. et al. Staphylococcus aureus spa type t267, clonal ancestor of bovine subclinical mastitis in India. J. Appl. Microbiol. 114, 1604–1615. https://doi.org/10.1111/jam.12186 (2013).
Sheela, P. et al. Characterisation by SPA typing of Staphylococcus aureus isolates originating from bovine and bubaline mastitis in southern India. Indian J. Comp. Microbiol. Immunol. Infect. Dis. 40, 21–20 (2019).
Venugopal, N. et al. Molecular detection and typing of methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci isolated from cattle, animal handlers, and their environment from Karnataka, Southern Province of India. Vet. World 12, 1760–1768. https://doi.org/10.14202/vetworld.2019.1760-1768 (2019).
Mahanti, A. et al. Characterization of methicillin-resistant and enterotoxins producing Staphylococcus aureus in bovine milk in India. J. Agric. Food Res. 2, 100017 (2020).
Gomes, F. & Henriques, M. Control of bovine mastitis: Old and recent therapeutic approaches. Curr. Microbiol. 72, 377–382. https://doi.org/10.1007/s00284-015-0958-8 (2016).
NAAS. Mastitis management in dairy animals. Report No. Policy paper No. 61, 12 (National Academy of Agricultural Sciences, 2013).
Bansal, B. K. & Gupta, D. K. Economic analysis of bovine mastitis in India and Punjab—Review. Indian J. Dairy Sci. 62, 337–345 (2009).
Christy, R. J. Estimation of direct economic loss due to clinical mastitis in Villupuram district of Tamil Nadu. Int. J. Adv. Dr. Res. 3, 22–24 (2014).
Jingar, S. et al. Economic loss due to clinical mastitis in cross-bred cows. Int. J. Livest. Res. 8, 254–263 (2018).
Sharma, N. et al. Bovine mastitis: An Asian perspective. Asian J. Anim. Vet. Adv. 7, 454–476 (2012).
Tamizhkumaran, J., Sudeepkumar, N. K. & Thilakar, P. Incidence and economics of mastitis in Tamil Nadu. Asian J. Res. Anim. Vet. Sci. 3, 1–4 (2019).
Monistero, V. et al. Staphylococcus aureus isolates from bovine mastitis in eight countries: Genotypes, detection of genes encoding different toxins and other virulence genes. Toxins 10, 247. https://doi.org/10.3390/toxins10060247 (2018).
Olde Riekerink, R. G., Barkema, H. W., Scholl, D. T., Poole, D. E. & Kelton, D. F. Management practices associated with the bulk-milk prevalence of Staphylococcus aureus in Canadian dairy farms. Prev. Vet. Med. 97, 20–28. https://doi.org/10.1016/j.prevetmed.2010.07.002 (2010).
Sudhan, N. A., Singh, R., Singh, M. & Soodan, J. S. Studies on prevalence, etiology and diagnosis of subclinical mastitis among cross-bred cows. Indian J. Anim. Res. 39, 127–130 (2005).
Budd, K. E. et al. Extensive genomic diversity among bovine-adapted Staphylococcus aureus: Evidence for a genomic rearrangement within CC97. PLoS ONE 10, e0134592. https://doi.org/10.1371/journal.pone.0134592 (2015).
Aires-de-Sousa, M. et al. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine and caprine milk collected in Rio de Janeiro state. Appl. Environ. Microbiol. 73, 3845–3849 (2007).
Hata, E. et al. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Clin. Microbiol. 48, 2130–2139. https://doi.org/10.1128/JCM.01940-09 (2010).
Hwang, S. Y., Park, Y. K., Koo, H. C. & Park, Y. H. spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea. J. Vet. Sci. 11, 125–131. https://doi.org/10.4142/jvs.2010.11.2.125 (2010).
Said, K. B. et al. Regional profiling for determination of genotype diversity of mastitis-specific Staphylococcus aureus lineage in Canada by use of clumping factor A, pulsed-field gel electrophoresis, and spa typing. J. Clin. Microbiol. 48, 375–386. https://doi.org/10.1128/JCM.01768-09 (2010).
Song, M. et al. Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 195, 1–8. https://doi.org/10.1016/j.ijfoodmicro.2014.11.020 (2015).
Chandrasekaran, D. et al. Pattern of antibiotic resistanc mastitis in dairy cows. Vet. World 7, 389–394 (2014).
Gopal, S. & Divya, K. C. Can methicillin-resistant Staphylococcus aureus prevalence from dairy cows in India act as potential risk for community-associated infections?: A review. Vet. World 10, 311–318. https://doi.org/10.14202/vetworld.2017.311-318 (2017).
Kumar, R., Yadav, B. R. & Singh, R. S. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. J. Biosci. 36, 175–188. https://doi.org/10.1007/s12038-011-9004-6 (2011).
Kutar, K., Verma, A. K., Sharma, B., Kumar, A. & Yadav, S. K. Analysis of mecA gene and antibiotic resistance in Staphylococcus aureus isolates from bovine mastitis. Indian J. Comp. Microbiol. Immunol. Infect. Dis. 36, 22–27 (2015).
Chambers, H. F. Methicillin resistance in staphylococci: Molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10, 781–791. https://doi.org/10.1128/CMR.10.4.781-791.1997 (1997).
Kim, C. et al. The mechanism of heterogeneous beta-lactam resistance in MRSA: Key role of the stringent stress response. PLoS ONE 8, e82814. https://doi.org/10.1371/journal.pone.0082814 (2013).
Madiraju, M. V., Brunner, D. P. & Wilkinson, B. J. Effects of temperature, NaCl, and methicillin on penicillin-binding proteins, growth, peptidoglycan synthesis, and autolysis in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 31, 1727–1733. https://doi.org/10.1128/aac.31.11.1727 (1987).
Uddin, M. J. & Ahn, J. Associations between resistance phenotype and gene expression in response to serial exposure to oxacillin and ciprofloxacin in Staphylococcus aureus. Lett. Appl. Microbiol. 65, 462–468. https://doi.org/10.1111/lam.12808 (2017).
Cuny, C., Wieler, L. H. & Witte, W. Livestock-associated MRSA: The impact on humans. Antibiotics (Basel) 4, 521–543. https://doi.org/10.3390/antibiotics4040521 (2015).
Fessler, A. et al. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65, 619–625. https://doi.org/10.1093/jac/dkq021 (2010).
Vanderhaeghen, W. et al. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol. 144, 166–171. https://doi.org/10.1016/j.vetmic.2009.12.044 (2010).
Baba, K., Ishihara, K., Ozawa, M., Tamura, Y. & Asai, T. Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int. J. Antimicrob. Agents 36, 352–354. https://doi.org/10.1016/j.ijantimicag.2010.06.040 (2010).
Larsen, J. et al. Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS ONE 7, e31245. https://doi.org/10.1371/journal.pone.0031245 (2012).
Li, T. et al. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front. Cell Infect. Microbiol. 7, 127. https://doi.org/10.3389/fcimb.2017.00127 (2017).
Neela, V. et al. Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47, 4138–4140. https://doi.org/10.1128/JCM.01363-09 (2009).
Chuang, Y. Y. & Huang, Y. C. Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: An emerging issue?. Int. J. Antimicrob. Agents 45, 334–340. https://doi.org/10.1016/j.ijantimicag.2014.12.007 (2015).
DastmalchiSaei, H. & Panahi, M. Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from dairy ruminants: Differences in the distribution of clonal types between cattle and small ruminants. Arch. Microbiol. 202, 115–125. https://doi.org/10.1007/s00203-019-01722-z (2020).
Garcia-Alvarez, L. et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 11, 595–603. https://doi.org/10.1016/S1473-3099(11)70126-8 (2011).
Zoppi, S. et al. Livestock-associated methicillin-resistant Staphylococcus aureus and related risk factors in holdings of veal calves in Northwest Italy. Microb. Drug Resist. https://doi.org/10.1089/mdr.2020.0226 (2021).
Feltrin, F. et al. A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl. Environ. Microbiol. 82, 816–821. https://doi.org/10.1128/AEM.02854-15 (2016).
El-Ashker, M. et al. Antimicrobial resistance pattern and virulence profile of S. aureus isolated from household cattle and buffalo with mastitis in Egypt. Vet. Microbiol. 240, 108535. https://doi.org/10.1016/j.vetmic.2019.108535 (2020).
El-Ashker, M. et al. Staphylococci in cattle and buffaloes with mastitis in Dakahlia Governorate, Egypt. J. Dairy Sci. 98, 7450–7459. https://doi.org/10.3168/jds.2015-9432 (2015).
Hasman, H. et al. Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 141, 326–331. https://doi.org/10.1016/j.vetmic.2009.09.025 (2010).
Luini, M. et al. Methicillin-resistant Staphylococcus aureus (MRSA) is associated with low within-herd prevalence of intra-mammary infections in dairy cows: Genotyping of isolates. Vet. Microbiol. 178, 270–274. https://doi.org/10.1016/j.vetmic.2015.05.010 (2015).
Rabello, R. F. et al. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 56, 1505–1511. https://doi.org/10.1099/jmm.0.47357-0 (2007).
Sarrou, S. et al. Dissemination of methicillin-susceptible CC398 Staphylococcus aureus strains in a rural Greek area. PLoS ONE 10, e0122761. https://doi.org/10.1371/journal.pone.0122761 (2015).
Sheet, O. H., Grabowski, N. T., Klein, G., Reich, F. & Abdulmawjood, A. Characterisation of mecA gene negative Staphylococcus aureus isolated from bovine mastitis milk from Northern Germany. Folia Microbiol. (Praha) 64, 845–855. https://doi.org/10.1007/s12223-019-00698-z (2019).
Smith, E. M. et al. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43, 4737–4743. https://doi.org/10.1128/JCM.43.9.4737-4743.2005 (2005).
Vandendriessche, S. et al. High genetic diversity of methicillin-susceptible Staphylococcus aureus (MSSA) from humans and animals on livestock farms and presence of SCCmec remnant DNA in MSSA CC398. J. Antimicrob. Chemother. 69, 355–362. https://doi.org/10.1093/jac/dkt366 (2014).
Asai, T. et al. Presence of Staphylococcus aureus ST398 and ST9 in Swine in Japan. Jpn. J. Infect. Dis. 65, 551–552. https://doi.org/10.7883/yoken.65.551 (2012).
Yang, X. et al. Multilocus sequence typing and virulence-associated gene profile analysis of Staphylococcus aureus isolates from retail ready-to-eat food in China. Front. Microbiol. 9, 197. https://doi.org/10.3389/fmicb.2018.00197 (2018).
Dan, M. et al. Antimicrobial resistance, virulence gene profile and molecular typing of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, northwest China. J. Glob. Antimicrob. Resist. 16, 98–104. https://doi.org/10.1016/j.jgar.2018.08.024 (2019).
Badua, A. T., Boonyayatra, S., Awaiwanont, N., Gaban, P. B. V. & Mingala, C. N. Antibiotic resistance and genotyping of mecA-positive methicillin-resistant Staphylococcus aureus (MRSA) from milk and nasal carriage of dairy water buffaloes (Bubalus bubalis) in the Philippines. J. Adv. Vet. Anim. Res. 7, 397–406. https://doi.org/10.5455/javar.2020.g434 (2020).
Guo, D., Liu, Y., Han, C., Chen, Z. & Ye, X. Phenotypic and molecular characteristics of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolated from pigs: Implication for livestock-association markers and vaccine strategies. Infect. Drug Resist. 11, 1299–1307. https://doi.org/10.2147/IDR.S173624 (2018).
Havaei, S. A. et al. Detection of mecA and enterotoxin genes in Staphylococcus aureus isolates associated with bovine mastitis and characterization of Staphylococcal cassette chromosome mec (SCCmec) in MRSA strains. Iran J. Microbiol. 7, 161–167 (2015).
Khaji, L. & Shahreza, M. H. S. SCCmec types in methicillin-resistant Staphylococcus aureus strains of various types of milk. Electron. J. Biol. 13, 1 (2016).
Suleiman, A. B., Umoh, V. J., Kwaga, J. K. P. & Shaibu, S. J. Prevalence and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) isolated from bovine mastitis milk in Plateau State, Nigeria. Intern. Res. J. Microbiol. 2, 264–270 (2012).
Kamberovic, F. et al. mecA-positive methicillin-sensitive Staphylococcus aureus clinical isolates in Zenica-Doboj Canton, Bosnia and Herzegovina. J. Chemother. 27, 330–336. https://doi.org/10.1179/1973947814Y.0000000207 (2015).
Mistry, H. et al. Prevalence and characterization of oxacillin susceptible mecA-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in India. PLoS ONE 11, e0162256. https://doi.org/10.1371/journal.pone.0162256 (2016).
Saeed, K. et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection 42, 843–847. https://doi.org/10.1007/s15010-014-0641-1 (2014).
Saeed, K., Dryden, M. & Parnaby, R. Oxacillin-susceptible MRSA, the emerging MRSA clone in the UK?. J. Hosp. Infect. 76, 267–268. https://doi.org/10.1016/j.jhin.2010.03.004 (2010).
Lange, C., Cardoso, M., Senczek, D. & Schwarz, S. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 67, 127–141. https://doi.org/10.1016/s0378-1135(99)00031-0 (1999).
Brakstad, O. G., Aasbakk, K. & Maeland, J. A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30, 1654–1660. https://doi.org/10.1128/JCM.30.7.1654-1660.1992 (1992).
Shopsin, B. et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37, 3556–3563. https://doi.org/10.1128/JCM.37.11.3556-3563.1999 (1999).
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J. & Spratt, B. G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015 (2000).
Bauer, A. W., Kirby, W. M., Sherris, J. C. & Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496 (1966).
CLSI. 156 (Clinical and Laboratory Standards Institute, 2018).
Stegger, M. et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect. 18, 395–400. https://doi.org/10.1111/j.1469-0691.2011.03715.x (2012).
McClure-Warnier, J. A., Conly, J. M. & Zhang, K. Multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Vis. Exp. https://doi.org/10.3791/50779 (2013).
Lina, G. et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132. https://doi.org/10.1086/313461 (1999).
Mehrotra, M., Wang, G. & Johnson, W. M. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38, 1032–1035. https://doi.org/10.1128/JCM.38.3.1032-1035.2000 (2000).
Acknowledgements
This work was supported through a grant (BT/PR11245/ADV/90/165/2014) to NRH and SI, and a Research Associate Fellowship to MA, from the Department of Biotechnology, Ministry of Science and Technology, Government of India. The authors are thankful to all the researchers who generously donated isolates.
Author information
Authors and Affiliations
Contributions
The work was ideated by NRH and SI. Re-confirmation of the micro-organisms and curation was done by MA, PS and SS. Laboratory work and data acquisition was carried out by MA, PS, SS, PG, PJ, MG and SM. Data was analysed by MA, PS, SI and NRH. MA and NRH wrote and revised the manuscript. All the authors reviewed the submitted versions of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Annamanedi, M., Sheela, P., Sundareshan, S. et al. Molecular fingerprinting of bovine mastitis-associated Staphylococcus aureus isolates from India. Sci Rep 11, 15228 (2021). https://doi.org/10.1038/s41598-021-94760-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94760-x
This article is cited by
-
Isolation and Characterization of Staphylococcus aureus in Bovine Milk from Rewa, India
Indian Journal of Microbiology (2024)
-
Genome sequencing and comparative genomic analysis of bovine mastitis-associated Staphylococcus aureus strains from India
BMC Genomics (2023)
-
Effects of the natural antimicrobial peptide aureocin A53 on cells of Staphylococcus aureus and Streptococcus agalactiae involved in bovine mastitis in the excised teat model
World Journal of Microbiology and Biotechnology (2023)
-
Prevalence and molecular characteristics of Staphylococcus aureus in raw milk and milk products in Ethiopia: a systematic review and meta-analysis
International Journal of Food Contamination (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.