Abstract
The increasing severity and frequency of natural disturbances requires a better understanding of their effects on all compartments of biodiversity. In Northern Fennoscandia, recent large-scale moth outbreaks have led to an abrupt change in plant communities from birch forests dominated by dwarf shrubs to grass-dominated systems. However, the indirect effects on the belowground compartment remained unclear. Here, we combined eDNA surveys of multiple trophic groups with network analyses to demonstrate that moth defoliation has far-reaching consequences on soil food webs. Following this disturbance, diversity and relative abundance of certain trophic groups declined (e.g., ectomycorrhizal fungi), while many others expanded (e.g., bacterivores and omnivores) making soil food webs more diverse and structurally different. Overall, the direct and indirect consequences of moth outbreaks increased belowground diversity at different trophic levels. Our results highlight that a holistic view of ecosystems improves our understanding of cascading effects of major disturbances on soil food webs.
Similar content being viewed by others
Introduction
Natural disturbances, such as fires, droughts, or insect outbreaks, are key drivers of ecosystem dynamics and community structure1. Global change could exacerbate their severity and frequency worldwide with potential extensive impacts on biodiversity, ecosystems and human societies2,3. Understanding the effect of disturbances on the dynamics and structure of biodiversity is therefore more than ever a crucial issue in ecology. Yet, the high variability of local biodiversity trends in response to global changes asks for more integrative analyses, going beyond mere measures of species richness and accounting for the multiple components of the ecosystems4,5. Particularly, soil organisms are rarely included when synthesizing biodiversity trends in the face of disturbances, despite their recognized and well documented influence on multiple ecosystem functions (e.g. nutrient cycling) and nature contributions to people (e.g. carbon storage or depollution)6,7,8,9.
Most studies quantifying the effect of disturbances on biodiversity have focused on a single trophic or taxonomic group, often directly affected by the disturbance, like plants9. However, much less is known on how the effects propagate across trophic levels ultimately affecting the entire ecosystem. Plants and soil organisms are tightly linked through direct and indirect interactions, including mutualism, parasitism or predation, which promote the exchange and supply of nutrients and ensure multiple ecosystem processes6,7. Ignoring these trophic interactions and how resource deprivation in one trophic level can cascade to other levels may obscure the true consequences of disturbances for ecosystems10. Furthermore, misleading conclusions could be drawn if resulting disturbance effects differ between trophic levels11. Most natural disturbances cause immediate fluctuations in the quantity and quality of available soil resources1. Extreme winds can remove or deposit organic matter on the forest floor, while insect outbreaks increase soil nutrient inputs through defoliation and insect faeces and corpses. These local changes in basal resource availability can have important consequences on the abundance and diversity of primary producers (e.g. plants or nitrifying bacteria) and primary consumers (e.g. decomposers or herbivores), but also subsequently on the whole soil food web through bottom-up cascading effects12,13,14. Predicting whether the effects would vanish or amplify remains challenging due to the complexity of soil food webs in real ecosystems. Stoichiometry-based studies have provided numerous evidences that such indirect effects propagate across soil food webs from the microfauna to the macrofauna in terms of composition and biomass15,16,17. However, these approaches don’t include the microbial part of the soil food web, and often lack resolution or breadth for the micro and macro fauna when describing the diversity and composition of these complex communities. In addition, changes in the abundance and diversity of organisms across the food web are likely to induce structural changes in the entire interaction network, potentially leading to alternative ecosystem states8,18,19. Thus, quantifying cascading effects of disturbances on ecosystems requires a holistic view of biodiversity with not only exhaustive sampling methods to capture all-in-end biodiversity, but also a suitable analytic approach to analyze changes in trophic levels and interactions.
To meet this challenge, we combined the power of environmental DNA metabarcoding (eDNA)20 to obtain a nearly complete view of the belowground biodiversity, with a food web approach and network theory. Grouping species with the same trophic position (i.e. shared predators and preys/resources) in ecological networks facilitates the study of complex multitrophic communities21,22,23. In such an approach, the focus is not on species, but rather on trophic groups and trophic interactions. The definition of the trophic groups depends both on the resolution of the observation units (e.g. the taxonomic resolution) and the information available on their diet or trophic position24,25,26, and is also related to the ecological question. When studying the large-scale consequences of disturbances on biodiversity, there is a trade-off between sufficiently fine resolution to reliably and meaningfully measure cascading effects22,27, and sufficiently broad resolution to avoid knowledge gaps and cope with heterogeneity of taxonomic resolution in the data25,28. Once a food web is built, diversity can be measured within trophic groups (e.g. species diversity) and between trophic groups (e.g. trophic diversity or diversity of interactions), allowing the integration of ecological processes occurring at different dimensions of the food web (e.g. competition and predation)29,30. For this, network theory provides appropriate metrics to describe and compare the diversity and structure of ecological networks, accounting for both group abundances and interactions31,32.
Here, we study the effect of moth outbreaks on soil food webs of subarctic birch forests in Northern Fennoscandia. These forests have experienced moth outbreaks of unprecedented scale and severity in recent decades, which have led to a sudden and persistent vegetation change -from birch forests with understory dominated by dwarf shrubs to grass-dominated systems associated with high tree mortality- that was still visible 8 years after the disturbance33,34,35,36 (Fig. 1). Moth outbreaks is a good model for assessing the cascading effects of disturbance on soil food webs, as the larvae only attack the foliage of the dominant primary producers, i.e. the birch tree (Betula pubescens), and some abundant species of erect and dwarf shrubs in the understory layer (e.g. Betula nana, Empetrum nigrum, Vaccinium spp.). In parallel, soil organic matter is enriched through dead plants and N addition from larval faeces and corpses37,38. We can therefore expect that impacts on the whole soil food web arise from bottom-up effects from changes in the vegetation and basal resources to the other trophic compartments12. Drastic shifts in the composition of biological communities following defoliation have been already reported in these nutrient-limited soils where the dominance of the allelopathic dwarf shrub Empetrum nigrum in the understory leads to regressive succession that may inhibit soil microbial activity, organic matter decomposition, and thus nutrient availability39,40,41. These shifts correspond to a replacement of Empetrum nigrum by the grass Avenella flexuosa34 with subsequent effects on the diversity and abundance of organisms directly relying on plants, including vertebrate herbivores33, birds42, saproxylic beetles38, and fungal communities43,44. However, we still ignore whether moth outbreaks induced indirect effects across the soil food web, whether these effects are of comparable magnitude to those observed for vegetation, and finally, whether these effects have significant consequences on trophic interactions and ultimately on the whole soil food web structure.
Sampling design in undamaged and defoliated forests. Map of the study location in the Northeastern Norway (a), in the Varanger region (b). Red areas (b) represent birch forest that experienced severe defoliation during the most recent moth outbreak. Yellow stars indicate the stations that were sampled in this study, at each extreme of two pre-established transects (black dots) going from highly defoliated forests stands to undamaged stands. Soil sampling was conducted in each station along an L-shaped transect (c). Photos illustrate the stations from undamaged (d) and defoliated (e) forests. Red flags in the photos indicate the sampling points represented in (c). Undamaged forests were characterized by living birch trees (Betula pubescens) and a shaded understory dominated by ericaceous shrubs (e.g. Empetrum nigrum). Defoliated forests were characterized by dead birch trees, patches of remaining ericaceous shrubs and a soil covered by light-tolerant grass and herbs including the dominant Avenella flexuosa. Photo credits: Heidy Schimann. Map (a) was created using ArcGIS® software 10.4.1 by Esri (www.esri.com). Map (b) was modified from38 (https://doi.org/10.1371/journal.pone.0099624.g001).
We used eDNA data obtained from 86 soil samples from two well-studied areas in northeastern Norway (i.e., Tana and Kirkenes). This study design allowed for appropriate pairwise comparisons between coupled undamaged and defoliated forest based on well-documented defoliation patterns from both remote sensing and field methods (Fig. 1). The sampling design aimed at capturing the environmental heterogeneity at different spatial scales of the landscape within these areas. We then classified both microorganisms and macroinvertebrates into 9 broad trophic classes and 37 finer trophic groups to build metawebs45 at two levels of resolution for the study area (Fig. 2). The metawebs were then used to infer local soil food webs based on taxa detected locally in each soil sample. The trophic class resolution corresponds to what is commonly used in soil food web ecology (e.g.22,27), but we additionally included the trophic group resolution because a finer resolution is needed to capture specific effects of disturbance on groups that are hidden at a coarser resolution. For instance, different types of mycorrhizal fungi like arbuscular mycorrhizal fungi and ectomycorrhizal fungi may have opposite responses to tree defoliation, the former increasing and the later decreasing in their proportion following disturbances46.
Methodology used to build the metaweb from soil eDNA. First, eDNA was extracted and processed from the 86 soil samples to obtain a list of taxa for the study area. Second, using an extensive collection of trophic knowledge from databases, literature and experts, taxa were assigned to broad trophic classes and then to finer trophic groups, which separate distant phylogenetic groups or groups that differ in their resources acquisition strategy. Main trophic links were collected from literature and current knowledge to build the metaweb at two levels of resolution (a,b). The colours correspond to the trophic classes (a) that are refined and split in the highly resolved metaweb (b).
Using this approach, we tested three hypotheses about the cascading effects of moth defoliation on the local soil food webs at different levels of organization. First, (H1) moth defoliation changes the diversity in MOTUs (Molecular Operational Taxonomic Unit) and the relative abundances of most trophic groups. We expected positive effects on most decomposers and their consumers through the impulse in soil resources availability47,48 from both moth outbreaks and the decreased abundance of the allelopathic species Empetrum nigrum. In parallel, we expected negative effects on e.g., ectomycorrhizal and ericoid mycorrhizal fungi, as the result of the decline of birch and ericaceous shrub roots. Second, (H2) the magnitude of the effect differs among trophic groups across the soil food web. We expected the effect of defoliation to be stronger for primary consumers and decomposers that are directly affected by changes in basal resources availability and plant composition, and then to decrease toward higher trophic levels (attenuation of the effects). Third, (H3), moth defoliation changes the overall structure of the local soil food webs10,49. We expected to observe differences in the trophic groups and links diversity and composition of the local food webs between defoliated and undamaged forests.
Results and discussion
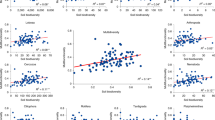
Fitting a multilevel linear model for each trophic group, we found that moth defoliation increased MOTU diversity and the relative abundances of most trophic groups (Fig. 3). This is consistent with H1 and food web theory predictions, i.e. the effect of disturbances should propagate up the food web levels when resources are enriched through bottom-up processes14,50. Overall, diversity and relative abundance followed similar trends within trophic groups (Fig. 3a,b).
Effect of defoliation on diversity and relative abundance of trophic groups. Effect size of defoliation on MOTU diversity (a) and relative abundance of reads (b) for each trophic group with 90% credible intervals. The barplot (c) shows the total number of reads (logarithmic scale) of each trophic group in the overall dataset. A multilevel linear model was fitted individually for each trophic group with a dummy variable for defoliation as predictor and a random factor accounting for the nested sampling design. MOTU diversity was standardized by the maximum value observed within each trophic group to obtain comparable effect sizes between groups. The colours correspond to the trophic class definitions (see Fig. 2).
The basal groups directly linked to plants or basal resources (e.g., soil organic matter and light), i.e., mycorrhizal fungi, phytophagous or plant parasites, decomposers and primary producers, were expected to respond to changes in the composition of plant communities and nutrient enrichment following the outbreak. Here, comparing undamaged and defoliated forests, we observed a radical shift from ectomycorrhizal to arbuscular mycorrhizal fungal communities. This is consistent with the reduction of birch fine woody roots in defoliated forests, which are obligate hosts for most ectomycorrhizal fungi, and with the increases of herb and grass roots that are mostly associated with arbuscular mycorrhizal fungi43,44,51 (Supplementary Fig. 2). The increased diversity and relative abundance of slugs, snails and plant pathogen protists could be in part explained by the increased palatability of the plant assemblages. Indeed, grasses like Avenella flexuosa, which is dominant in the defoliated forests, are more palatable as compared to allelopathic species like Empetrum nigrum33,52. Photosynthetic protists diversity and relative abundance also increased in defoliated sites which are more open, hence allowing more light to reach the soil (Fig. 1).
Among the decomposers, defoliation led to an increase in the diversity of heterotrophic bacteria, protists, saprotroph-plant pathogen fungi and wood saprotroph fungi. Similarly, the relative abundance of protists, saprotroph-plant pathogen fungi and enchytraeids increased. Differences in plant litter chemistry between undamaged and defoliated forests (Supplementary Fig. 2) might drive the communities of decomposers53 and could explain these changes. For instance, the litter produced by Empetrum nigrum, which dominates undamaged forests, releases of phenolic compounds52 that can strongly reduce plant species diversity40,41. Such detrimental effects might also hold true for the diversity and abundance of most decomposers. Soils from defoliated forests had lower C/N ratios, suggesting that defoliation promote more labile, easily decomposable organic matter inputs (Supplementary Fig. 3) but more precise soil nutrient measurements would be needed to confirm this.
Contrary to our expectation, the magnitude of the effect of defoliation did not decrease further up the food web (Fig. 3a,b), but was instead equally important at all trophic levels. This result did not depend on the number of sequences obtained for each group (Fig. 3c). This rejects the hypothesis of a mitigation of the effects of the disturbance when moving up to higher trophic levels in the soil food web (H2). For example, the indirect effect of defoliation on the diversity of copiotrophic bacteria was as strong as the effect on their protist predators, and as strong as the effect on nematodes feeding on protists. In addition, the effect of defoliation on animal parasites, which are at the top of the soil food web, was similar to the effect on mycorrhizal fungi. Our findings are consistent with other studies pointing out that species-poor ecosystems, like subarctic birch forests, could be more prone to the propagation of bottom-up disturbances along food webs54. Furthermore, while some groups were affected by defoliation, other groups within the same trophic class were not (e.g. herbivore mite vs. plant pathogens protists, or ectomycorrhizal vs. ericoid mycorrhizal fungi). Other studies have highlighted the challenge of predicting the effect of an environmental stressor on overall biodiversity due to the variety of responses that organisms can have, associated with attributes such as dispersal abilities or resistance structures (e.g. cysts in protists)55,56. This is particularly important in soil food webs consisting of organisms with large differences in body size, life-span and life history strategies, and therefore in their response time to disturbance, which can vary from seconds to decades7,48,56. This complexity hampers our ability to detect consistent patterns when studying soil food webs at fixed sampling times.
We then examined how changes in trophic groups relative abundances influenced the network structure of local soil food webs, using network diversity indices31. Following H3, moth defoliation significantly altered the whole soil food web structure in terms of node and link abundances, both for the trophic class and group resolutions (Supplementary Fig. 4). An increase in local diversity (α-diversity) of trophic groups and links in defoliated forests partially explained the changes in food web structure (Fig. 4). When zooming out to trophic classes, differences in the α-diversity of soil food webs were less obvious but food webs were nevertheless slightly more diverse for defoliated forests (Supplementary Fig. 5). This reflects that within a trophic class, trophic groups can have opposite responses (Fig. 3) that are averaged out when only considering trophic class, and highlights the importance of using a finer trophic resolution than what is often used in the literature to understand the variability of cascading effects in the different components of the soil food web. On average, we observed a decrease in the proportion of most classes of primary consumers (i.e. plant mycorrhiza, herbivores/plant pathogens, decomposers) within the soil food webs in defoliated forests, and an increase in the proportion of higher trophic level classes (i.e. bacterivores, omnivores, predators), which were rare in the undamaged forests (Fig. 5). These changes in relative abundance proportions within the soil food web are not to be confounded with the individual changes in the relative abundances of the trophic groups (Fig. 3b). For instance, a decrease in the proportion of some classes might be related to weaker increase in average of the relative abundance of the groups within the class from undamaged to defoliated forests, compared to a stronger average increase for classes in higher trophic levels.
Topology and α-diversity of local food webs in undamaged vs. defoliated forests. The values represent the α-diversity of the soil food webs for each area and category of defoliation at the trophic group resolution: A1(p) is the diversity in trophic group abundances (nodes) and A1(L) the diversity in trophic links abundances (edges) using Shannon diversity. Nodes of the local food webs corresponded to the local relative abundances of the groups varying from 0 (when the group was absent) to 1 (when the group was at its maximum observed abundance). Links were binary links (i.e. present or absent) assuming an interaction when the two groups concerned were present. For the visualization, four local soil food webs (with an average value of A1(p)) were selected to highlight the differences in diversity between undamaged and defoliated forests of each area. The colours correspond to the trophic classes and the nodes are distributed vertically based on their trophic level from the bottom (basal levels) to the top (higher levels).
Structural differences among the local soil food webs from undamaged to defoliated forests at the trophic class resolution. Orange colour represents an increase, and purple colour a decrease in the relative abundance proportion within the local food webs of trophic classes (nodes) and link probability between classes (edges) from undamaged to defoliated forest. Relative abundance proportion corresponds to the sum of the relative abundances of the trophic groups inside the trophic class normalized within the local food web to sum one. Link probability corresponds to the probability of interaction between two classes given the links between their respective trophic groups and the relative abundances of these groups. The widths of the edges are scaled by the square root of the changes in link probability. Size of the nodes are proportional to the value of change in relative abundances proportion within the soil food web, indicated with numbers. Nodes are distributed vertically based on their trophic level from the bottom (basal levels) to the top (higher levels).
The observed shifts in the structure of soil food webs could translate into impacts on multiple ecosystem functions, including carbon and nutrient fluxes, and plant productivity21,22. Recent studies have observed a slowdown in soil C and N cycles following severe outbreaks in these forests and have related this result to the decrease in the below-ground C-allocation to the rhizosphere and the decrease of ectomycorrhizal fungi37,51. An interesting avenue would be to relate how other components of the food web diversity (e.g. decomposer channel) contribute to the C:N stoichiometry to derive predictions on the long-term effects of these important disturbances on biogeochemical cycles.
The spatial extent of the study was limited to two landscape areas of ca 20 km extent, and we acknowledge that further monitoring would be required to assess the full extent of soil food webs responses to moth outbreaks in subarctic birch forests. Previous studies have found that the effect of moth outbreaks on biological communities can vary depending on local productivity and climatic conditions (as represented by the two areas Tana and Kirkenes)33,34,42. We found, however, a consistent response for most soil organisms across the two areas that translated into significant local changes in the whole soil food web diversity and composition. The consistency and strength of the effects of defoliation on the different facets of local soil food webs point to general conclusions on the bottom-up cascading effects of moth outbreaks on soil communities in these subarctic birch forests, despite the heterogeneity in environmental condition of the studied system.
Conclusion
The multitrophic approach used in this study, which combines an exhaustive diversity sampling (here eDNA data) with current trophic knowledge, an extended soil food web approach and ecological network theory, allows understanding the cascading effects of disturbances on soil biodiversity. We demonstrated that recent moth outbreaks in birch forests of Northern Fennoscandia caused major local shifts in the diversity and relative abundance of most trophic groups, ultimately changing the structure of the soil food web. We found more diverse soil food webs in defoliated forests compared to undamaged forests, accompanied by an increase in the proportion of groups in higher trophic levels.
We emphasize the need to consider different levels of resolution to ensure the robustness of conclusions and improve our understanding of how soil diversity responds to disturbances. Highly resolved food webs allow to map the cascading effects by revealing the variability of organisms’ responses. In contrast, low resolution food webs provide a general picture on how these changes affect the food web structure. Our study opens new prospects in understanding the response of complex and diverse food webs to disturbance.
Material and methods
Sampling
The study took place in the Varanger region at approximately 70° N, 29° E, Northeastern Norway. This region is located in the transition between subarctic deciduous forests and the arctic tundra. Periodic outbreaks of the autumn moth (Epirrita autumnata) and more recently the winter moth (Operophtera brumata) have occurred in the region with a 9–10-years frequency approximately. Recently, the consecutive episodes by the two species caused a severe mortality of birch trees35. Sampling was replicated in two areas located approximately 70 km apart, both at the border of the outbreak range, but with slight differences in the defoliation year: Tana (70°03′ N, 27°45′ E.), defoliated during 2006–2007, and Kirkenes (69°46′ N, 29°20′ E) defoliated during 2007–2009. Differences in the forest characteristics between these two areas allow to control for the influence of the initial forest characteristic on the effect of defoliation, that has been proved to be non-negligible in past studies34,38,42. In each area, stations along a linear transect were previously established from highly impacted forest stands to undamaged stands38,42. In order to maximise the differences between defoliated and undamaged forests we selected the two stations at one extreme of the transect corresponding to defoliated forest, i.e. almost all tree stems dead or heavily damaged, and the two stations at the other extreme of the transect corresponding to undamaged forest, i.e. all trees alive, based on the damage-scores measured in38,42 (Fig. 1). The two adjacent stations, separated from at least 2 km within defoliated or undamaged forests, were considered as local replicates and were surrounded by a large area of forest in their same condition, i.e. defoliated or undamaged. Defoliated and undamaged stations within an area were ca. 20 km apart. In July 2017, we sampled in each station 15 soil cores along an L-shaped transect with 10 m distances between neighbouring cores, corresponding to the biological replicates at the plot scale and aiming to account for microhabitat heterogeneity. This sampling design allowed to account for the local heterogeneity at different spatial scales (from meters to kilometres) and it was a good compromise for covering sufficiently local diversity across groups of varying spatial distributions57, while already minimizing spatial autocorrelation as it has been shown for earthworms and bacteria (> 5 m between soil samples58,59). Soil corers were cleaned and flame sterilized between each sample collection. Extracellular DNA was then extracted from 15 g as described in60,61. Botanical surveys were conducted and consisted of annotating the species present in the vicinity (1 m2) of each soil core.
Laboratory analyses
DNA extractions were conducted at the field on a mobile field unit. PCR, sequencing and soil physico-chemical analyses were performed at the Laboratoire d’Écologie Alpine (LECA) in Grenoble, France. Physicochemical soil properties were quantified from soil cores, including soil organic matter content (%), pH, soil moisture and C (%), N (%) and P content.
DNA extraction, PCR and sequencing negative controls were included in the experiment and used to identify potential contaminants and to control for false positives caused by tag‐switching events. In order to set extracellular DNA (eDNA) free from clay and silica particles, each sample was rotatively shaken for 15 min in a 15 ml saturated phosphate buffer solution (Na2HPO4; 0.12 M; pH ≈ 8). Two ml of sediment/buffer mixture were then sampled and centrifuged for 10 min at 10,000 g. A 400 µl aliquot of supernatant was recovered and used as starting material for eDNA extraction using NucleoSpin® Soil extraction kit (Macherey–Nagel GmbH, Düren, Germany), following manufacturer’s instructions except skipping the lysis cell step60. After elution, DNA extracts were diluted 10 times before being used as template for amplification. Eight negative extraction controls were also performed.
DNA amplification and sequencing
To assign the sequence reads to their relevant samples after high-throughput sequencing, we added unique eight base-long tags (with at least five differences between each other) to the 5’ end of each primer (modified from62,63). DNA amplifications were carried out in a final volume of 20 μl containing 2 μl of DNA sample, 10 μl of AmpliTaq Gold 360 Master Mix 2X (Applied Biosystems™, Foster City, CA, USA), 2 μl of primers mix at initial concentration of 5 μM of each primer and 0.16 μl of Bovine Serum Albumin. A total of 10 PCR negative and six positive PCR controls were included. Each sample (including all controls) was amplified in quadruplicate. Eukaryotes, Fungi and Protists were targeted using the respective DNA markers: Euka02 (18S rRNA gene), Fung02 (ITS1) and Bact01 (16S rRNA gene) described in20. PCR thermocycling conditions were as follow: after an initial step of 10 min at 95 °C, the mixtures underwent 45 cycles of 30 s at 95 °C, 30 s at 57–55–45 °C (Bact01, Fung02, Euka02, respectively) and 60 s at 72 °C, followed by a final elongation at 72 °C for 7 min. The amplification success was checked using capillary electrophoresis (QIAxcel System; Qiagen). PCR products were mixed in an equi-volume way (15 µl each) and 8 aliquots of 100 µl of the resulting mix were then purified using MinElute Purification kit (Qiagen GmbH, Hilden, Germany). Purified products were then pooled together before sequencing. This later was performed by pair-end sequencing on Illumina HiSeq 2000 platform (2*125 for Euka02, and 2*250 for both Bact01 and Fung02) at Fasteris, Geneva, Switzerland.
Bioinformatics
Sequences from the three libraries were pre-processed using the OBITools software64. Forward and reverse paired-end reads were assembled based on their overlapping 3’-end sequences, demultiplexed and dereplicated. We then removed sequences with low paired‐end alignment scores, singletons, short sequences and sequences containing ambiguous bases, as well as PCR errors using the obiclean command. Molecular Operational Taxonomic Units were built by clustering sequences at 97% of similarity using SUMACLUST65. Taxonomic annotations were performed with the SILVAngs pipeline (Quast et al. 2013), using the SILVA version 132 for Bact02 and Euka01. For Fung02 and Euka01 (only metazoa), we used the ecotag command from the OBITools, and the EMBL database version 136. Taxonomic annotations with > 75% identities were retained. Cross-sample contaminations and reagent contaminants were removed on the basis of negative and empty controls, and dysfunctional PCRs were detected and removed following the procedures described in66 with the metabaR R package67,68. For each marker, non-targeted taxa were eliminated. For Euka01 marker, we also excluded MOTUs identified as fungi, plants, and non-soil animals. After curation, PCR replicates were pooled together into samples. Only remaining common samples between the three MOTU tables were retained (n = 86). Number of reads, MOTUs, PCR replicates and samples before and after the curation process are available in Supplementary Table 2.
Soil food webs
Using current knowledge on soil organisms, we classified the MOTUs, based on their taxonomic annotations, into 9 broad trophic classes, using a classic soil food web backbone (e.g.22,27). These trophic classes included primary consumers, decomposers, phytophagous or plant parasites, mycorrhizal fungi, bacterivores, fungivores, omnivores, predators and animal parasites (Fig. 2a). Next, we defined 37 finer trophic groups by separating phylogenetic distant groups that could have a different set of prey/predators (e.g., bacterivore mites and bacterivore nematodes) or groups differing in their resources acquisition strategy (e.g. different types of mycorrhiza and saprotrophs). The definition on the trophic groups was made in accordance with the information available and the taxonomic resolution of the marker (Fig. 2b, Supplementary Fig. 1, Supplementary Table 1). For example, we kept collembola as a unique trophic group because the marker Euka02 was not resolutive enough to assign the MOTUs of this group to the family level, which was needed to a finer trophic classification. We kept both levels of resolution for the analyses, i.e., trophic class and trophic group. The databases used for the taxonomic assignment were FUNGuild69 for fungi, FAPROTAX70 for bacteria, NEMAguild69 and Nemaplex (http://nemaplex.ucdavis.edu/) for nematodes, and the main references used included71 for protists (and72 for cercozoa), and73 for heterotrophic bacteria (i.e. copiotrophic and oligotrophic classification). The main taxonomic clades composing the trophic classes and groups are in Supplementary Table 3. Specific criteria used to define the trophic classes and groups for each kingdom are in Supplementary Table 4. A table for each kingdom including the list of taxa, the trophic groups assignment, the taxonomic level of assignment and the references or databases used is available on Supplementary files.
The MOTU diversity of each trophic group was estimated per sample using the Shannon diversity (i.e. the exponential of the Shannon entropy) since this is a relevant measure for eDNA data74. In eDNA metabarcoding studies, changes in the abundance/biomass of an individual taxon may be inferred, in some extents, from changes in their relative abundances across samples, although this correspondence can be noised by different biological or technical factors (reviewed in20). However, some taxon can exhibit higher gene copies than others, making these changes in relative abundance more difficult to compare across groups contrary to other abundance standardized measures such as biomass. Relative abundances were thus estimated using a double-transformation. First, the total read counts of each trophic group were converted to proportions within a sample, and second, the resulting proportions were standardized by the largest observed proportion across all samples for each trophic group. Relative abundance of each group varied from 0 (absent) to 1 (largest observed diversity/proportion), allowing to obtain comparable measures across groups. Relative abundances of trophic classes were calculated by summing the relative abundances of the trophic group included in the trophic class31.
The metaweb, which contains the potential trophic interactions of the soil food webs of the system under study, was built for trophic classes and trophic groups45. Trophic links between trophic groups and trophic classes were added based on the main feeding preferences. Some constraints were added when assigning the trophic links between trophic groups based on (1) the organisms size, i.e. predators fed only on smaller preys, with some exceptions like animal parasites and omnivore nematodes that can eat larger preys, and macroorganisms did not interact with microorganisms, (2) habitat differentiation, i.e. strict plant endoparasites (i.e. protists) were not considered as prey of other free living predators, and (3) feeding preferences, e.g. fungivores fed only on saprotrophic fungi and Ectomycorrhizal, which are preferred to arbuscular mycorrhizal fungi75. The complete metaweb of trophic groups can be found in Supplementary Fig. 1 and the adjacency matrix is available in Supplementary files. Resource nodes were added to the food web representations with a structural purpose and corresponded to the main resources of the soil food web, i.e., sunlight, organic matter and plants, but were excluded from the diversity analyses, because the aim was to quantify the diversity of organisms within the soil food web. Differences in resources and plant composition between undamaged and defoliated forests were evaluated aside with multivariate analyses (see below). The metaweb was then used to characterize the composition and structure of the local soil food webs based on the trophic classes or groups detected locally in each soil sample (n = 86), assuming that classes or groups present locally interact as in the metaweb. For the local soil food webs at the trophic group resolution, nodes corresponded to the local relative abundance of the groups and links were binary (i.e., present or absent) assuming an interaction when the two groups concerned were present. For the trophic class resolution, nodes corresponded to the sum of the relative abundances of the trophic groups inside the trophic class and the links were weighted by the probability of interaction between two classes given the links between their respective trophic groups and the relative abundances of these groups as a proxy for the probability of an encounter31.
Statistical analyses
Differences in resources and plant composition between undamaged and defoliated forests were evaluated with multivariate analyses. A correspondence analysis was run to evaluate the differences in plant community composition. Plant communities from undamaged forests were mostly associated with ericaceous dwarf shrubs such as Empetrum nigrum and Vaccinium spp., but also of other shrubs and herbs in Kirkenes, e.g., Salix sp., Betula nana, Equisetum sp. (Supplementary Fig. 2). In defoliated forests plant composition was more variable among samples, including several species of grass and herbs, such as the dominant Avenella flexuosa. For the soil physico-chemical characteristics that we measured, the first two axes of a Principal Component Analysis explained 74.7% of the variance. The first axis was related to soil organic matter (SOM) and the second axis was related to the litter quality (measured with the C/N ratio) and inversely to soil acidity (i.e. pH) (Supplementary Fig. 3). Samples from defoliated forests were related to higher values of SOM, C, N, P and pH and lower C/N values.
To assess the effect of moth defoliation on MOTU diversity and the relative abundance of the trophic groups, a multilevel linear model was applied separately to each trophic group using the function ‘stan_lmer’ from the R package Rstanarm76 with the default priors. In each model, a fixed effect for defoliation was included as a dummy variable (0 corresponding to the undamaged forest and 1 to the defoliated forests). To account for the structure of the sampling design, i.e. soil cores clustered within stations and stations clustered within areas, we added a nested random term for stations within area to the intercept, where station was a factor with 8 levels and area a factor with 2 levels. Note that random factors allowed for borrowing information from each station and area, and that using a Bayesian approach led to non-zero estimates of area and station random effects, contrary to approaches using REML. Even if we suspected that the effect of defoliation could vary between the areas due to the contrasting habitat characteristics of Kirkenes and Tana, preliminary analyses showed that the effect was similar for both areas (i.e. the coefficient of the interaction between area and defoliation was small and 95% CI widely overlapped with 0 for most groups). MOTU diversity was standardized by the largest diversity observed across samples for each trophic group, to obtain comparable effect sizes across groups. A Yeo-Johnson transformation was applied to the relative abundances to improve the distribution of the residuals. Each model was run with 4 parallel MCMC chains with 15,000 iterations each. Model convergence was assessed visually and by checking Rhat < 1.10 for all the parameters. The normality of residuals was evaluated visually by using quantile–quantile (Q–Q) plots, and residuals were plotted against fitted values to assess outliers or influential values.
To study changes in the structure of local food webs, we estimated network diversity indices using the R package econetwork31. It allows computing several diversity indices on groups and link abundances using a viewpoint parameter that control the importance given to low vs. high relative abundances. We used a measure of dissimilarity of node and link compositions at different resolutions (trophic group and trophic class) to analyse whether there was a change in the structure of local soil food webs due to defoliation. A mixed multivariate distance matrix regression was then run using the dissimilarity matrix as the response, including a dummy variable for defoliation as a predictor and accounting for the nested sampling design as a random effect using the R package MDMR77. Local diversity (α-diversity) was estimated as the generalised mean of local diversity within each category of defoliation (i.e. defoliated forest and undamaged forest) within each area (Tana and Kirkenes). Both network local diversity and dissimilarity were computed using 1 as viewpoint parameter (eta in the package). Using this value of parameter, local diversity is the exponential of Shannon entropy. Figures 2, 3, 4, 5 were made using the R software (R 3.6.3)68.
References
Pickett, S. T. A. & White, P. S. The Ecology of Natural Disturbance and Patch Dynamics (Academic Press, 1985).
IPBES. Global assessment report on biodiversity and ecosystem services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES Secretariat, 2019).
Brun, P. et al. Large-scale early-wilting response of Central European forests to the 2018 extreme drought. Glob. Change Biol. 00, 1–15 (2020).
Cardinale, B. J., Gonzalez, A., Allington, G. R. H. & Loreau, M. Is local biodiversity declining or not? A summary of the debate over analysis of species richness time trends. Biol. Conserv. 219, 175–183 (2018).
Vellend, M. et al. Global meta-analysis reveals no net change in local-scale plant biodiversity over time. Proc. Natl. Acad. Sci. U.S.A. 110, 19456–19459 (2013).
Bardgett, R. D. & Wardle, D. A. Aboveground-Belowground Linkages: Biotic Interactions, Ecosystem Processes, and Global Change (Oxford University Press, 2010).
Bardgett, R. D. & van der Putten, W. H. Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014).
Bardgett, R. D. & Caruso, T. Soil microbial community responses to climate extremes: Resistance, resilience and transitions to alternative states. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190112 (2020).
Thom, D. & Seidl, R. Natural disturbance impacts on ecosystem services and biodiversity in temperate and boreal forests: Disturbance impacts on biodiversity and services. Biol. Rev. 91, 760–781 (2016).
van der Putten, W. H. et al. Trophic interactions in a changing world. Basic Appl. Ecol. 5, 487–494 (2004).
Lafferty, K. D. & Suchanek, T. H. Revisiting Paine’s 1966 sea star removal experiment, the most-cited empirical article in the American Naturalist. Am. Nat. 188, 365–378 (2016).
Scherber, C. et al. Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature 468, 553–556 (2010).
Barnes, A. D. et al. Direct and cascading impacts of tropical land-use change on multi-trophic biodiversity. Nat. Ecol. Evol. 1, 1511–1519 (2017).
Barbier, M. & Loreau, M. Pyramids and cascades: A synthesis of food chain functioning and stability. Ecol. Lett. 22, 405–419 (2019).
Mancinelli, G. & Mulder, C. Chapter three—detrital dynamics and cascading effects on supporting ecosystem services. In Advances in ecological research Vol. 53 (eds Woodward, G. & Bohan, D. A.) 97–160 (Academic Press, 2015).
Mulder, C., Vonk, J. A., Hollander, H. A. D., Hendriks, A. J. & Breure, A. M. How allometric scaling relates to soil abiotics. Oikos 120, 529–536 (2011).
Allen, A. P. & Gillooly, J. F. Towards an integration of ecological stoichiometry and the metabolic theory of ecology to better understand nutrient cycling. Ecol. Lett. 12, 369–384 (2009).
de Ruiter, P. C., Neutel, A.-M. & Moore, J. C. Energetics, patterns of interaction strengths, and stability in real ecosystems. Science 269, 1257–1260 (1995).
Estes, J. A. et al. Trophic downgrading of planet earth. Science 333, 301–306 (2011).
Taberlet, P., Bonin, A., Zinger, L. & Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring (Oxford University Press, 2018).
Gravel, D., Albouy, C. & Thuiller, W. The meaning of functional trait composition of food webs for ecosystem functioning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150268 (2016).
Barnes, A. D. et al. Energy flux: The link between multitrophic biodiversity and ecosystem functioning. Trends Ecol. Evol. 33, 186–197 (2018).
Elton, C. S. Animal Ecology 1–256 (Macmillan Co., 1927). https://doi.org/10.5962/bhl.title.7435.
Bohan, D. A. et al. Next-generation global biomonitoring: Large-scale, automated reconstruction of ecological networks. Trends Ecol. Evol. 32, 477–487 (2017).
Roslin, T. & Majaneva, S. The use of DNA barcodes in food web construction—terrestrial and aquatic ecologists unite!. Genome 59, 603–628 (2016).
Cohen, J. E. et al. Improving food webs. Ecology 74, 252–258 (1993).
Buzhdygan, O. Y. et al. Biodiversity increases multitrophic energy use efficiency, flow and storage in grasslands. Nat. Ecol. Evol. 4, 393–405 (2020).
Martinez, N. D. Effects of resolution on food web structure. Oikos 66, 403 (1993).
Thompson, R. M. et al. Food webs: Reconciling the structure and function of biodiversity. Trends Ecol. Evol. 27, 689–697 (2012).
Kardol, P., Throop, H. L., Adkins, J. & de Graaff, M.-A. A hierarchical framework for studying the role of biodiversity in soil food web processes and ecosystem services. Soil Biol. Biochem. 102, 33–36 (2016).
Ohlmann, M. et al. Diversity indices for ecological networks: A unifying framework using Hill numbers. Ecol. Lett. 22, 737–747 (2019).
Pellissier, L. et al. Comparing species interaction networks along environmental gradients. Biol. Rev. 93, 785–800 (2017).
Jepsen, J. U. et al. Ecosystem impacts of a range expanding forest defoliator at the forest-tundra ecotone. Ecosystems 16, 561–575 (2013).
Karlsen, S. R., Jepsen, J. U., Odland, A., Ims, R. A. & Elvebakk, A. Outbreaks by canopy-feeding geometrid moth cause state-dependent shifts in understorey plant communities. Oecologia 173, 859–870 (2013).
Jepsen, J. U., Hagen, S. B., Ims, R. A. & Yoccoz, N. G. Climate change and outbreaks of the geometrids Operophtera brumata and Epirrita autumnata in subarctic birch forest: Evidence of a recent outbreak range expansion. J. Anim. Ecol. 77, 257–264 (2008).
Vindstad, O. P. L., Jepsen, J. U., Ek, M., Pepi, A. & Ims, R. A. Can novel pest outbreaks drive ecosystem transitions in northern-boreal birch forest?. J. Ecol. 107, 1141–1153 (2019).
Sandén, H. et al. Moth outbreaks reduce decomposition in subarctic forest soils. Ecosystems 23, 151–163 (2019).
Vindstad, O. P. L. et al. Numerical responses of saproxylic beetles to rapid increases in dead wood availability following geometrid moth outbreaks in sub-arctic mountain birch forest. PLoS ONE 9, e99624 (2014).
Nilsson, M.-C. & Wardle, D. A. Understory vegetation as a forest ecosystem driver: Evidence from the northern Swedish boreal forest. Front. Ecol. Environ. 3, 421–428 (2005).
Bråthen, K. A. & Ravolainen, V. T. Niche construction by growth forms is as strong a predictor of species diversity as environmental gradients. J. Ecol. 103, 701–713 (2015).
Bråthen, K. A., Gonzalez, V. T. & Yoccoz, N. G. Gatekeepers to the effects of climate warming? Niche construction restricts plant community changes along a temperature gradient. Perspect. Plant Ecol. Evol. Syst. 30, 71–81 (2018).
Vindstad, O. P. L., Jepsen, J. U. & Ims, R. A. Resistance of a sub-arctic bird community to severe forest damage caused by geometrid moth outbreaks. Eur. J. For. Res. 134, 725–736 (2015).
Parker, T. C. et al. Slowed biogeochemical cycling in sub-arctic birch forest linked to reduced mycorrhizal growth and community change after a defoliation event. Ecosystems 20, 316–330 (2017).
Saravesi, K. et al. Moth outbreaks alter root-associated fungal communities in subarctic mountain birch forests. Microb. Ecol. 69, 788–797 (2015).
Dunne, J. A. The network structure of food webs. In Ecological Networks: Linking Structure to Dynamics in Food Webs (eds Pascual, M. & Dunne, J. A.) 27–86 (Oxford University Press, 2006).
Rodriguez-Ramos, J. C. et al. Changes in soil fungal community composition depend on functional group and forest disturbance type. New Phytol. 00, 1–13 (2020).
Decaëns, T. Macroecological patterns in soil communities. Glob. Ecol. Biogeogr. 19, 287–302 (2010).
Bardgett, R. D., Yeates, G. W. & Anderson, J. M. Patterns and determinants of soil biological diversity. In Biological Diversity and Function in Soils (eds Hopkins, D. et al.) 100–118 (Cambridge University Press, 2005).
Worm, B. & Duffy, J. E. Biodiversity, productivity and stability in real food webs. Trends Ecol. Evol. 18, 628–632 (2003).
Ponsard, S., Arditi, R. & Jost, C. Assessing top-down and bottom-up control in a litter-based soil macroinvertebrate food chain. Oikos 89, 524–540 (2000).
Kristensen, J. Å., Rousk, J. & Metcalfe, D. B. Below-ground responses to insect herbivory in ecosystems with woody plant canopies: A meta-analysis. J. Ecol. 108, 917–930 (2020).
González, V. T. et al. Batatasin-III and the allelopathic capacity of Empetrum nigrum. Nord. J. Bot. 33, 225–231 (2015).
Veen, G. F. et al. The role of plant litter in driving plant-soil feedbacks. Front. Environ. Sci. 7, 168 (2019).
Calizza, E., Rossi, L., Careddu, G., Sporta Caputi, S. & Costantini, M. L. Species richness and vulnerability to disturbance propagation in real food webs. Sci. Rep. 9, 19331 (2019).
Antiqueira, P. A. P., Petchey, O. L., dos Santos, V. P., de Oliveira, V. M. & Romero, G. Q. Environmental change and predator diversity drive alpha and beta diversity in freshwater macro and microorganisms. Glob. Change Biol. 24, 3715–3728 (2018).
Hedlund, K. et al. Trophic interactions in changing landscapes: Responses of soil food webs. Basic Appl. Ecol. 5, 495–503 (2004).
Ettema, C. H. & Wardle, D. A. Spatial soil ecology. Trends Ecol. Evol. 17, 177–183 (2002).
O’Brien, S. L. et al. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 18, 2039–2051 (2016).
Jiménez, J. J., Decaëns, T., Lavelle, P. & Rossi, J.-P. Dissecting the multi-scale spatial relationship of earthworm assemblages with soil environmental variability. BMC Ecol. 14, 26 (2014).
Taberlet, P. et al. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 21, 1816–1820 (2012).
Zinger, L. et al. Extracellular DNA extraction is a fast, cheap and reliable alternative for multi-taxa surveys based on soil DNA. Soil Biol. Biochem. 96, 16–19 (2016).
Binladen, J. et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2, e197 (2007).
Valentini, A. et al. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Mol. Ecol. Resour. 9, 51–60 (2009).
Boyer, F. et al. obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182 (2016).
Mercier, C., Boyer, F., Bonin, A. & Coissac, E. SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. in Programs and Abstracts of the SeqBio 2013 workshop. Abstract 27–29 (Citeseer, 2013).
Zinger, L. et al. DNA metabarcoding—Need for robust experimental designs to draw sound ecological conclusions. Mol. Ecol. 28, 1857–1862 (2019).
Zinger, L. et al. metabaR : an R package for the evaluation and improvement of DNA metabarcoding data quality. https://doi.org/10.1101/2020.08.28.271817 (2020).
R Core Team. A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2019).
Nguyen, N. H. et al. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016).
Louca, S., Parfrey, L. W. & Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277 (2016).
Adl, S. M. et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019).
Fiore-Donno, A. M. et al. Functional traits and spatio-temporal structure of a major group of soil protists (Rhizaria: Cercozoa) in a temperate grassland. Front. Microbiol. 10, 1332 (2019).
Ho, A., Lonardo, D. P. D. & Bodelier, P. L. E. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93, 6 (2017).
Calderón-Sanou, I., Münkemüller, T., Boyer, F., Zinger, L. & Thuiller, W. From environmental DNA sequences to ecological conclusions: How strong is the influence of methodological choices?. J. Biogeogr. 47, 193–206 (2020).
Antunes, P. M. & Koyama, A. Chapter 9 - Mycorrhizas as Nutrient and Energy Pumps of Soil Food Webs: Multitrophic Interactions and Feedbacks. in Mycorrhizal Mediation of Soil Fertility, Structure, and Carbon Storage (eds. Johnson, N. C., Gehring, C. & Jansa, J.) 149–173 (Elsevier, 2017).
Goodrich, B., Gabry, J., Ali, I. & Brilleman, S. rstanarm: Bayesian applied regression modeling via Stan. (R package version 2.21.1, 2020).
McArtor, D. B., Lubke, G. H. & Bergeman, C. S. Extending multivariate distance matrix regression with an effect size measure and the asymptotic null distribution of the test statistic. Psychometrika 82, 1052–1077 (2017).
Acknowledgements
This research was funded by the French Agence Nationale de la Recherche (ANR) through the GlobNet (ANR-16-CE02-0009) and EcoNet (ANR-18-CE02-0010) projects. We thank Clement Lionnet and Nicolas Leguillarme for support with the development of bioinformatic pipelines, Cindy Arnoldi for the soil physicochemical analyses and Julien Renaud for providing the map in Fig.1b. Jane Jepsen et Ole Petter Vindstad contributed to the study design and information about the defoliation and ecosystem changes.
Author information
Authors and Affiliations
Contributions
W.T. and N.G.Y. conceptualize the overall idea, and together with I.C.S. and T.M. conceived the research aims and analyses. L.Z., H.S., L.G. and N.G.Y. conducted the field sampling and L.G. ran the laboratory procedures. I.C.S., L.Z., H.S., M.H. and S.S.M. conducted the trophic data collection. I.C.S. conducted the bioinformatics and statistical analyses and led the writing with significant contributions from all co‐authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Calderón-Sanou, I., Münkemüller, T., Zinger, L. et al. Cascading effects of moth outbreaks on subarctic soil food webs. Sci Rep 11, 15054 (2021). https://doi.org/10.1038/s41598-021-94227-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94227-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.