Abstract
Chilo suppressalis (Walker, 1863) is a serious stem borer of rice and water-oat plants, and has phenotypically diverged into rice and water-oat populations. Insect gut microbiota plays an important role in the host life and understanding the dynamics of this complicated ecosystem may improve its biological control. The effect of diet and gut compartments on the gut microflora of divergent populations of C. suppressalis is not fully clear. Herein, we characterized the gut microbiota of C. suppressalis populations fed on two hosts (i.e., water-oats fruit pulps and rice seedlings), by sequencing the V3–V4 hypervariable region of the 16S rRNA gene using the Illumina MiSeq platform. Gut bacterial communities showed variation in relative abundance among C. suppressalis populations fed on water-oats fruit pulps or rice seedlings. Proteobacteria and Firmicutes became the predominant phyla, and Enterobacteriaceae, Enterococcaceae and Halomonadaceae were the predominant family in all C. suppressalis populations. The highest bacteria diversity was found in the midgut of the rice population fed on water-oat fruit pulps. Bacterial communities in the midgut were more diverse than those in the hindgut. The bacterial genera distribution showed great differences due to diet types and gut compartments among populations. Our results demonstrated that the host plants tested had a considerable impact on gut bacterial composition of C. suppressalis populations. Additionly, the unique gut morphology and physiological conditions (viz., oxygen content, enzymes) also contributed to variation in microbiomes. In conclusion, our study provided an important insight into investigation of insect-bacteria symbioses, and biocontrol of this species and other related lepidopterans.
Similar content being viewed by others
Introduction
Chilo suppressalis (Walker, 1863) is one of the destructive generalists of rice in Asia, southern Europe, and northern Africa1,2,3. The larvae bore into the rice stems and feed on tender tissues, resulting in ‘‘dead heart’’, ‘‘white heads’’ and ‘‘dead sheath’’ of the infested plants4. Thus severe yield and economic losses are caused per year, particularly in China due to the large cultivation of rice3,4,5,6,7,8. In recent years, an aquatic vegetable ‘water-oat’ is winning a warm praise from grower and customers, because of its economic and nutritional benefit9. It is grown year-round, facilitated by global warming, variety alternative, cropping system, and expansion of the greenhouse, thus increase the damage of this pest species. The intercropping pattern (rice is planted in a mosaic fashion under a crop rotation system with the water-oat) facilitates a transfer of C. suppressalis from rice plant to water-oat plant. Both the rice and water-oat belong to the tribe Oryzeae (family Gramineae) and possess the similar habitat, biology and ecology, but their nutrients and allelochemicals are markedly different9,10,11,12,13. Although C. suppressalis can complete their life cycles on rice or water-oat, those feeding on water-oat fruit pulps possess higher survival rate, pupal weight and shorter developmental duration than those feeding on rice10,11,14,15,16,17. After a long period of adaptation, C. suppressalis has diverged phenotypically into rice population and water-oat population11,16,18,19,20,21. The divergent populations exhibit significant phenotypic differences in morphology10,15,16, behaviors11,19,20,22,23, biochemical and physiological indexes6,11,19,20,22,23,24, emergence peak14,15,25, genetic differences19,26, ecological and biological characters10,14,17,20,22,27. Despite these variations were revealed after host shift, the mechanisms underlying host plant adaptation remains unclear.
The insects’ gut is a tube opening from the mouth to the anus, and is divided into three distinct regions, the foregut, midgut and hindgut. Food is usually stored in the foregut; useful materials (nutrients) are absorbed in the midgut, and partial nutrients and water are reabsorbed in the hindgut. The gut is a desirable, nutrient-rich ecological niche where multiple microbial taxa flourish and reproduce. The anterior hindgut is the most densely symbiont-inhabited site, due to the available, partially digested food being from the midgut, and the excretions from the Malpighian tubules28. The microbial taxa perform nutritional role to their hosts by providing nutrients lacking in hosts’ diets. Additionly, some species of the bacteria contribute to the other various functions, including immune, development, survival, reproduction, detoxification29,30,31,32,33,34,35,36 and population differentiation37. Therefore, they can help the insects adapt to host plants successfully. This is especially obvious in phytophagous insect, because of many secondary materials existed in host plants.
Over a long period of coevolution, a symbiosis has been formed between gut bacteria and their insect hosts. Gut bacteria exhibit some plasticity, and their communities alter with the change of insect diet38,39. Such an adaptiveness supplies a solid foundation for the development of host-associated differentiation40. Further, it has demonstrated that the population divergence is associated with microorganisms, and the symbiotic bacteria are important factors that promote evolution in the genus Nasonia37 and fruit fly Drosophila melanogaster41. Microbial community associated with insects feeding on alternative host plants were commonly reported in other lepidopterans38,40,42,43,44,45,46. However, the dynamics interactions between C. suppressalis populations and their gut microbiota are far from being understood, excepting for association of insecticides and gut bacteria47,48. Thus, a complete characterization of host-associated variation in bacteria composition is indispensable for an overall understanding of insect-bacteria symbioses of this species, and for the development of novel biocontrol management strategies. Herein, data from 16S rRNA next-generation amplicon sequencing were used, with the aiming to characterize the gut microbial communities of phenotypically divergent populations of larval C. suppressalis, as well as to explore ecological questions related to the host and gut compartment.
Methods
Specimen collection and rearing
In order to obtain representative populations of water-oat and rice, larvae of C. suppressalis were collected from the water-oat field in Lishui and the rice field in Yuyao, Zhejiang, China in 2018, where rice TN1 and water-oat Zizania latifolia are exclusively planted. During the experiment in our laboratory, rice-seedlings and water-oat pulps were collected from our institutional experimental field. The use of plant parts in the present study complies with institutional guidelines. The C. suppressalis larvae from the water-oat and rice fields were reared respectively with fresh water-oat fruit pulps and rice seedlings. C. suppressalis larvae were kept in an insectarium at 28 ± 1 °C, with a photoperiod of 16 h: 8 h (light/dark), and a relative humidity > 80%. All the larvae were maintained in the laboratory for three generations before dissection.
We analyzed the 16S rRNA gene to estimate the gut bacterial composition in the midgut and hindgut of larvae feeding on water-oat fruit pulps and rice seedlings. Treatments, abbreviations, and locations of the samples were provided in Table 1. C. suppressalis fed with their original hosts were named as original populations, and those fed with non-original hosts were named as cross-rearing populations.
Experimental design
Schematic diagram of the fully factorial experimental design used in the present study. C. suppressalis from water-oat field were respectively reared on water-oat fruit pulps (J) and rice seedlings (j); and those from rice field were respectively reared on rice seedlings (R) and water-oat fruit pulps (r). All the groups were reared for three continuous generations before examining the effects of host plant, population origin and gut compartment on the gut microbial communities (Fig. 1).
Schematic diagram of the fully factorial experimental design used in the present study. Chilo suppressalis from water-oat field and rice field were reared on water-oats fruit pulps or rice seedlings for three continuous generations to examine the effects of host plant and population origin on the gut microbiota. There were two cross-rearing populations (i.e., non-original populations) comprised of water-oat population individuals reared on rice seedlings (j) and rice population individuals reared on water-oat fruit pulps (r). The corresponding original populations were water-oat population individuals reared on water-oat fruit pulps (J) and rice population individuals reared on rice seedlings (R).
C. suppressalis dissection and gut sample collection
Healthy, uniformly developed individuals of the same batch of C. suppressalis were collected. Each individual was anesthetized by placed on ice and externally sterilized with 75% ethanol and rinsed 3 times with sterilized water. The gut were dissected out with a sterilized fine-tip forcep and washed twice with sterile 0.9% NaCl solution quickly. The midgut and hindgut were carefully separated and placed in different sterile microcentrifuge tubes, synchronously. Midguts and hindguts of 50 individuals of each population were collected as one sample, and three samples were taken for each population. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C for DNA isolation.
DNA isolation, 16S rDNA amplification
Total bacterial genomic DNA was extracted from eight sets of sample groups using a Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s instructions. The DNA was finally eluted with TE buffer (Tris–EDTA buffer). DNA purity and concentration were measured using the NanoDrop 2000 spectrophotometer (Nano-drop Technologies, Wilmington, DE, USA). The total DNA was stored at − 70 °C until use.
The bacterial 16S rRNA variable V3−V4 regions were used to identify bacteria. Two universal primers (341F and 806R) containing the specific barcode sequence were used for the amplification of the V3−V4 regions (341F: 5’-CCTAYGGGRBGCASCAG-3’, 806R: 5’-GGACTACNNGGGTATCTAAT-3’). The Polymerase Chain Reaction (PCR) reaction was performed in triplicate 20.0 µL mixture containing 4.0 μL 5 × FastPfu Buffer, 2.0 μL 2.5 mM dNTPs, 0.8 μL of each Primer (5.0 μM), 0.4 μL FastPfu Polymerase, and 10 ng of template DNA. The amplification procedure was as follows: 95 °C for 2 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s and a final extension at 72 °C for 5 min.
Illumina MiSeq sequencing
Amplicons were extracted from 2% agarose gels and purified using a AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) following the manufacturer’s protocols and quantified using QuantiFluor™-ST (Promega, U.S.). Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) on an Illumina Novaseq6000 platform according to the standard instructions.
Processing of sequencing data
Raw fastq files were demultiplexed, quality-filtered using QIIME (version 1.17) with the following criteria: (i) The 250 bp reads were truncated at any site receiving an average quality score < 20 over a 10 bp sliding window, discarding the truncated reads that were shorter than 50 bp; (ii) exact barcode matching, 2 nucleotide mismatch in primer matching, reads containing ambiguous characters were removed; (iii) only sequences that overlap longer than 10 bp were assembled according to their overlap sequence. Reads which could not be assembled were discarded.
Operational Units (OTUs) were clustered using UPARSE (version 7.1 http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The phylogenetic affiliation of each 16S rRNA gene sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the silva (SSU115) 16S rRNA database.
Results
General structure of gut
The gut of C. suppressalis was a continuous tube running from the mouth to the anus. It was structurally divided into foregut, midgut and hindgut. The foregut (Fg) was a slender, elongate tube, expanding posteriorly and constricting at its ends. The midgut (Mg) was a well-developed saclike tube beginning from the end of the foregut and extending to the long, narrow hindgut (Hg). The freshly dissected foregut was translucent, the midgut was opaque white, and the hindgut was yellowish-brown (Fig. 2).
Analysis of bacterial 16S rDNA gene sequences
Illumina sequencing obtained 861,370 sequences clustering into 3234 operational taxonomic units (OTUs) (Table 2). Chao1 estimator and Shannon Index were calculated for the determination of the richness and homogeneity of the community. The relative bacterial abundance of 18 phyla differed significantly across the eight samples (Kruskal–Wallis test, p < 0.0001). The midgut and hindgut of the rice population feeding on water-oat fruit pulps (rMG, rHG), possessed the highest bacteria diversity. The bacteria in the midgut were more diverse than those in the hindgut (Table 2).
Microbial diversity of gut microbiota
A total of 49 and 62 OTUs were observed in the midguts and hindguts, respectively (Fig. 3A,B). The core OTUs identified belonged to the phyla Proteobacteria, Firmicutes, Actinobacteria, Saccharibacteria, and Bacteroidetes (S1 Fig). The OTUs detected from the midgut were grouped into 31 families, of which five families were abundant: Enterobacteriaceae (24.6%), Halomonadaceae (20.2%), Enterococcaceae (31.4%), Bacillaceae (11.4%), and Streptococcaceae (6.9%) (S1A Table; S2 Table and S3 Table). However, the OTUs coming from the hindgut were grouped into 28 families. The predominent families were Enterobacteriaceae (66.4%), Enterococcaceae (11.2%), Bacillaceae (5.0%), Streptococcaceae (3.0%), Xanthomonadaceae (2.2%) and Flavobacteriaceae (1.7%) (S1B Table; S2 Table and S3 Table). The rMG and rHG had the maximum number of unique OTUs, whereas the midgut and hindgut of the water-oat population feeding on rice seedlings (jMG, jHG) possessed the minimum number.
Venn diagram of OTUs from unique species owned by each sample and common species shared by two or more samples. (A) Midgut samples of cross-rearing populations and original populations. (B) Hindgut samples of cross-rearing populations and original populations. (C) Midgut and hindgut samples of original populations. (D) Midgut and hindgut samples of cross-rearing populations. JMG midguts of water-oat population feeding on water-oat fruit pulps, RMG midguts of rice population feeding on rice seedlings, jMG midguts of water-oat population feeding on rice seedlings, rMG midguts of rice population feeding on water-oat fruit pulps, JHG hindguts of water-oat population feeding on water-oat fruit pulps, RHG hindguts of rice population feeding on rice seedlings, jHG hindguts of water-oat population feeding on rice seedlings, rHG hindguts of rice population feeding on water-oat fruit pulps.
A total of 44 and 66 OTUs were observed in the guts of original and cross-rearing populations, separately (Fig. 3C,D). These OTUs were pooled into 26 families for the midgut of original populations. The relative abundances of five families were Enterobacteriaceae, Halomonadaceae, Bacillaceae, Enterococcaceae and Streptococcaceae (S1C Table; S2 Table and S3 Table). However, in the hindgut of cross-rearing populations, the OTUs were grouped to 35 families, of which the abundant ones were Enterobacteriaceae, Enterococcaceae, Streptococcaceae, Xanthomonadaceae and Halomonadaceae (S1D Table; S2 Table and S3 Table).
Taxonomic distribution of gut bacteria
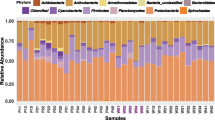
Taxonomic classification yielded 122 families belonging to 18 bacterial phyla (Fig. 4; S3 Table and S4 Table), and the predominant phyla were Proteobacteria (16.0–96.4%), followed by Firmicutes (2.3–78.9%). At the family level, Enterobacteriaceae (8.0–78%) was the most predominant taxa, followed by Enterococcaceae (1.7–64.2%) and Halomonadaceae (0.3–69.8%) (S3 Table). The family Bacillaceae was only found in the water-oat population and two cross-rearing populations, although it was low relative abundance (Fig. 4; S3 Table). It was enriched in the midgut of the water-oat population (JMG ) (17.9–33.1%), followed by the midgut of the rice population feeding on water-oat fruit pulps (rMG) (17.0–26.8%) and the hindgut of the water-oat population (JHG) (4.6–15.1%). They exhibited a high variation of relative abundance associated with diet and gut compartment, though the most abundant taxa were identified in the midgut and hindgut of all populations.
Compositions of gut microbiota at the family level of original and cross-rearing populations of C. suppressalis. The Y-axis represents the proportion of each taxon. JMG1–JMG3 midguts of water-oat population feeding on water-oat fruit pulps, RMG1–RMG2 midguts of rice population feeding on rice seedlings, jMG1–jMG3 midguts of water-oat population feeding on rice seedlings, rMG1–rMG3 midguts of rice population feeding on water-oat fruit pulps, JHG1–JHG3 hindguts of water-oat population feeding on water-oat fruit pulps, RHG1–RHG3 hindguts of rice population feeding on rice seedlings, jHG1–jHG3 hindguts of water-oat population feeding on rice seedlings, rHG1–rHG3 hindguts of rice population feeding on water-oat fruit pulps; Original populations: C. suppressalis collected from water-oat field and reared on water-oat fruit pulps; or C. suppressalis collected from rice field and reared on rice seedlings. Cross-rearing populations: C. suppressalis collected from water-oat field but reared on rice seedlings; or C. suppressalis collected from rice field but reared on water-oat fruit pulps. Abbreviations for each sample are explained in Tables 1 and 2.
Regardless of diet, a more homogeneous phylum distribution was found in the hindguts of all original populations and cross-rearing populations (i.e., hindguts of the water-oat population feeding on water-oat fruit pulps (JHG) and rice seedlings (jHG), hindguts of the rice population feeding on rice seedlings (RHG) and water-oat fruit pulps (rHG)): Proteobacteria (71.5–80.9%), Firmicutes (9.0–27.7%), Bacteroidetes (0.1–8.8%), Actinobacteria (0.1–2.8%) and Saccharibacteria (0.6%), respectively (Fig. 4; S1 Figure; S3 Table and S4 Table). However, community of the Firmicutes and Proteobacteria was changed in the midgut of the water-oat population feeding on water-oat fruit pulps (JMG) (40.3–71.8%, 27.3–58.6%), midgut of the water-oat population feeding on rice seedlings (jMG) (50.6–82.0%, 17.8–49.0%) and midgut of the rice population feeding on water-oat fruit pulps (rMG) (72.1–87.3%, 10.5–24.8%). Four bacterial phyla in the midgut of the rice population feeding on rice seedlings (RMG) were more homogeneous in richness: Proteobacteria (96.2–96.6%), Firmicutes (2.2–2.3%), Bacteroidetes (0.6–0.9%) and Actinobacteria (0.3–0.5%).
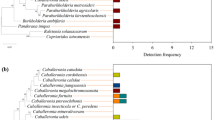
The bacterial genera from original populations showed distinct distribution according to diet types and gut compartments (Fig. 5; S5 Table). Halomonas (69.9%) and Klebsiella (70.1%) were dominant in the midgut and hindgut of the rice population feeding on rice seedlings (RMG and RHG); but Bacillus (26.9%) and Klebsiella (35.14%) were prevailed in the midgut of the water-oat population feeding on water-oat fruit pulps (JMG), Citrobacter (40.8%) was enriched in the hindgut of the water-oat population feeding on water-oat fruit pulps (JHG). Enterococcus was dominant in the midguts of the two cross-rearing populations (jMG (64.8%) and rMG (45.9%)), and Citrobacter was prevailed in the hindguts of the two cross-rearing populations (jHG (43.7%) and rHG (37.1%)). However, the bacteria in cross-rearing populations showed different genus distributions based on diet types. The Klebsiella (27.6%) and Bacillus (18.7%) were the relative dominance in jMG and rMG, whereas the Enterococcus (18.9%, 6.7%) and Klebsiella (20.4%, 11.1%) were relatively prevalent in jHG and rHG.
Compositions of gut microbiota at the genus level of original and cross-rearing populations of C. suppressalis. The composition of each sample was based on the taxonomic assignment of the 16S rDNA sequences. The Y-axis represented the proportion of each taxon. JMG1–JMG3 midguts of the water-oat population feeding on water-oat fruit pulps, RMG1–RMG2 midguts of rice population feeding on rice seedlings, jMG1–jMG3 midguts of the water-oat population feeding on rice seedlings, rMG1–rMG3 midguts of rice population feeding on water-oat fruit pulps, JHG1–JHG3 hindguts of the water-oat population feeding on water-oat fruit pulps, RHG1–RHG3 hindguts of rice population feeding on rice seedlings, jHG1–jHG3 hindguts of the water-oat population feeding on rice seedlings, rHG1–rHG3 hindguts of rice population feeding on water-oat fruit pulps; Original populations: C. suppressalis collected from water-oat field and reared on water-oat fruit pulps; or C. suppressalis collected from rice field and reared on rice seedlings. Cross-rearing populations: C. suppressalis collected from water-oat field but reared on rice seedlings; or C. suppressalis collected from rice field but reared on water-oat fruit pulps. Abbreviations for each sample are explained in Tables 1 and 2.
Diet- and compartment-related variations in the gut microbial composition
In all populations, there were significant differences in the relative abundances at the family level (p < 0.0001, Kruskal–Wallis test). 95 bacterial taxa were identified at the genus level. Influence of compartment sampling proved significant with a well-defined cluster formed by the midguts of all original and cross-rearing populations (i.e., JMG, jMG, rMG and RMG). By contrast, bacteria from the hindguts of all populations (i.e., RHG, JHG, jHG, rHG) were more heterogeneous for constituting four different clusters (Fig. 6). All the midguts and hindguts exhibited a significant difference in bacteria abundance of three families: Enterobacteriaceae, Enterococcaceae and Bacillaceae. Enterobacteriaceae was dominant in the hindgut (66.4%), but was decreased to 24.6% in the midgut. In comparison, Enterococcaceae was less abundant in the hindgut (11.2%), whereas increased to 31.4% in the midgut; Bacillaceae (5.0%) resided in the hindgut was increased to 11.4% in the midgut (Fig. 6; Table S3).
Abundance and composition of gut microbiota of all populations. Heatmap represent the proportions of OTUs at the family level. The X-coordinate represents the sample of each population, and the Y-coordinate represents the taxon. The color code indicates relative abundance, ranging from blue (low abundance) to yellow to red (high abundance). JMG1–JMG3 midguts of the water-oat population feeding on water-oat fruit pulps, RMG1–RMG2 midguts of rice population feeding on rice seedlings, jMG1–jMG3 midguts of the water-oat population feeding on rice seedlings, rMG1–rMG3 midguts of rice population feeding on water-oat fruit pulps, JHG1–JHG3 hindguts of the water-oat population feeding on water-oat fruit pulps, RHG1–RHG3 hindguts of rice population feeding on rice seedlings, jHG1–jHG3 hindguts of the water-oat population feeding on rice seedlings, rHG1–rHG3 hindguts of rice population feeding on water-oat fruit pulps; Original populations: C. suppressalis collected from water-oat field and reared on water-oat fruit pulps; or C. suppressalis collected from rice field and reared on rice seedlings. Cross-rearing populations: C. suppressalis collected from water-oat field but reared on rice seedlings; or C. suppressalis collected from rice field but reared on water-oat fruit pulps.
The differences at the family level were (Fig. 6): (1) a higher abundance of Enterobacteriaceae in the hindguts (55.8%) than in the midguts of the rice population feeding on water-oat fruit pulps (8.6%) and the water-oat population feeding on water-oat fruit pulps (35.9%); (2) a higher presence of Enterococcaceae in the midgut of the water-oat population feeding on rice seedlings (64.8%) than in the midgut and hindgut of the water-oat population feeding on water-oat fruit pulps (12.4%%, (10.0%), midgut of the rice population feeding on water-oat fruit pulps (45.9%) and the hindgut of the water-oat population feeding on rice seedlings (18.9%); (3) and a higher presence of Halomonadaceae in the midguts of two original populations (RMG: 69.9%; JMG: 4.9%) and the rice population feeding on water-oat fruit pulps (6.0%), hindguts of the water-oat population feeding on water-oat fruit pulps and rice seedlings (3.4%, 0.3%). However, the Bacillaceae was higher in the midgut and hindgut of the water-oat population feeding on water-oat fruit pulps (26.9%, 11.3%) and midgut of the rice population feeding on water-oat fruit pulps (18.6%), than that in the midgut of the water-oat population feeding on rice seedlings (0.6%) and hindgut of the rice population feeding on water-oat fruit pulps (1.2%).
A non-metric multidimensional scaling (NMDS) analysis was performed to analyze the influence of diet and compartment on the microbiota (Fig. 7A–D). The analysis revealed a clear separation of samples in accordance to the gut regions and a closer association among samples of the same gut region. At the midgut, the clusters were well defined and the highest variability was found in the RMG (i.e., midgut of rice population feeding on rice seedlings) cluster. The RMG and JMG (i.e., midgut of the water-oat population feeding on water-oat fruit pulps) clusters exhibited the most different taxa composition, followed by the rMG (i.e., midgut of the rice population feeding on water-oat fruit pulps) and jMG (i.e., midgut of the water-oat population feeding on rice seedlings) clusters, showing an intermediate composition (Fig. 7A). At the hindgut, there were clearly separated clusters: the RHG (i.e., hindgut of rice population feeding on rice seedlings) clusters exhibited a higher inter-sample variation; the JHG (i.e., hindgut of the water-oat population feeding on water-oat fruit pulps) cluster showed an intermediate composition respecting to the RHG, jHG (i.e., hindgut of the water-oat population feeding on rice seedlings) and rHG (i.e., hindgut of rice population feeding on water-oat fruit pulps) clusters (Fig. 7B). The midguts and hindguts clusters from all original populations (JMG, JHG, RMG, RHG) were well-defined, and the JMG, RHG clusters had similar homogeneity level (Fig. 7C). RMG was the most heterogeneous, followed by JMG and RHG. Clusters of cross-rearing populations were better defined than those of original populations. rMG was the most heterogeneous in taxa composition, followed by the jHG.
NMDS of the gut microbiota of C. suppressalis. (A) NMDS of the taxon distribution of midgut samples. The samples were clustered by diets and represented with different colors: jMG (red, circles), rMG (green, rhombus), JMG (blue, squares) and RMG (saffron yellow, triangle). (B) NMDS of the taxon distribution of the hindgut samples. The samples were clustered by diets and represented with different colors: jHG (red, circles), rHG (green, rhombus), JHG (blue, squares) and RHG (saffron yellow, triangle). (C) NMDS of the taxon distribution of the midgut and hindgut samples from the water-oat and rice populations. The samples were clustered by diets and represented with different colors: JHG (red, circles), RHG (green, rhombus), JMG (blue, squares) and RMG (saffron yellow, triangle). (D) NMDS of the taxon distribution of the midgut and hindgut samples from cross-rearing populations. The samples were clustered by diets and represented with different colors: jHG (red, circles), rHG (green, rhombus), jMG (blue, squares) and rMG (saffron yellow, triangle). The ellipses represent the standard error of the centroid for each group of samples with a confident limit of 95%.
Discussion
To date, there were few documents on how gut microbial communities differ across divergent insect populations based on diet and gut compartments. Gut bacterial diversity overall was notably greater in the rice population feeding on water-oat fruit pulps compared to the water-oat population or rice population feeding on rice seedlings. Bacterial communities resided in the midgut were more diverse and variable than those in the hindgut. Only bacteria of Citrobacter, Enterococcus, Halomonas, and Klebsiella were shared by original populations of C. suppressalis, and they were core microbiota based on their relative distribution. The core bacteria was able to colonize in different gut regions49, and might have evolved in closely related to hosts and were potential symbiont or beneficial bacteria50,51,52. Since rice seedlings and water-oat were very different in nutritional ingredient and secondary compounds, it was probable that the bacteria inhabited in C. suppressalis gut were beneficial to their hosts.
The gut bacterial composition and richness exhibited significant differences in the midgut and hindgut of different populations of C. suppressalis. Halomonas and Klebsiella dominated in the midgut and hindgut of rice seedlings-fed rice population, and Klebsiella and Citrobacter were prevailed in the midgut and hindgut of water-oat-fed water-oat population. Enterococcus were enriched in the midgut of cross-rearing populations, whereas Citrobacter was found exclusively in the hindgut of cross-rearing populations. According to Shao et al. And Zhang et al., the Enterococcus was associated with insecticide and pathogen resistances53,54, and the presence of this genus in C. suppressalis may enhance the immune of this pest during it host shift.
Our findings also showed that a remarkable different bacteria composition in the RMG and JMG and a intermediate bacteria composition in the rMG and jMG. The inter-individual variability was previously documented in honey bees Apis mellifera55, Anopheles56, and cockroaches Blattella germanica38,57, Shelfordella lateralis58 and Periplaneta americana59. The divergence in taxon composition may reflect divergent functional roles in specific resource use. Gut harbored bacteria community of the water-oat population and rice population feed on their original hosts is closely adapted to the C. suppressalis’s diets. With the change of diets, (i.e., water-oat population feeding on rice seedlings, and the rice population feeding on water-oat fruit pulps), the compositional change could be partly responsible for undergoing a recombination of the bacteria, accordingly. Curtis and Sloan suggested that the variation could be attributed to acquire microorganisms from a greatly diverse environmental reservoir microflora, randomly60. However, as the populations of C. suppressalis were reared many generations in identical laboratory conditions, such a variability could be ascribed to host genetics and population divergence, as was suggested by Sullam et al.61 Cluster analysis showed the jHG and rHG samples formed the most well-defined clusters, suggesting stable microbial profiles. The inter-individual differences suggested that SSB gut microbiome profiles may serve as useful biomarkers for bio-control in population-based studies.
The oligophagous diet of stem borers provided suitable ecological niches for harboring bacteria in compared with monophagous lepidopterans55. As the phyla Proteobacteria were reported to be involved in carbohydrate degradation, such as starches and hemicellulose62, and can be involved in pectin-degrading63 and nitrogen64. Firmicutes was suggested to take part in energy absorption from the diet and may influence the development65. The present results illuminated the abundance of two dominant phyla (i.e., Proteobacteria and Firmicutes) and the difference of three families (Enterobacteriaceae, Enterococcaceae and Halomonadaceae) in C. suppressalis populations. As the representative of the oligophagous, C. suppressalis feeding either on water-oat fruit pulps or rice seedlings. Both host plants shared the same family Gramineae, but their biochemical components and secondary substances were very different9. Our findings suggested that the rapid fluctuation of bacterial flora in larval gut was probably influenced by the biochemical components and secondary substances coming from the host plants; and the diet was an important factor in modulating the bacteria community, as was documented for other insect species39,66,67,68,69,70,71,72,73.
The gut bacterial genera were also varied, due to the difference of diets in C. suppressalis: in original populations, Halomonas was dominant in the RMG, Klebsiella was prevailed in the RHG and JMG, and Citrobacter was enriched in the JHG; in cross-rearing populations, Enterococcus was abundant in the midgut, and Citrobacter was predominant in the hindgut. Since diet and host taxonomy modulated bacteria community71,74, the successful expansion of bacteria over time probably in turn suppressed the bacteria growth from other phyla in the same habitat66. Therefore, we inferred that the different bacteria dominance might be related to successful reproduction of some bacteria genus and suppression of others. Whether the bacteria of Citrobacter, Enterococcus, Halomonas, and Klebsiella detected in the gut of original populations of C. suppressalis was truly associated with the host defense merits further investigation.
One interesting and unexpected result concerned the two compartments chosen for analysis, as we found that variability in microbial composition was higher in the midgut than in the hindgut, independently of diet. The obvious community difference indicated that only some specific groups of microorganisms were able to survive and colonize in the hindgut. However, Kacaniova et al. reported that the hindgut contained a higher number of anaerobic microorganisms than the midgut of honeybee75. Although the midgut and hindgut were alkaline, the unique morphology, favorable physiological conditions (viz., oxygen content, lack of unfavorable enzymes), and the availability of partially digested food could become a benign site for maintaining a special bacteria and quick proliferation in the hindgut of C. suppressalis. Indeed, this may be a controversial issue, and the different richness and colonization efficiency of the host symbiont indicated that further investigation should be done to understand their drivers.
Conclusion
We investigated the gut microbial communities of two phenotypically divergent populations of C. suppressalis. The comparison of the midgut and hindgut microbia of C. suppressalia fed on the same diet provided insights into the compartment changes in the gut microbiota of SSB. Analysis of microbial community supplied an initial step toward improving our understanding of the mechanisms underlying C. suppressalis adaptation to host plants at the microbiological level. The results showed that the highest bacteria diversity was found for the midgut of the rice population feeding on water-oat fruit pulps. The most dominant phyla were Proteobacteria and Firmicutes; and the enriched families were Enterobacteriaceae, followed by Enterococcaceae and Halomonadaceae. The microbial communities were highly diverse at the genera level due to diet types or gut compartments among populations. The bacterial community composition was driven mainly by diet types, and affected by other factors including gut compartments. These findings provided an important insight into investigation of insect-bacteria symbioses, and biocontrol of this species and other lepidopterans.
Data availability
The raw reads were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. SRP116573).
Abbreviations
- SSB:
-
Striped stem borer
- NMDS:
-
Non-metric multidimensional scaling
- PCR:
-
Polymerase chain reaction
- OTUs:
-
Operational units
- JMG:
-
Midgut of water-oat population
- JHG:
-
Hindgut of water-oat population
- RMG:
-
Midgut of rice population
- RHG:
-
Hindgut of rice population
- jMG:
-
Midgut of water-oat population feeding on rice seedlings
- jHG:
-
Hindgut of water-oat population feeding on rice seedlings
- rMG:
-
Midgut of rice population feeding on water-oat fruit pulps
- rHG:
-
Hindgut of rice population feeding on water-oat fruit pulps
- NCBI:
-
National center for biotechnology information
- SRA:
-
Sequence read archive
References
Jiang, M. & Cheng, J. Interactions between the striped stem borer Chilo suppressalis (Walk.) (Lep., Pyralidae) larvae and rice plants in response to nitrogen fertilization. J. Pest Sci. 76, 124–128 (2003).
Alfaro, C., Navarrollopis, V. & Primo, J. Optimization of pheromone dispenser density for managing the rice striped stem borer, Chilo suppressalis (Walker), by mating disruption. Crop Prot. 28, 567–572 (2009).
Chen, M., Shelton, A. & Ye, G. Insect-resistant genetically modified rice in China: From research to commercialization. Annu. Rev. Entomol. 56, 81–101 (2011).
Muralidharan, K. & Pasalu, I. C. Assessments of crop losses in rice ecosystems due to stem borer damage (Lepidoptera: Pyralidae). Crop Prot. 25, 409–417 (2006).
Guo, H., Li, S., Peng, J. & Ke, W. Zizania latifolia Turcz. cultivated in China. Genet. Resour. Crop. Evol. 54, 1211–1217 (2007).
Hou, M., Han, Y. & Lin, W. Influence of soil moisture on supercooling capacity and associated physiological parameters of overwintering larvae of rice stem borer. Entomol. Sci. 12, 155–161 (2009).
He, Y. et al. Regression analysis of dynamics of insecticide resistance in field populations of Chilo suppressalis (Lepidoptera: Crambidae) during 2002–2011 in China. J. Econ. Entomol. 106, 1832–1837 (2013).
Wang, Y. N. et al. Comparison of three transgenic Bt rice lines for insecticidal protein expression and resistance against a target pest, Chilo suppressalis (Lepidoptera: Crambidae). Insect Sci. 23, 78–87 (2016).
Guo, L. et al. A host plant genome (Zizania latifolia) after a century-long endophyte infection. Plant J. 83, 600 (2015).
Chen, J., Yu, X. & Zhen, X. Biological performances of the striped stem borer, Chilo suppressalis Walker fed on Jiaobai, Zizania caduciflora, and rice plants. Acta Agricult. Zhejiangensis. 15, 139–143 (2003).
Quan, W. et al. Do differences in life-history traits and the timing of peak mating activity between host-associated populations of Chilo suppressalis have a genetic basis?. Ecol. Evol. 6, 4478–4487 (2016).
Cho, M. J. et al. Symbiotic adaptation of bacteria in the gut of Reticulitermes speratus: Low endo-β-1, 4-glucanase activity. Biochem. Bioph. Res. Co. 395, 432–435 (2010).
Xu, Z. et al. Relationship between biological quality and nutrition of rice and damage of Chilo suppressalis. Acta Phytophyl. Sin. 38, 139–146 (2011).
Maki, Y. & Yamashita, M. Ecological difference of rice stem borer, Chilo suppressalis Walker in the various host plants. Bull. Hyogo. Pref. Agric. Exp. Sta. 3, 47–50 (1956).
Tsuchida, K. & Ichihashi, H. Estimation of monitoring range of sex pheromone trap for the rice stem borer moth, Chilo suppressalis (Walker) (Lepidoptera: Pyralidae) by male head width variation in relation to two host plants, rice and water-oats. Appl. Entomol. Zool. 30, 407–414 (1995).
Matsukura, K., Hoshizaki, S., Ishikawa, Y. & Tatsuki, S. Morphometric differences between rice and water-oats population of the striped stem borer moth, Chilo suppressalis (Lepidoptera: Crambidae). Appl. Entomol. Zool. 41, 529–535 (2006).
Ding, N. et al. A comparison of the larval overwintering biology of the striped stem borer, Chilo suppressalis (Lepidoptera: Crambidae), in rice and water-oat fields. Appl. Entomol. Zool. 48, 147–153 (2013).
Zibaee, A., Sendi, J. J., Ghadamyari, M., Alinia, F. & Etebari, K. Diazinon resistance in different selected strains of Chilo suppressalis (Lepidoptera: Crambidae) in northern Iran. J. Econ. Entomol. 102, 1189–1196 (2009).
Ishiguro, N., Yoshida, K. & Tsuchida, K. Genetic differences between rice and water-oat feeders in the rice stem borer, Chilo suppressalis (Walker) (Lepidoptera: Crambidae). Appl. Entomol. Zool. 41, 585–593 (2006).
Ueno, H., Furukawa, S. & Tsuchida, K. Difference in the time of mating activity between host-associated populations of the rice stem borer, Chilo suppressalis (Walker). Entomol. Sci. 9, 255–259 (2006).
Yang, C., Yang, X., Fu, Q., Xu, K. & Lu, B. R. Limited divergence among populations of rice striped stem borer in southeast China caused by gene flow: Implications for resistance management. J. Syst. Evol. 50, 443–453 (2012).
Konno, Y. & Tanaka, F. Mating time of the rice-feeding and water-oat-feeding strains of the rice stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). Appl. Entomol. Zool. 40, 245–247 (1996).
Samudra, I. M., Emura, K., Hoshizaki, S., Ishikawa, Y. & Tatsuki, S. Temporal difference in mating behavior between rice- and wateroats-populations of the striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Crambidae). Appl. Entomol. Zool. 37, 257–262 (2002).
Yu, X. et al. Differentiation of striped stem borer (SSB), Chilo suppressalis Walker from rice and Zizania caduciflora Habitats. Acta Ecol. Sin. 22, 341–345 (2002).
Matsukura, K., Hoshizaki, S., Ishikawa, Y. & Tatsuki, S. Differences in timing of the emergence of the overwintering generation between rice and water-oats populations of the striped stem borer moth, Chilo suppressalis (Lepidoptera: Crambidae). Appl. Entomol. Zool. 44, 485–489 (2009).
Zhong, H., Li, F., Chen, J., Zhang, J. & Li, F. Comparative transcriptome analysis reveals host-associated differentiation in Chilo suppressalis (Lepidoptera: Crambidae). Sci. Rep. 7, 13778 (2017).
Han, Y., Hao, L. & Hou, M. Comparison of overwintered bionomics of Chilo suppressalis larvae from paddy-rice field with those from water-oat field in North China. Chin. J. Eco-Agric. 17, 541–544 (2009).
Crotti, E. et al. Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microb. 76, 6963–6970 (2010).
Buchner, P. Endosymbiosis of animals with plant microorganisms (Interscience, 1965).
Dale, C. & Moran, N. Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465 (2006).
Behar, A., Yuval, B. & Jurkevitch, E. Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J. Insect Physiol. 54, 1377–1383 (2008).
Chung, S. H. et al. Herbivore exploits orally secreted bacteria to suppress plant defenses. P. Natl. Acad. Sci. USA 110, 15728 (2013).
Ceja-Navarro, J. A. et al. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 6, 1–9 (2015).
Cheng, D. et al. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5, 13 (2017).
Emery, O., Schmidt, K. & Engel, P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 26, 2576–2590 (2017).
Lee, J. B. et al. Gut symbiotic bacteria stimulate insect growth and egg production by modulating hexamerin and vitellogenin gene expression. Dev. Comp. Immunol. 69, 12–22 (2017).
Brucker, R. M. & Bordenstein, S. R. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667 (2013).
Pérez-Cobas, A. E. et al. Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol. Ecol. 91, 1–14 (2015).
Su, L. et al. Variation in the gut microbiota of termites (Tsaitermes ampliceps) against different diets. Appl. Biochem. Biotech. 181, 32–47 (2017).
Lü, J. et al. Host plants influence the composition of the gut bacteria in Henosepilachna vigintioctopunctata. PLoS ONE 14, e0224213 (2019).
Luo, H. et al. The relationship between phenolics metabolism and browning and lignification of fresh-cut Zizania latifolia. Food Sci. Technol. 40, 51–55 (2015).
Wu, B. et al. Effect of environmental factors on secondary substances of rice planting and relationship with resistance to brown planthopper (Homoptera: Delphacidae). Southwest China J. Agric. Sci. 29, 2371–2378 (2016).
Konno, Y. Mating-choice and host preference tests in the rice-feeding and water-oat-feeding types of the rice stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). Annu. Rep. Plant Protect. North Japan. 49, 102–104 (1998) ((in Japanese)).
Sharon, G. et al. From the cover: commensal bacteria play a role in mating preference of Drosophila melanogaster. P. Natl. A. Sci. USA 107, 20051–20056 (2010).
Robinson, C. J., Schloss, P., Ramos, Y., Raffa, K. & Handelsman, J. Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb. Ecol. 59, 199–211 (2010).
Belda, E. et al. Microbial diversity in the midguts of field and lab-reared populations of the European corn borer Ostrinia nubilalis. PLoS ONE 6, e21751 (2011).
Feng, W., Wang, X., Zhou, W., Liu, G. & Wan, Y. Isolation and characterization of lipase-producing bacteria in the intestine of the silkworm, Bombyx mori, reared on different forage. J. Insect Sci. 11, 125 (2011).
Tang, X. et al. Complexity and variability of gut commensal microbiota in polyphagous Lepidopteran larvae. PLoS ONE 7, e36978 (2012).
Bereded, N. K. et al. Metabarcoding analyses of gut microbiota of nile tilapia (Oreochromis niloticus) from Lake awassa and lake chamo, ethiopia. Microorganisms 8, 1040 (2020).
Berg, R. D. The indigenous gastrointestinal microflora. Trends Microbiol. 4, 430–435 (1996).
Martinson, V. G. et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 20, 619–628 (2011).
Koch, H., Abrol, D. P., Li, J. & Schmid-Hempel, P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 22, 2028–2044 (2013).
Zhang, J., He, Y. & Chen, J. Diversity analysis of bacterial community in midguts of larvae of the striped stem borer, Chilo suppressalis (Lepidoptera: Crambidae), with different levels of resistance to insecticides. Acta Entomol. Sin. 56, 1075–1082 (2013).
Shao, Y., Ariascordero, E., Guo, H., Bartram, S. & Boland, W. In vivo Pyro-SIP assessing active gut microbiota of the cotton leafworm. Spodoptera littoralis. Plos One 9, e85948 (2014).
Moran, N. A., Hansen, A. K., Powell, J. E. & Sabree, Z. L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 7, 2 (2012).
Osei-Poku, J., Mbogo, C. M., Palmer, W. J. & Jiggins, F. M. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 21, 5138–5150 (2012).
Carrasco, P. et al. Succession of the gut microbiota in the cockroach Blattella germanica. Int. Microbiol. 17, 99–109 (2014).
Schauer, C., Thompson, C. & Brune, A. Pyrotag sequencing of the gut microbiota of the cockroach Shelfordella lateralis reveals a highly dynamic core but only limited effects of diet on community structure. PLoS ONE 9, 2 (2014).
Sabree, Z. L. et al. Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. Appl. Environ. Microbiol. 78, 204–210 (2012).
Curtis, T. P. & Sloan, W. T. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7, 221–226 (2004).
Sullam, et al. Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. Isme J. 9, 1508 (2015).
Adams, A. S. et al. Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. Isme J. 5, 1323–1331 (2011).
Engel, P., Martinson, V. G. & Moran, N. A. Functional diversity within the simple gut microbiota of the honey bee. P. Natl. A. Sci. 109, 11002–11007 (2012).
Pinto-Tomas, A. A. et al. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326, 1120–1123 (2009).
Chen, B. et al. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6, 29505 (2016).
Strano, C. O., Malacrinò, A., Campolo, O. & Palmeri, V. Influence of host plant on Thaumetopoea pityocampa gut bacterial community. Microb. Ecol. 75, 487–494 (2018).
Harris, E. V., de Roode, J. C. & Gerardo, N. M. Diet–microbiome–disease: Investigating diet’s influence on infectious disease resistance through alteration of the gut microbiome. PLoS Pathog. 15, e1007891 (2019).
Jones, A. G., Mason, C. J., Felton, G. W. & Hoover, K. Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci. Rep. 9, 2792 (2019).
Mason, C. J. et al. Diet influences proliferation and stability of gut bacterial populations in herbivorous lepidopteran larvae. PLoS ONE 15, e0229848 (2020).
Ley, R. The evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Colman, D. R., Toolson, E. C. & Takacs-Vesbach, C. D. Do diet and taxonomy influence insect gut bacterial communities?. Mol. Ecol. 21, 5124–5137 (2012).
Yun, J. H. et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264 (2014).
Kim, J. M. et al. Effects of diet type, developmental stage, and gut compartment in the gut bacterial communities of two Cerambycidae species (Coleoptera). J. Microbiol. 55, 21–30 (2017).
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A. & Kopp, A. Bacterial communities of diverse Drosophila species: Ecological context of a host-microbe model system. Plos Genet. 7, 2 (2011).
Kacaniova, M., Chlebo, R., Kopernicky, M. & Trakovicka, A. Microflora of the honeybee gastrointestinal tract. Folia. Microbiol. 49, 169–171 (2004).
Acknowledgements
The authors are grateful to Pan Xiaoting for rearing the striped stem Chilo suppressalis borer. We sincerely thank anonymous reviewers for their critical review and providing valuable comments to this manuscript. Mingke Biotechnology (Hangzhou, China) Co., Ltd. is also thanked for supplying technical assistance. This work is financially supported by the Zhejiang Provincial Natural Science Foundation of China (Grant Nos. LY16C140006, LQ19C140003), China Agriculture Research System of MOF and MARA (Grant No. CARS-24-G-06).
Author information
Authors and Affiliations
Contributions
J.C. conceived and designed the study; H.Z. and J.C. designed the experiments and discussed the results; H.Z. performed the experiments, analyzed the data and drafted the manuscript; J.Z. and F.L. and collected and reared insect populations. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhong, H., Zhang, J., Li, F. et al. Gut microbial communities associated with phenotypically divergent populations of the striped stem borer Chilo suppressalis (Walker, 1863). Sci Rep 11, 15010 (2021). https://doi.org/10.1038/s41598-021-94395-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94395-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.