Abstract
Skeletal muscle growth plays a critical role during porcine muscle development stages. Genome-wide transcriptome analysis reveals that long intergenic non-coding RNAs (lincRNAs) are implicated as crucial regulator involving in epigenetic regulation. However, comprehensive analysis of lincRNAs in embryonic muscle development stages remain still elusive. Here, we investigated the transcriptome profiles of Duroc embryonic muscle tissues from days 33, 65, and 90 of gestation using RNA-seq, and 228 putative lincRNAs were identified. Moreover, these lincRNAs exhibit the characteristics of shorter transcripts length, longer exons, less exon numbers and lower expression level compared with protein-coding transcripts. Expression profile analysis showed that a total of 120 lincRNAs and 2638 mRNAs were differentially expressed. In addition, we also performed quantitative trait locus (QTL) mapping analysis for differentially expressed lincRNAs (DE lincRNAs), 113 of 120 DE lincRNAs were localized on 2200 QTLs, we observed many QTLs involved in growth and meat quality traits. Furthermore, we predicted potential target genes of DE lincRNAs in cis or trans regulation. Gene ontology and pathway analysis reveals that potential targets of DE lincRNAs mostly were enriched in the processes and pathways related to tissue development, MAPK signaling pathway, Wnt signaling pathway, TGF-beta signaling pathway and insulin signaling pathway, which involved in skeletal muscle physiological functions. Based on cluster analysis, co-expression network analysis of DE lincRNAs and their potential target genes indicated that DE lincRNAs highly regulated protein-coding genes associated with skeletal muscle development. In this study, many of the DE lincRNAs may play essential roles in pig muscle growth and muscle mass. Our study provides crucial information for further exploring the molecular mechanisms of lincRNAs during skeletal muscle development.
Similar content being viewed by others
Introduction
Skeletal muscle is an important component of the body in mammals, mainly involved in the growth and development of the body. Skeletal muscle abnormalities can lead to physical dysfunction such as Muscular dystrophy, idiopathic inflammatory myopathies and cardiomyopathy1,2,3,4. During the past decades of molecular biology study, great progress has been made on the molecular mechanism underlying the growth and development of porcine skeletal muscle5, for example, MyoD, MyF5 and MRF4 are involved in myogenesis and differentiation6,7,8. Additionally, studies have found that insulin-like growth factors (IGF1) can act as an activator of MAPK/ERK and PI3K/Akt signaling pathways to promote the proliferation and differentiation of muscle cells9, and IGF1 mediated pathway that the IGF1–Akt–mTOR pathway has been found to participate in positive regulation of muscle growth10,11. In recent years, the emergence of long intergenic non-coding RNA has become a new research hotspot in the molecular biological field, which provides a new way to advance the research on the mechanism of skeletal muscle development.
Long intergenic non-coding RNAs, which are a new class of RNA molecules longer than 200 nucleotides with little or no protein-coding capacity12. Recent evidences have established that lincRNAs have a significant role in regulating gene expression at epigenetic, transcriptional and post transcriptional levels13,14, they can perform essential functions during basic biological processes, such as chromatin modification15, imprinting16,17 , maintenance of pluripotency18. With the emergence and widespread application of high-throughput sequencing technology, thousands of lincRNAs have been identified in genome-wide analysis, more and more lincRNAs have been functionally validated. A recent study indicated that lincRNA-p21 is involved in regulating the proliferation and apoptosis of vascular smooth muscle cells by enhancing the activity of P53, providing a new target for the treatment of atherosclerosis19. Currently, studies on lincRNAs in porcine embryo development are less well understood, therefore, our analysis in the differences of lincRNAs at embryonic development stages will provide a good model for studying the mechanisms that regulate skeletal muscle development.
In the present study, we applied RNA sequencing to characterize global gene expression patterns of muscle tissues from Duroc on days 33, 65, and 90 and systematically analyzed the muscle expression profile during porcine skeletal muscle development20. We identified 228 putative lincRNAs and found that many lincRNAs differentially expressed. Moreover, we predicted the potential target genes of DE lincRNAs by cis or trans ways. Gene Ontology and pathways enrichment analysis showed that lincRNAs potentially regulated the process of protein-coding genes. An interactive network was performed to elucidate the interplay between DE lincRNAs and their potential target genes. This study of skeletal muscle of transcriptome profiles will provide a useful resource to further explore the understanding of mechanisms, besides, elucidating the underlying mechanisms of skeletal muscle growth and development will be helpful for the improvement of production benefits of pig.
Summary of RNA-seq data mapping and transcripts assembly in Duroc
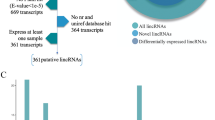
In this study, we downloaded 9 RNA-seq libraries which contained 647,779,568 paired-end reads from the NCBI during three embryonic muscle developmental stages of Duroc. Samples were named separately 33d_1, 33d_2, 33d_3, 65d-1, 65d_2, 65d_3, 90d-1, 90d_2, 90d_3. After trimming and filtering, a total 636,985,862 clean reads were mapped to the annotated Sscrofa11.1 genome using HISAT2, we founded that approximately 95% of the quality-filtered reads were mapped and over 70% of the reads could be uniquely mapped to the genome (Table 1). Based on the result, StringTie was used to reconstruct the transcripts and merge into a file that obtained 70,869 transcripts. The RNA-seq process for identifying lincRNAs was shown in Fig. 1. Finally, 228 putative lincRNAs were identified, there were 191 lincRNAs that have been annotated in the pig reference genome database, and these known lincRNAs were distributed throughout all chromosomes. The remaining 37 lincRNAs have no overlap with the pig annotation database, they were separately distributed on chromosomes 3 to 8 and 11 to 18, and chromosome 17 was found to have the highest novel lincRNAs density.
Characteristics analysis of identified lincRNAs
Previous study showed that the difference of lncRNAs with protein-coding genes in pig21. However, sequence characteristic of lincRNAs during embryonic muscle development remains unclear. Based on the annotated information for the pig reference genome, we examined the characteristic of putative lincRNAs in transcript length, exon length, exon numbers and expression level compared with protein-coding genes. As a result, we observed that the average transcript length of known lincRNAs, novel lincRNAs and protein-coding genes were about 1377 bp, 1203 bp and 3296 bp, respectively. It followed that novel lincRNAs were similar to known lincRNAs and shorter than protein-coding genes in transcript length (Fig. 2A). In addition, the average exon length of known lincRNAs, novel lincRNAs and protein-coding genes were 515 bp, 505 bp and 284 bp, respectively. Although the average transcript length of lincRNAs was shorter, the average exon length of lincRNAs was longer than that of protein-coding genes (Fig. 2B). In exon numbers, our result showed that the exon numbers of lincRNAs were gathered at 2–5, while the average exon numbers of protein-coding genes were 11.6, we noticed that this result was consistent with the above two research (Fig. 2C). In normalized read counts expression level (FPKM), the average value of known lincRNAs, novel lincRNAs and protein-coding genes were 1.2, 0.9 and 4.7, respectively. We concluded that lincRNAs had a lower expression level compared with protein-coding genes. In general, lincRNAs were shorter in transcript length, but longer in exon length, had fewer exon, and were expressed at lower level compared with protein-coding genes (Fig. 2D). Which were highly consistent with previous reports22,23.
Differential expression analysis of lincRNAs
To evaluate the differences in gene expression patterns during three developmental stages, DEseq2 was used to identify differentially expressed lincRNAs and protein-coding genes between two paired samples (D33 vs. D65; D65 vs. D90; D33 vs. D90). when |fold change|≥ 1 and adjusted p -value ≤ 0.05, there were 66 DE lincRNA genes including 50 upregulated and 16 downregulated identified between Day 33 and 65 (Fig. 3A), 29 DE lincRNA genes including 12 upregulated and 17 downregulated identified between Day 65 and 90 (Fig. 3B), 74 DE lincRNA genes including 48 upregulated and 26 downregulated identified between Day 33 and 90 (Fig. 3C). All DE lincRNAs in three groups were distributed in Fig. 3D. In addition, when |fold change|≥ 2 and adjusted p-value ≤ 0.01, a total 2638 DE protein-coding genes were identified (Fig. 3E).
Heat map of differential expression analysis of lincRNAs and protein-coding genes during three developmental stages. (A) Heat maps of differentially expressed lincRNAs in D33 vs. D65 group; (B) Heat maps of differentially expressed lincRNAs in D65 vs. D90 group; (C) Heat maps of differentially expressed lincRNAs in D33 vs. D90 group; (D) Histogram of differentially expressed lincRNAs among the three groups; (E) Heat maps of differentially expressed protein-coding genes among the three groups.
QTL localization and functional enrichment
QTL is closely associated with many traits. To explore the relationship between differentially expressed lincRNAs and QTL traits, we performed a correlation analysis by mapping DE lincRNAs to the QTL regions related to pig traits, the pig QTL database contains 31,455 QTLs, representing 695 different traits24. Our analysis results showed that 113 of 120 DE lincRNAs were located in 2200 QTL, which corresponded to 331 traits, 27 trait types, 4 trait classes. The greatest number of QTLs were associated with the trait “Meat and Carcass Traits”, accounting for about 59% of the total QTLs. The second highest number of QTL traits “Production Traits” accounted for 11% of the total QTLs (Fig. 4A). We statistically analyzed localization in QTLs associated with muscle, obesity, and growth traits, and found that most DE lincRNAs were targeted at the three trait types. Notably, 100 of 113 DE lincRNAs were closely associated with growth and 86 DE lincRNAs were located in muscle related traits, from this we hypothesized that DE lincRNAs could have an important effect on muscle growth and development (Fig. 4B). Furthermore, we examined the distribution of these QTLs on chromosomes, and found that QTLs were distributed on all chromosomes. Interestingly, the greatest number of QTLs for the three traits were located on chromosome 4 and chromosome 6 (Fig. 4C).
Prediction of potential target genes of DE lincRNAs
Previous studies have shown that lincRNAs can regulate the expression of target genes by cis or trans via, and participate in the functional regulation of some organisms25,26. Firstly, we predicted potential target genes of DE lincRNAs in cis regulation to determine the possible function of DE lincRNAs by searching for protein-coding genes around 100 kb upstream and downstream of DE lincRNAs. We found 303 protein-coding genes were close to DE lincRNAs. GO enrichment analysis showed that 65 of 303 protein-coding genes were assigned to 9 GO terms which mainly involved in the biological processes of transcriptional regulation (Table 2). Furthermore, we performed Pearson correlation analysis revealed that 37 potential target genes (PTGs) were highly correlated with 29 DE lincRNAs (r ≥ 0.8, p-value ≤ 0.01). Among them, 12 of 37 PTGs differentially expressed. Meanwhile, most DE lincRNAs were positively correlated with their PTGs. MSTRG.6732 and MSTRG.2061 were significantly correlated with ERGIC1 and HMGB1. Besides, MSTRG.4842 and MSTRG.14169 could regulate their PTGs in two ways: positive regulation and negative regulation. The potential target genes for DE lincRNAs regulation were shown in Table 3.
Functional enrichment analysis of PTGs associated with DE lincRNAs
Furthermore, we also predicted the potential target genes from DE lincRNAs in trans regulation, and acquired 4609 PTGs corresponding to 50 DE lincRNAs (r ≥ 0.96, adjusted p-value ≤ 0.01). Among these genes, 548 PTGs were differentially expressed in groups as DEPTGs. Which suggested that most of lincRNAs regulated gene expression through trans regulation. GO enrichment analysis showed that 4609 PTGs were enriched in 547 biological processes and 548 DEPTGs were enriched in 287 biological processes. In cases of biological process. Some GO terms were significantly associated with muscle development and energy metabolism, such as skeletal muscle tissue development, muscle contraction, cell proliferation, protein catabolic process, insulin receptor signaling pathway and regulation of glucose transport (Fig. 5A; Fig. 5C). Besides, 4609 PTGs and 548 DEPTGs were enriched in 64 pathways and 28 pathways, respectively. KEGG pathways were involved in Wnt signaling pathway, ECM-receptor interaction, MAPK, calcium signaling, ErbB signaling pathway and TGF-beta signaling pathway (Fig. 5B; Fig. 5D). The results indicated that DE lincRNAs had an important role in regulating their potential target genes regulated composition and growth and development of muscle cells by muscle cells proliferate and differentiate, substance metabolism energy transport and conversion.
Gene ontology and pathway analysis of PTGs of DE lincRNAs. (A) Biological processes analysis associated with muscle growth of PTGs of DE lincRNAs; (B) Pathway analysis of associated with muscle growth of PTGs of DE lincRNAs; (C) Biological processes analysis associated with muscle growth of DEPTGs of DE lincRNAs; (D) Pathway analysis of associated with muscle growth of DEPTGs of DE lincRNAs.
Co expression network analysis of DE lincRNAs and DE potential target genes
To understand the relationship of expression between DE lincRNAs and their DEPTGs. The expression regulation relationship between 50 DE lincRNAs and 548 DEPTGs was analyzed, we calculated the interaction of DE lincRNAs and DEPTGs. Pearson correlation analysis results were presented that 860 pairs between DE lincRNAs and DEPTGs with positive correlation and 86 pairs with negative correlation were identified (Fig. 6A). We selected DE lincRNAs and DEPTGs related to skeletal muscle growth and development pathways to construct co-expression networks, and 24 DE lincRNAs exhibited a high co-expression relationship with 48 DEPTGs. Noticeably, DE lincRNA MSTRG.388, MSTRG.4602 and MSTRG.7020 were involved in the regulation of several DEPTGs (Fig. 6B). In order to further explore the function of DE lincRNAs, we investigated nine DEPTGs involving in muscle development related pathways corresponding to 13 DE lincRNAs, we found that SHH targeted by lincRNA MSTRG.27 and MSTRG.388 played an important role in myogenesis (Fig. 6C), and SHH had an essential inductive function in the early activation of the myogenic regulatory factors Myf‐5 and MyoD27,28. Besides, lincRNA MSTRG.4602, MSTRG.98 and MSTRG.243 regulated MYOZ1 that encoded calsarcin-2 protein participated in the expression of PPAR-Y2 in skeletal muscle29.
Correlation expression regulation analysis of DE lincRNA genes and their potential target genes. (A) Co-expression network diagram between DE lincRNA genes and DEPTGs; (B) Co-expression network diagram between DE lincRNA genes and DEPTGs enriched in skeletal muscle development related pathways; (C) The interaction of major DE lincRNA genes and DEPTGs enriched in muscle-related pathways.
Validation of lincRNA expression levels through qRT-PCR
According to the previous RNA-seq results, we selected nine pairs of DE lincRNA genes and their potential target genes and analyzed their expression levels by qRT-PCR (MSTRG.98 vs. CA4, MSTRG.98 vs. MYOZ1, MSTRG.243 vs. MYOZ1, MSTRG.4602 vs. MYOG, MSTRG.4602 vs. TGFB2, MSTRG.4602 vs. MAPK14, MSTRG.4602 vs. FOXO3, MSTRG.17803 vs. FAIM2, MSTRG.4034 vs. CA4) (Fig. 7). The experimental results showed that the correlation (r2) between DE lincRNAs and potential target genes were at above 0.86 and the p-values were less than 0.01. The experimental results of the qRT-PCR have a similar tend to the original Pearson correlation coefficient between DE lincRNAs and potential target genes.
Linear regression of DE lincRNAs and their DEPTGs expression. The r0 and p0 represent the Pearson correlation coefficient and p-value of each pair of differentially expressed lincRNA and its potential target gene, the r and p were calculated by qRT-PCR verification experiment. (A) MSTRG.98 vs. CA4. (B) MSTRG.98 vs. MYOZ1. (C) MSTRG.243 vs. MYOZ1. (D) MSTRG.4602 vs. MYOG. (E) MSTRG.4602 vs. TGFB2. (F) MSTRG.4602 vs. MAPK14. (G) MSTRG.4602 vs. FOXO3. (H) MSTRG.17803 vs. FAIM2. (I) MSTRG.4034 vs. CA4.
Discussion
Skeletal muscle growth and development are a complex process, which directly determine the meat production and quality in the pig industry. Skeletal muscle is mainly composed of muscle fibers, basement membrane, muscle satellite cells and nerves. Study found that the numbers of muscle fiber have been fixed before the pigs were born, indicating that muscle fiber development is mainly determined during the embryonic period30,31. Muscle fiber development takes place in two waves in pig embryonic, the first wave of muscle fiber formation occurs from 30 to 60 days and the second wave occurs from 45 to 90 days32,33. In our study, we investigated lincRNAs expression profile in days 33, 65, and 90, which included the primary, second, and final waves of muscle fiber development33. Even though the previous studies have showed lincRNAs associated with muscle growth in pig, the dynamic process of expression profile of lincRNAs in embryonic muscle fibers is rare, and our study provides theoretical basis for new exploration in the future.
Based on RNA-seq data published in NCBI, we compared whole gene expression profile in muscle tissue from Duroc in differential development periods. Through a series of transcriptome pipeline analysis, there were 228 putative lincRNAs identified using RNA-seq sequencing, we predicted 37 novel lincRNAs that were not annotated from the nine muscle libraries, which enrich the pig lincRNA annotation and the specific features need to be further investigated in the future. Moreover, we performed a characteristic analysis of putative lincRNAs, involving in transcript and exon length, exon numbers and FPKM, the results showed that the similar characteristic of shorter transcript length, longer exon length, fewer exons, and lower expression levels compared with previous reports34,35. Meanwhile, the reliability of the analysis is further improved.
We identified 120 DE lincRNAs and 2638 DE protein-coding genes based on a designed pipeline. Previous studies have shown that there were a large number of lncRNAs located within known QTL regions36. To understand the relation between DE lincRNAs and QTLs, we also performed QTL localization analysis for differentially expressed lincRNAs. Some QTLs were involved in large regions, so that multiple genes were located on the same QTL, or multiple QTLs had the same gene location. In among, the specific mechanism may need to be verified by subsequent experiments.
To explore the potential function of DE lincRNAs, we investigated the regulation of lincRNAs on gene expression through cis and trans regulation26. For the cis-regulation of DE lincRNAs, we observed that nearby target genes of differentially expressed lincRNAs were related to regulation of transcription, previous studies have confirmed that the porcine lincRNAs were more likely to be enriched in adjacent protein-coding genes that mediate transcriptional regulation, our study was in accordance with Zhao’s report37. In addition, some genes have been shown to be associated with muscle cell proliferation and fat deposition. For example, DLK1 was a critical factor in regulating skeletal muscle development and regeneration through Notch dependent38. Previous studies found that PPARA was involved in the regulation of fat deposition in porcine subcutaneous fat and longissimus dorsi muscle39,40. Besides, myofibrillar structural protein myomesin-3 (MYOM3) was not only associated with muscular dystrophy related proteins and muscle strength, which could be a potential biomarker for monitoring of muscular dystrophy, but was hypermethylated in ischemic cardiomyopathy (ICM)41,42. Among them, DEL-MSTRG. 31,882 and its potential target gene with patatin-like phospholipase domain containing 4 (PNPLA4) showed significant positive correlation at the expression level. Therefore, we inferred that DE lincRNAs modulated differences at different stages of development by regulating their potential target genes.
In this study, we identified many target genes of DE lincRNAs that play critical roles in skeletal muscle development. According to these results, we observed that numerous target genes significantly related to DE lincRNAs were involved in the biological processes of skeletal muscle development, such as ACTC1, FOXP1, IGF2BP3, MYOG, MYOZ1, MEF2A, SP1, TGFB2. Previous study reported ACTC1 was implicated in skeletal muscle fiber contraction43. In the study by Yang et al., we found that SP1 could act as a central regulator to coordinate skeletal muscle development. IGF2BP3 was a target of SP1 and considered to be a candidate gene for displaying DNA methylation and mRNA expression levels during skeletal muscle development44. In addition to the MRFs family, some studies reported MEF2 family could directly regulate myogenesis and morphogenesis45. It is noteworthy that SHH targeted by MSTRG.27 and MSTRG.388 is a key transcription factor that regulates the expression of myogenesis and genes related to muscle development. Studies have shown that SHH has significant impact on maintaining MYF5 gene expression and early muscle development46.
Subsequently, we investigated Gene Ontology and KEGG pathways analysis of potential target genes of DE lincRNAs, and found that skeletal muscle organ and tissue development processes, muscle contraction, striated muscle cell development were some of significantly enriched GO terms. These results suggest that identified DE lincRNAs have an important impact on the skeletal muscle. Regulation of glucose import, regulation of glucose transport, and insulin receptor signaling pathway also significantly enriched, which were important ways of obtaining and transporting energy, we deduce that DE lincRNAs could participate in the regulatory mechanism of skeletal muscle development through mediating cellular energy responses. Our KEGG pathway analysis showed that significantly enriched pathways including MAPK signaling pathway, TGF-beta signaling pathway, Wnt signaling pathway, ECM − receptor interaction, regulation of kinase activity. Previous studies have confirmed that TGF-beta signaling pathway contributes to muscle development in mice47. Moreover, the extracellular matrix (ECM) is a network of structures surrounding muscle fibers, providing a close connection with cell proliferation, differentiation and metabolism. Therefore, we infer that DE lincRNAs could contribute to the differences in skeletal muscle development. In addition, some cardiac diseases, such as viral myocarditis, dilated cardiomyopathy, and hypertrophic cardiomyopathy, were also significantly enriched, these results suggest that DE lincRNAs may have an important effect on myocardial development.
Conclusion
In the study, we identified 228 putative lincRNAs and analyzed the characteristics of lincRNAs transcriptome compared with protein-coding genes in embryonic muscle tissue of Duroc. We observed numerous differentially expressed lincRNAs and protein-coding genes during differential development stages. Functional enrichment analysis of potential target genes by DE lincRNAs revealed that many lincRNAs participated in muscle growth and development related processes and pathways. Co-expression networks indicated the functional relationship between protein-coding genes and lincRNAs. In summary, our work provides a valuable resource for future research into the potential functions of pig growth and development and is expected to promote the progress of pig production.
Materials and methods
Data sources
RNA-seq sequencing data containing nine samples was obtained from the NCBI SRA database. The accession numbers and reads of the RNA-seq data were shown in Table 1. In this study, total male samples were strictly collected from the embryonic muscle tissue of Duroc, and were grouped into three developmental stages (days 33, 65 and 90, three replicates for each stage)20. We identified 228 putative lincRNAs and found that many lincRNAs differentially expressed. Moreover, we predicted the potential target genes of DE lincRNAs by cis or trans ways. Gene Ontology and pathways enrichment analysis showed that lincRNAs potentially regulated the processes of protein-coding genes. An interactive network was performed to elucidate the interplay between DE lincRNAs and their potential target genes. This study of skeletal muscle of transcriptome profile will provide a useful resource to further explore the understanding of mechanisms. Besides, elucidating the underlying mechanisms of skeletal muscle growth and development will be helpful for the improvement of production benefits of pig.
RNA-seq reads mapping and transcriptomic assembly
To ensure the reliability of RNA reads and suitability for the subsequent analysis. FastQC (version 0.11.9) tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was run to quality control checks on raw sequences data and the sequences of poor quality were trimmed and filtered with Trimmomatic (version 0.36) software to obtain clean reads48. The high-quality filtered reads were aligned against the porcine reference genome (Sscrofa11.1 ) using HISAT2 (version 2.0.3) with default parameters49. The pig reference genome file was downloaded from Ensembl (ftp://ftp.ensembl.org/pub/release-99/gtf/sus_scrofa/). Next, SAM format files which obtained by mapping were converted to BAM format files with SAMtools (version 0.1.19). After that, StringTie (version 1.3.4) was used to assemble transcripts into nine GTF files, then transcripts of all the samples were combined by the Merge parameter of StringTie into a non-redundant transcript set to produce a uniform transcript GTF50. As a result of assembly produced a large amount of novel transcripts, which were mapped to reference annotation file using the GffCompare to discovery novel transcripts information49.
The pipeline lincRNAs identification and analysis
To identify porcine lincRNAs, we performed the following screening of the transcripts obtained after GffCompare, transcripts which the class-code annotated as ‘U’, were more than 200 bp in length and contained at least 2 exons were retained51. Next, all remaining transcripts were scored with CPC to determine their coding potential, transcripts of CPC < 0 were considered unable to encode proteins52. Then, we translated transcripts sequences into possible protein domains with Transeq2 and excluded transcripts that were matched in the Pfam database (E-value < 1e-5)53. Furthermore, transcripts that contained similar known proteins in non-redundant reference sequence (NR) database and UniRef90 database were discarded by BLASTX tool (E-value < 1e-5)54. Finally, we performed normalization on the transcript by calculating the ‘fragments per kilo-base of exon model per million mapped reads’ (FPKM) using StringTie with the parameter ‘-B’, and transcripts were retained while FPKM was greater than 0.5 in at least a sample49.
Comparison of identified lincRNAs and protein-coding transcripts
At present, the Ensembl database contains comprehensive genetic information for many species. We downloaded the pig reference annotation file that contained 45,788 protein-coding transcripts corresponding to 23,422 protein-coding genes in order to compare the characteristic differences between identified lincRNAs and protein-coding genes. LincRNAs annotation information was downloaded from the ALDB database, we acquired about 12,103 known lincRNA transcripts corresponding to 7,381 lincRNA genes, identified lincRNAs and protein-coding genes were aligned to the corresponding reference annotation files to obtain their detailed information, respectively55.
Differential expression analysis of lincRNAs
We used the python package called ‘HTseq-count’ to calculate the numbers of reads from nine samples56, and the resulting count files were used to evaluate the differential expression levels between different groups by the DEseq2 package in R57. By screening, lincRNA transcripts with |log2 fold change |≥ 1 and adjusted p-value ≤ 0.05 were identified differentially expressed, protein-coding transcripts with |log2 fold change |≥ 2 and adjusted p-value ≤ 0.05 were identified differentially expressed.
QTL location analysis of differentially expressed lincRNAs
To further explore the function of differentially expressed lincRNAs (DE lincRNAs), a correlation analysis was performed between DE lincRNAs with quantitative trait locus (QTL). The pig QTL reference file was downloaded from https://www.animalgenome.org/cgi-bin/QTLdb/SS/index, and the parameter ‘intersectBed’ was used to acquire DE lincRNAs to capture the QTL traits associated with lincRNAs.
Prediction of potential target genes
We predicted the molecular functions of protein-coding genes regulated by RNA in cis and trans. Firstly, the neighboring protein-coding genes nearby DE lincRNAs (< 100 kb) were identified based on cis-prediction principles using Bedtools. For trans regulation of DE lincRNAs, we calculated the Pearson correlation coefficient (r) between DE lincRNAs and protein coding genes, we selected protein-coding genes that Pearson correlation coefficient ≥ 0.96, adjusted p-value ≤ 0.05 as the potential target genes of DE lincRNAs.
Functional enrichment analysis GO and KEGG
The list of potential target genes was performed to predict biological process and potential signaling pathway based on gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) in DAVID website (http://david.abcc.ncifcrf.gov/home.jsp)58,59. The GO terms and KEGG pathways with p-value ≤ 0.05 were considered to be significantly enriched60,61,62,63,64. Because of the poor annotation of the pig reference genome, the protein-coding gene IDs were converted into human homologous gene IDs using BioMart from Ensembl.
Network correlation analysis of DE lincRNA genes and DEPTGs
Network interaction graph can intuitively reflect the relationship between DE lincRNAs and their potential target genes. We select PTGs that Pearson correlation coefficient ≥ 0.96 and adjusted p-value ≤ 0.05 differentially expressed in groups were defined as differentially expressed PTGs (DEPTGs), the highly correlated relationship between DE lincRNAs and the underlying potential target genes were established and visualized by Cytoscape (version 3.4).
Validation of differentially expressed lincRNAs
To verify our analysis results, qRT-PCR was carried out to test the expression between five DE lincRNAs and seven potential target genes which were randomly selected. There were 16 samples from embryonic muscle tissue used for the experiments (each experiment contained three biological replicates). Total RNA was extracted using Trizol reagent according to the manufacturer’s protocols, and reverse transcribed to cDNA using PrimeScript RT reagent Kit with gDNA Eraser r (Takara, Dalian, China). Quantitative PCR was performed using SYBR Premix Ex Taq II on Bio-Rad CFX-96 system (Bio-Rad Laboratories, Hercules). The relative expression levels of all genes were calculated by the 2–∆∆CT method. All primers were designed with Primer 5 program (Table S7).
Data availability
All the raw data involved in this study could be obtained from public database. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/?term=SRP216286.
Ethics statement
All the experiments were done in accordance with the relevant guidelines and regulations of animal care and use committee and the study was approved by The Scientific Ethic Committee of Huazhong Agricultural University, Hubei province.
References
Li, R. et al. Exploring the lncRNAs related to skeletal muscle fiber types and meat quality traits in pigs. Genes https://doi.org/10.3390/genes11080883 (2020).
Ciciliot, S., Rossi, A. C., Dyar, K. A., Blaauw, B. & Schiaffino, S. Muscle type and fiber type specificity in muscle wasting. Int. J. Biochem. Cell Biol. 45, 2191–2199. https://doi.org/10.1016/j.biocel.2013.05.016 (2013).
Rayavarapu, S., Coley, W., Kinder, T. B. & Nagaraju, K. Idiopathic inflammatory myopathies: pathogenic mechanisms of muscle weakness. Skelet Muscle 3, 13–13. https://doi.org/10.1186/2044-5040-3-13 (2013).
Petchey, L. K. et al. Loss of Prox1 in striated muscle causes slow to fast skeletal muscle fiber conversion and dilated cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 111, 9515–9520. https://doi.org/10.1073/pnas.1406191111 (2014).
Braun, T. & Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12, 349–361. https://doi.org/10.1038/nrm3118 (2011).
Valdez, M. R., Richardson, J. A., Klein, W. H. & Olson, E. N. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 219, 287–298. https://doi.org/10.1006/dbio.2000.9621 (2000).
Montarras, D. et al. Developmental patterns in the expression of Myf5, MyoD, myogenin, and MRF4 during myogenesis. New Biol. 3, 592–600 (1991).
Hernández-Hernández, J. M., García-González, E. G., Brun, C. E. & Rudnicki, M. A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin Cell Dev. Biol 72, 10–18. https://doi.org/10.1016/j.semcdb.2017.11.010 (2017).
Fuentes, E. N. et al. IGF-I/PI3K/Akt and IGF-I/MAPK/ERK pathways in vivo in skeletal muscle are regulated by nutrition and contribute to somatic growth in the fine flounder. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300, R1532-1542. https://doi.org/10.1152/ajpregu.00535.2010 (2011).
Schiaffino, S., Dyar, K. A., Ciciliot, S., Blaauw, B. & Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 280, 4294–4314. https://doi.org/10.1111/febs.12253 (2013).
Schiaffino, S. & Mammucari, C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle 1, 4. https://doi.org/10.1186/2044-5040-1-4 (2011).
Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. https://doi.org/10.1126/science.1138341 (2007).
Ponting, C. P., Oliver, P. L. & Reik, W. Evolution and functions of long noncoding RNAs. Cell 136, 629–641. https://doi.org/10.1016/j.cell.2009.02.006 (2009).
Lee, J. T. Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439. https://doi.org/10.1126/science.1231776 (2012).
Sarkar, D., Leung, E. Y., Baguley, B. C., Finlay, G. J. & Askarian-Amiri, M. E. Epigenetic regulation in human melanoma: past and future. Epigenetics 10, 103–121. https://doi.org/10.1080/15592294.2014.1003746 (2015).
Leighton, P. A., Ingram, R. S., Eggenschwiler, J., Efstratiadis, A. & Tilghman, S. M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375, 34–39. https://doi.org/10.1038/375034a0 (1995).
Pandey, R. R. et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246. https://doi.org/10.1016/j.molcel.2008.08.022 (2008).
Pauli, A., Rinn, J. L. & Schier, A. F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149. https://doi.org/10.1038/nrg2904 (2011).
Wu, G. et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 130, 1452–1465. https://doi.org/10.1161/circulationaha.114.011675 (2014).
Hong, L. et al. Genome-wide analysis of circular RNAs mediated ceRNA regulation in porcine embryonic muscle development. Front. Cell Dev. Biol. 7, 289. https://doi.org/10.3389/fcell.2019.00289 (2019).
Che, T. & Li, D. Long non-coding RNAs and mRNAs profiling during spleen development in pig. Plos ONE 13, e0193552. https://doi.org/10.1371/journal.pone.0193552 (2018).
Chen, G. et al. Transcriptome analysis reveals the effect of long intergenic noncoding RNAs on pig muscle growth and fat deposition. BioMed Res. Int. 2019, 2951427. https://doi.org/10.1155/2019/2951427 (2019).
Chen, L. et al. Transcriptome analysis suggests the roles of long intergenic non-coding RNAs in the growth performance of weaned piglets. Front. Genet. 10, 196. https://doi.org/10.3389/fgene.2019.00196 (2019).
Hu, Z. L., Park, C. A. & Reecy, J. M. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 44, D827-833. https://doi.org/10.1093/nar/gkv1233 (2016).
Luo, S. et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18, 637–652. https://doi.org/10.1016/j.stem.2016.01.024 (2016).
Yan, P., Luo, S., Lu, J. Y. & Shen, X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 46, 170–178. https://doi.org/10.1016/j.gde.2017.07.009 (2017).
Borycki, A. G. et al. Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Dev. (Cambridge, England) 126, 4053–4063 (1999).
Straface, G. et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J. Cell Mol. Med. 13, 2424–2435. https://doi.org/10.1111/j.1582-4934.2008.00440.x (2009).
Ma, J. et al. Swine PPAR-γ2 expression upregulated in skeletal muscle of transgenic mice via the swine Myozenin-1 gene promoter. Transgenic Res. 24, 409–420. https://doi.org/10.1007/s11248-014-9849-1 (2015).
Ashmore, C. R., Addis, P. B. & Doerr, L. Development of Muscle Fibers in the Fetal Pig. J. Anim. Sci. 36, 1088–1093. https://doi.org/10.2527/jas1973.3661088x (1973).
Swatland, H. J. & Cassens, R. G. Prenatal development, histochemistry and innervation of porcine muscle. J. Anim. Sci. 36, 343–354. https://doi.org/10.2527/jas1973.362343x (1973).
Wigmore, P. M. & Stickland, N. C. Muscle development in large and small pig fetuses. J. Anat. 137(Pt 2), 235–245 (1983).
Davoli, R., Braglia, S., Russo, V., Varona, L. & te Pas, M. F. Expression profiling of functional genes in prenatal skeletal muscle tissue in Duroc and Pietrain pigs. J. Anim. Breed. Genet. 128, 15–27. https://doi.org/10.1111/j.1439-0388.2010.00867.x (2011).
Zou, C. et al. Transcriptome analysis reveals long intergenic noncoding RNAs contributed to growth and meat quality differences between yorkshire and Wannanhua Pig. Genes https://doi.org/10.3390/genes8080203 (2017).
Yotsukura, S., duVerle, D., Hancock, T., Natsume-Kitatani, Y. & Mamitsuka, H. Computational recognition for long non-coding RNA (lncRNA): Software and databases. Brief. Bioinform. 18, 9–27. https://doi.org/10.1093/bib/bbv114 (2017).
Yu, L. et al. Comparative analyses of long non-coding RNA in lean and obese pig. Oncotarget 8, 41440–41450. https://doi.org/10.18632/oncotarget.18269 (2017).
Zhao, W. et al. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 5, 8957. https://doi.org/10.1038/srep08957 (2015).
Zhang, L. et al. Expression and functional analyses of Dlk1 in muscle stem cells and mesenchymal progenitors during muscle regeneration. doi:https://doi.org/10.3390/ijms20133269 (2019).
Stachowiak, M., Szczerbal, I. & Flisikowski, K. Investigation of allele-specific expression of genes involved in adipogenesis and lipid metabolism suggests complex regulatory mechanisms of PPARGC1A expression in porcine fat tissues. BMC Genet. 19, 107. https://doi.org/10.1186/s12863-018-0696-6 (2018).
Stachowiak, M. et al. Polymorphism in 3’ untranslated region of the pig PPARA gene influences its transcript level and is associated with adipose tissue accumulation. J. Anim. Sci. 92, 2363–2371. https://doi.org/10.2527/jas.2013-7509 (2014).
Rouillon, J. et al. Serum proteomic profiling reveals fragments of MYOM3 as potential biomarkers for monitoring the outcome of therapeutic interventions in muscular dystrophies. Hum. Mol. Genet. 24, 4916–4932. https://doi.org/10.1093/hmg/ddv214 (2015).
Glezeva, N. et al. Targeted DNA methylation profiling of human cardiac tissue reveals novel epigenetic traits and gene deregulation across different heart failure patient subtypes. Circ. Heart Fail. 12, e005765. https://doi.org/10.1161/circheartfailure.118.005765 (2019).
Zhan, S. et al. Dynamic transcriptomic analysis in hircine longissimus dorsi muscle from fetal to neonatal development stages. Funct. Integr. Genomics 18, 43–54. https://doi.org/10.1007/s10142-017-0573-9 (2018).
Yang, Y. et al. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 49, 1313–1329. https://doi.org/10.1093/nar/gkaa1203 (2021).
Black, B. L. & Olson, E. N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167–196. https://doi.org/10.1146/annurev.cellbio.14.1.167 (1998).
Teboul, L., Summerbell, D. & Rigby, P. W. The initial somitic phase of Myf5 expression requires neither Shh signaling nor Gli regulation. Genes Dev. 17, 2870–2874. https://doi.org/10.1101/gad.1117603 (2003).
Lee, S. J. Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2, e789. https://doi.org/10.1371/journal.pone.0000789 (2007).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT. StringTie and Ballgown. 11, 1650–1667. https://doi.org/10.1038/nprot.2016.095 (2016).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295. https://doi.org/10.1038/nbt.3122 (2015).
Zou, C. et al. Transcriptome analysis reveals long intergenic non-coding RNAs involved in skeletal muscle growth and development in pig. Sci. Rep. 7, 8704. https://doi.org/10.1038/s41598-017-07998-9 (2017).
Kong, L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345-349. https://doi.org/10.1093/nar/gkm391 (2007).
Prakash, A., Jeffryes, M., Bateman, A. & Finn, R. D. The HMMER web server for protein sequence similarity search. Curr. Protocols Bioinforma. https://doi.org/10.1002/cpbi.40 (2017).
Pirooznia, M., Perkins, E. J. & Deng, Y. Batch blast extractor: an automated blastx parser application. BMC Genom. https://doi.org/10.1186/1471-2164-9-s2-s10 (2008).
Li, A. et al. ALDB: a domestic-animal long noncoding RNA database. PLoS ONE 10, e0124003. https://doi.org/10.1371/journal.pone.0124003 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31, 166–169. https://doi.org/10.1093/bioinformatics/btu638 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. https://doi.org/10.1038/nprot.2008.211 (2009).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl. Acids Res. 37, 1–13. https://doi.org/10.1093/nar/gkn923 (2009).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. https://doi.org/10.1038/75556 (2000).
Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics (Oxford, England) 21, 3787–3793. https://doi.org/10.1093/bioinformatics/bti430 (2005).
Ogata, H. et al. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. https://doi.org/10.1093/nar/27.1.29 (1999).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545-d551. https://doi.org/10.1093/nar/gkaa970 (2021).
Acknowledgements
The work was supported by National Natural Science Foundation of China (NSFC, 31872322), the Fundamental Research Funds for the Central Universities (2662017PY030).
Author information
Authors and Affiliations
Contributions
C.L. conceived and designed the experiments and explained the data; W.Z. performed the analysis of this data with the help of Z.L.; Q.L. and S.X. contributed to the collection of samples; M.L. and Y.W. carried out the experiment of qRT-PCR; W.Z. wrote and revised the manuscript. All authors have reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, W., Li, Z., Liu, Q. et al. Analysis of long intergenic non-coding RNAs transcriptomic profiling in skeletal muscle growth during porcine embryonic development. Sci Rep 11, 15240 (2021). https://doi.org/10.1038/s41598-021-94014-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94014-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.