Abstract
The characteristics of IDH-wild-type lower-grade astrocytoma remain unclear. According to cIMPACT-NOW update 3, IDH-wild-type astrocytomas with any of the following factors show poor prognosis: combination of chromosome 7 gain and 10 loss (+ 7/− 10), and/or EGFR amplification, and/or TERT promoter (TERTp) mutation. Multiplex ligation-dependent probe amplification (MLPA) can detect copy number alterations at reasonable cost. The purpose of this study was to identify a precise, cost-effective method for stratifying the prognosis of IDH-wild-type astrocytoma. Sanger sequencing, MLPA, and quantitative methylation-specific PCR were performed for 42 IDH-wild-type lower-grade astrocytomas surgically treated at Kyoto University Hospital, and overall survival was analysed for 40 patients who underwent first surgery. Of the 42 IDH-wild-type astrocytomas, 21 were classified as grade 4 using cIMPACT-NOW update 3 criteria and all had either TERTp mutation or EGFR amplification. Kaplan–Meier analysis confirmed the prognostic significance of cIMPACT-NOW criteria, and World Health Organization grade was also prognostic. Cox regression hazard model identified independent significant prognostic indicators of PTEN loss (risk ratio, 9.75; p < 0.001) and PDGFRA amplification (risk ratio, 13.9; p = 0.002). The classification recommended by cIMPACT-NOW update 3 could be completed using Sanger sequencing and MLPA. Survival analysis revealed PTEN and PDGFRA were significant prognostic factors for IDH-wild-type lower-grade astrocytoma.
Similar content being viewed by others
Introduction
Glioma is a common tumour type originating in the human brain1. Glioblastoma, grade IV (GBM) is the most aggressive and major subtype of glioma, while diffuse astrocytoma, grade II (DA) and anaplastic astrocytoma, grade III (AA) are lower-grade astrocytomas. All these pathological entities had been classified mainly based on histology in the 2007 World Health Organization (WHO) classification of central nervous system tumours2. IDH mutation is widely recognised as a good predictor of survival among patients with glioma3, and codeletion of chromosome 1p and 19q (1p/19q codeletion) has been associated with oligodendroglioma and suggests longer survival4. Based on such findings, the WHO 2016 classification combined genetic profiling with a grading system based on the WHO 2007 classification. Oligodendroglioma was defined as a glioma with both IDH-mutation and 1p/19q codeletion, and the K27M mutation in H3F3A or less common HIST1H3B was included in the criteria for diffuse midline glioma, H3K27M-mutant5.
IDH-mutant and IDH-wild-type astrocytomas are well known to show distinct genetic profiles and prognosis3,6,7,8. However, the disease-defining genetic alterations of IDH-wild-type astrocytomas have not been revealed5,9, and diagnosis remains dependent on the histological findings. However, some studies have concluded that a substantial subset of IDH-wild-type lower-grade astrocytomas show a poor survival course similar to that of IDH-wild-type GBM7,9,10,11,12,13,14. Based on these studies, a previous report by the Consortium to Inform Molecular and Practical Approaches to CNS Tumour Taxonomy (cIMPACT-NOW) update 3 proposed that certain IDH-wild-type diffuse astrocytomas show a poor clinical course similar to IDH-wild-type glioblastomas, and that the characteristics of these tumours were a result of at least one of three genetic profiles: combined whole chromosome 7 gain and whole chromosome 10 loss (+ 7/− 10); and/or EGFR amplification; and/or TERT promoter (TERTp) mutation15. These three profiles have been recommended for inclusion in the next classification, with this specific type of astrocytoma to be diagnosed as “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”, referred to as “astrocytoma, grade 4” in the present study. In this manner, copy-number alterations (CNAs) as well as mutations have been studied to allow clear classification of IDH-wild-type astrocytoma corresponding to the clinical prognosis.
In general, the hot spot mutations of IDH1/2 and TERTp can be detected by Sanger sequencing3,16,17,18 and 1p/19q codeletion can be examined by fluorescence in situ hybridisation or multiplex ligation-dependent probe amplification (MLPA)19,20. MLPA can also detect copy-number alterations in EGFR and PTEN19,20, which are located on chromosome 7p and 10q, respectively. However, whole-chromosomal alterations in chromosomes 7 and 10 were difficult to detect using only one MLPA kit, and are reportedly better ascertained by single nucleotide polymorphism array11,14, DNA methylation array9,13, array-based comparative genomic hybridisation14,21, or next-generation sequencing12. Unfortunately, those methods are difficult to conduct in many local institutes and hospitals, and thus are less than ideal as global standards. MLPA can target multiple different sequences in a single PCR reaction and can be used on fragmented DNA for which only small quantities are available (> 20 ng per reaction)19,22. The only appliances needed to perform this method are a thermal cycler and a capillary sequencer. MLPA has the potential to detect subgroups of IDH-wild-type astrocytoma showing poor prognosis, such as “astrocytoma, grade 4”, but the feasibility of this method needs to be evaluated.
Results
Between August 19th, 2010, and December 19th, 2019, a total of 291 samples were resected from 257 patients in Kyoto University Hospital and cryopreserved or extracted into DNA before fixation. All samples received integrated diagnosis based on the WHO 2016 classification, and 42 tumours from 42 patients (31 men, 11 women) were classified as IDH-wild-type astrocytoma. The characteristics of these 42 tumours are described in Table 1. The tumours comprised 18 IDH-wild-type DAs and 24 IDH-wild-type AAs, and median age at resection was 55.5 years (range 5–85 years). No significant differences in age or sex were noted between tumour subtypes. Forty tumours were removed in first surgeries, comprising 18 DAs and 22 AAs, and no difference in age was evident between subtypes. However, initial treatments for AAs included chemoradiation therapy more frequently, whereas observation was selected more in DAs. The initial postsurgical treatments of these patients were 9 chemoradiation therapies and 1 chemotherapy for DAs, and 19 chemoradiation therapies and 1 chemotherapy for AAs, while observation with regular imaging without treatment was selected for 8 DAs and 2 AAs.
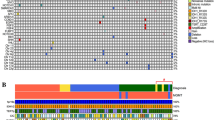
The detailed status of mutations and alterations in the 42 IDH-wild-type astrocytomas is shown in Fig. 1-A. TERTp mutation was detected in 18 IDH-wild-type astrocytomas (42.9%), comprising 9 DAs (50%) and 9 AAs (37.5%). Again, no significant difference was seen between these two tumour types. By examination using MLPA KIT probemix P105, EGFR amplification was seen in 8 IDH-wild-type astrocytomas (19.0%), comprising 1 DA (5.6%) and 7 AAs (29.2%), tending to be slightly more frequent in AAs than in DAs (p = 0.109). The combination of EGFR gain and PTEN loss (+ EGFR/− PTEN) was detected in 6 IDH-wild-type astrocytomas (14.3%), comprising 3 DAs (16.7%) and 3 AAs (12.5%), showing no significant difference. All DAs with EGFR amplification or + EGFR/-PTEN showed TERTp mutation, so all DAs diagnosed as “astrocytoma, grade 4” were equivalent to DAs with TERTp mutation, while 3 TERTp-wild type AAs showed EGFR amplification. Interestingly, no astrocytomas with + EGFR/− PTEN lacked the TERTp mutation.
(A) The cell plot shows characteristics of all 42 IDH-wild-type astrocytomas. (B) The schema of three step classification of IDH-wild-type astrocytomas. TERTp mutation was examined by Sanger sequencing, and EGFR amplification and combination of EGFR gain and PTEN loss were examined by multiplex ligation-dependent probe amplification (MLPA). The case with red square was diagnosed as “astrocytoma. grade 4”.
Classification of lower-grade astrocytomas, IDH-wild type
The results of classification of IDH-wild-type lower-grade astrocytomas are shown in Fig. 1-B. As the third step of our classification system, the + EGFR/− PTEN classification was used as a surrogate marker for tumours with + 7/− 10. TERTp mutation was detected in 18 of all 42 IDH-wild-type astrocytomas, and these 18 tumours were classified as “astrocytoma, grade 4” from Sanger sequence alone. In the 24 tumours without TERTp mutation, MLPA showed EGFR amplification in 3 AAs, and no tumours with + EGFR/− PTEN. As a result, 21 of all 42 tumours were classified as “astrocytoma, grade 4”, comprising 9 DAs and 12 AAs, in the first and second steps of our system. In our cohort, all instances of “astrocytoma, grade 4” were able to be diagnosed by the combination of Sanger sequence and MLPA.
Correlations with tumour profiles
Correlations between all pairs of the following factors were analysed: age at diagnosis, TERTp mutation, WHO grade, O6-methylguanine-DNA methyltransferase promoter (MGMTp) hyper methylation, and copy number alterations of EGFR, PTEN, CDKN2A, PDGFRA, MDM2, CDK4, NFKBIA, and TP53. TERTp mutation, EGFR amplification, CDKN2A homozygous loss, CDK4 gain or amplification (gain/amplification), CDK4 amplification, and MGMTp hypermethylation correlated with higher age at diagnosis (p = 0.0157, p = 0.0382, p = 0.0272, p = 0.0222, and p = 0.0045, respectively). Statistical correlations were detected between any two of TERTp mutation, EGFR gain/amplification and PTEN loss (TERTp and EGFR gain/amplification, odds ratio 5.91, p = 0.0236; TERTp mutation and PTEN loss, odds ratio 11, p = 0.004; EGFR gain/amplification and PTEN loss, odds ratio 9.38, p = 0.0304). Another correlation was seen between PDGFRA gain/amplification and CDKN2A homozygous loss (odds ratio, 7.78; p = 0.0195). WHO grade correlated significantly with MDM2 loss and MDM2 hemizygous loss (p = 0.0054 and p = 0.014, respectively). On the other hand, WHO grade showed no significance in the other profiles (Table 1).
Survival analysis of lower-grade astrocytomas, IDH-wild type
Clinical outcomes were calculated for the 40 cases in which the tumours were removed in first surgeries, including 18 DAs and 22 AAs. “Astrocytoma, grade 4” showed significantly shorter overall survival (OS) compared with other tumours in all astrocytomas (p = 0.0149) and DAs (p = 0.0036), but not in AAs (p = 0.1288) (Fig. 2-A). TERTp mutations were significantly associated with poor OS in all astrocytomas (p = 0.0228), and in DAs (p = 0.0036), but not in AAs (p = 0.0884) (Fig. 2-B). EGFR amplification was a significant factor for OS only in all astrocytomas (p = 0.0401), not in diffuse DAs (p = 0.7893) or AAs (p = 0.2877) (Fig. 2-C).
Survival analysis of 40 patients whose tumours were removed at their first surgery, were shown. The overall survival was analysed stratified by the following factors associated with the cIMPACT-NOW update 3 criteria; (A) the diagnosis of “astrocytoma, grade 4”, (B) TERTp mutation, or (C) EGFR amplification, in the groups of all IDH-wild type astrocytomas, IDH-wild type diffuse astrocytomas, and IDH-wild type anaplastic astrocytomas. The p values were calculated log-rank test and Wilcoxon test, and p < 0.05 was shown with red letters.
The relationship between WHO grade and OS was also analysed, and AAs showed poorer prognosis than DAs (p = 0.007). For cases of “astrocytoma, grade 4” or “non-astrocytoma, grade 4”, AA showed poorer survival curves compared with DA, and significant differences were identified with the Wilcoxon test (p = 0.04 each), but not with the log-rank test (p = 0.09 and p = 0.055, respectively) (Fig. 3-A).
Survival analysis of 40 patients whose tumours were removed at their first surgery, were shown. (A) The overall survival of IDH-wild type diffuse astrocytomas (grade II) and anaplastic astrocytomas (grade III) were compared in the groups of all IDH-wild type astrocytomas, “astrocytoma, grade 4”, and not-“astrocytoma, grade 4”. The overall survival was also analysed stratified by the following factors associated with factors not included in cIMPACT-NOW update 3 criteria; (B) PDGFR amplification, (C) PTEN status (intact, hemizygous loss, or homozygous loss), and (D) the combination status of PTEN loss and EGFR gain. The p values were calculated log-rank test and Wilcoxon test, and p < 0.05 was shown with red letters.
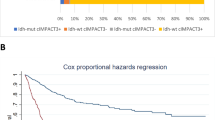
Log-rank testing showed significant correlations between OS and the following factors: age > 40 years, EGFR gain/amplification, PDGFRA amplification, PTEN homozygous loss and loss, CDK4 gain/amplification, MDM2 homozygous loss and alteration, and TP53 homozygous loss (Table 2). These factors and TERTp mutation, EGFR amplification and the diagnosis of “astrocytoma, grade 4”, as described above, were analysed by Cox proportional hazard modelling. After the stepwise procedure, three factors remained significant: PDGFRA amplification with the largest risk ratio (risk ratio 13.9, p = 0.0022), PTEN loss (risk ratio 9.75, p = 0.0003) with the most significance, and PTEN homozygous loss (risk ratio 6.97, p = 0.0329) (Table 3). PDGFRA amplification was detected in 4 astrocytomas, and survival analysis showed a significant result (p = 0.043) (Fig. 3-B). PTEN loss was detected in 11 astrocytomas and 3 were homozygous losses and 8 were hemizygous losses, 6 of which were coincided with EGFR gains. A significant difference in OS was seen between astrocytomas with intact PTEN and those with PTEN loss (p < 0.0001), while there was no difference in OS between PTEN homozygous and PTEN hemizygous astrocytomas (p = 0.1255) (Fig. 3-C). No difference in survival of PTEN-loss astrocytomas was seen between presence or absence of EGFR gain (p = 0.8133), and PTEN-loss astrocytomas showed poor prognosis compared with PTEN-intact astrocytomas irrespective of whether EGFR gain was present (with EGFR gain, p = 0.0001; without EGFR gain, p < 0.0001) (Fig. 3-D).
Validation of the status of chromosomes 7 and 10
As described above, MLPA KIT probemix P105 can detect CNA of EGFR at 7p.11.2 (EGFR(7p)) and PTEN at 10q.23.31 (PTEN(10q)). For validation of our classification system of the status of chromosomes 7 and 10 using MLPA, all samples were analysed using an additional two different MLPA KIT probemixes: chromosomes 7q by P370 and 10p by P303, respectively (Supplementary Fig. S1). Among six samples of showing + EGFR (7p)/− PTEN (10q) by P105 probemix, five samples showed both 7q gain by P370 and 10p loss by P303. We further examined all genome-level CNA analysis using a chromosomal microarray (CMA). A total of nine cases for which sufficient DNA was available were examined by CMA analysis to validate the results of CNA acquired by MLPA. Two cases showing both EGFR(7p)/7q gain and 10p/PTEN(10q) loss by MLPA system were confirmed to show + 7/− 10 by CMA (Supplementary Figs. S1, S2). One case that showed 7p(EGFR)/7q gain and 10q(PTEN) loss, but no 10p loss by MLPA system, was also confirmed to have + 7/− 10 by CMA (Supplementary Figs. S1, S2). These findings indicated that all 6 samples showing + EGFR (7p)/− PTEN (10q) by MLPA P105 probemix kit had features of + 7/− 10, suggesting that the status of + EGFR (7p)/− PTEN (10q) detected by MLPA system could be used as a surrogate marker for the + 7/− 10 phenotype.
Discussion
In the present study, the Sanger sequencing method could detect most instances of “astrocytoma, grade 4”, with the addition of MLPA successfully identifying all cases. Correlations between TERTp mutation and 7p gain or 10q loss have already been reported11. In the present study, correlation analysis showed that TERTp mutation was associated with EGFR gain and PTEN loss, and + EGFR/− PTEN astrocytoma always accompanied TERTp mutation. This fact may result from a relationship between TERTp mutation, gain of chromosome 7, and loss of chromosome 10. The database of The Cancer Genome Atlas (TCGA)6 included 86 IDH-wild-type grade II or III gliomas, and TERTp status was examined in 56 cases. TERTp mutation was detected in 37 cases, and EGFR amplification was shown in 15 out of 37 TERTp mutant gliomas and in 4 of 19 TERTp-wild-type gliomas. The status of + 7/− 10 was evaluated in 55 of the 56 cases of TERTp mutant grade II or III glioma, with 27 cases showing + 7/− 10. Among these 27 cases, 12 cases showed both TERTp mutation and EGFR amplification, 14 cases showed TERTp mutation alone, and the remaining one case showed EGFR amplification alone. In the present study, EGFR amplification was defined as different from EGFR gain, so + EGFR/− PTEN tumours never showed EGFR amplification. As in the present study, for IDH-wild-type grade II or III gliomas in TCGA database, TERTp status revealed almost all cases of “astrocytoma, grade 4” and the addition of EGFR status successfully identified all other cases of “astrocytoma, grade 4”. Sanger sequencing and MLPA were thus thought to be reasonable methods for classifying IDH-wild-type lower-grade gliomas based on the recommendations from cIMPACT-NOW update 315.
WHO grade offered a good marker of prognosis in the present study. IDH-wild-type AAs showed lower survival curve than IDH-wild-type DAs. Based on the WHO 2016 classification, glioma grade is partly affected by molecular factors including 1p/19q codeletion and histone mutations, but gliomas are mainly classified according to histological characteristics, which are almost the same as in the WHO 2007 classification. Although our cohort showed no differences in sex, age, or other prognostic factors detected in multivariate analysis, treatment selections did differ between DAs and AAs. Patients with AAs tended to be initially treated with strong chemoradiation therapies like the Stupp regimen23, and those with DAs tended to undergo observation alone more often; this might be one reason why the results of log-rank testing showing no statistical difference. The survival curve of DAs was definitely favourable compared with AAs in the early course, and the generalized Wilcoxon test showed a significant difference. Taking IDH-mutation status into consideration, WHO grade was reported as a significant factor for OS in lower-grade gliomas11,24,25, and one of the studies showed that WHO grade had higher prognostic value in IDH-wild-type astrocytomas compared with in IDH-mutant astrocytomas, with the authors proposing histological mitotic count as a significant predictor of prognosis24. This fact supports the importance of histological grading systems, especially for IDH-wild-type astrocytoma.
The survival analysis showed TERTp mutation as a prognostic factor for OS in the group of all IDH-wild-type astrocytomas and IDH-wild-type DAs, and the diagnosis of “astrocytoma, grade 4” with EGFR amplification was significant only in all IDH-wild-type astrocytomas. TERTp mutation and EGFR amplification have been reported as characteristics of IDH-wild-type GBM and as unfavourable prognostic factors in IDH-wild-type astrocytomas in many studies10,11,12,26, although a few studies have reported no significance14,27. In our study, EGFR amplification was slightly more frequent in AAs than in DAs, while TERTp showed no difference between subtypes. TERTp mutation, EGFR amplification, and diagnosis of “astrocytoma, grade 4” were significant factors in the group of all IDH-wild-type astrocytomas.
As mentioned above, WHO grade, TERTp mutation, EGFR amplification and diagnosis of “astrocytoma, grade 4” were good predictors of IDH-wild-type astrocytomas in Kaplan–Meier analysis, but Cox proportional hazard modelling detected no significance for OS in these factors. According to the Cox proportional hazard model of our cohort, copy number alteration of PTEN and PDGFRA amplification were significant predictors of OS.
CNA of PTEN was a strong predictor of prognosis, as demonstrated by both the Kaplan–Meier method and Cox proportional hazard modelling. No difference in OS was evident between PTEN hemizygous loss astrocytoma and PTEN homozygous loss astrocytoma, with both showing shorter OS than PTEN-intact astrocytoma. In addition, whether combined with EGFR gain or not, PTEN loss resulted in a significant difference in OS. PTEN loss is one of the typical genetic alterations in GBM, observed in about 30–40%28,29. Some studies of prognostic factors in GBM patients have been published, but the significance of PTEN loss has been controversial27,30,31. However, for IDH-wild-type lower-grade astrocytoma, some papers have stated that PTEN loss is associated with poor prognosis27,32, potentially because PTEN is a tumour suppressor gene27,33 and inactivation of PTEN signalling is thus important to malignant progression to glioblastoma34. The present study indicated PTEN loss as a strong predictor of poor prognosis in IDH-wild-type astrocytomas.
PDGFRA amplification showed a strong risk ratio in the present study, but only 4 AAs were included in the present study. PDGFRA amplification was also recognised as a characteristic of proneural GBM, which shows relatively good prognosis29,35. The frequency of PDGFRA amplification in lower-grade glioma has only been reported from studies of small numbers of low-grade gliomas36,37, and the evidence was insufficient to reach conclusions about the prognostic value. Strum et al. reported about subgrouping of GBMs based on the methylation profiles and compared them with other profiles of mutation and copy number status35. PDGFRA amplification was more common in a methylation cluster, “RTK I”, than in the other four clusters. “RTK I” cluster also showed CDKN2A loss frequently. In the present study, a correlation between PDGFRA gain/amplification and CDKN2A homozygous loss was seen, and might imply that astrocytoma with alteration of PDGFR is associated with “RTK I” GBM. In our cohort, no PDGFRA amplification was seen in DAs, but no difference in the frequency of this CNA was evident between DAs and AAs because of the small number with PDGFRA amplification. Further studies are required to clarify the prognostic value of PDGFRA.
After cIMPACT-NOW update 3, genetic analyses such as copy number analysis have been extensively studied for lower-grade glioma, and it has become clearer that several genetic markers are surely prognostic and need to be incorporated into clinical practice. To examine CNA at the whole-genome level, microarray systems or next-generation sequencing are generally used. However, these diagnostic systems require special equipment that carries a higher running cost. Those poor accessibility to them is usually unfavourable. In this context, our study showed important implications by showing that such prognostic stratification can be performed by direct sequencing and MLPA with simple methods at reasonable cost. The results of our validation study, all 6 tumours with + EGFR/− PTEN as determined by MLPA with P105 probemix showed + 7/− 10 in CMA or with additional MLPA methods. In these cases, one showed different results for 10p. In this case, CMA revealed 10p loss, but the copy number ratio as calculated by MLPA was 0.84, slightly higher than the threshold for chromosomal loss. Two cases with intact CNA as determined by CMA also revealed normal CNA by MLPA. Although further studies are essential regarding the results of MLPA and CMA for CNA in chromosome 7q or 10p, the results of CNA for EGFR and PTEN as detected by the MLPA system were suggested to offer good indicators for + 7/− 10.
Several limitations to the present study need to be considered. First, the study population was small. Second, the results of MLPA analysis were not able to be confirmed by other methodologies. Comparison of the results of MLPA and CMA were performed in nine cases due to the small amounts of DNA extracted from tumours. Third, the present study used only fresh or cryopreserved specimens, to obtain a sufficient quality and quantity of DNA and to avoid the influence of the paraffin embedding process to obtain precise results. Sanger sequencing and MLPA are generally available for the analysis of DNA extracted from FFPE samples. However, extraction of a sufficient quality of DNA from FFPE samples is well recognised as being not always easy and the quality of fragmented DNA in FFPE samples sometimes makes molecular analysis difficult. Such issues should be addressed in future research.
Conclusion
The present study showed that the combination of Sanger sequencing and MLPA was sufficient to identify a subgroup of patients with poorer prognosis in IDH-wild-type lower-grade astrocytoma. These patients were safely considered to have “astrocytoma, grade 4” according to the cIMPACT-NOW update 3 criteria. Our data also identified PTEN loss and PDGFRA amplification as significant prognostic factors, and these genetic alterations are good candidates for an upcoming new classification. WHO grade is still useful to predict the clinical course of patients with IDH-wild-type gliomas.
Methods
Ethics approval and consent to participate
This study was carried out in accordance with the principles of the Declaration of Helsinki, and approval was obtained from the institutional review board at Kyoto University Hospital (approval number: G1124). Informed consent was obtained from the patients or the parents/ legally authorized representatives of subjects that are under 18 for inclusion in this study.
Subjects
The purpose of the present study was to assess the feasibility of combining Sanger sequencing and MLPA in classifying IDH-wild-type lower-grade astrocytomas, as diagnosed by the WHO 2016 classification, into a new classification recommended by cIMPACT-NOW update 3, and to reveal prognostic factors for IDH-wild-type lower-grade astrocytoma.
The targets of the present study were IDH-wild-type astrocytomas surgically treated in Kyoto University Hospital. Inclusion criteria were as follows: (1) tumour samples after removal were stored as frozen or fresh specimens to maintain sufficient quality and quantity of DNA for extraction; (2) initial diagnosis was WHO grade II or grade III glioma; (3) Sanger sequence revealed no hot-point mutations in IDH1/2, H3F3A, or HIST1H3B; (4) MLPA showed no 1p/19q codeletion; and (5) informed consent was obtained.
Sanger sequencing
Tumour DNA was extracted from tumour specimens using NucleoSpin Tissue (MACHEREY–NAGEL, Düren, Germany). Regions of interest for driver genes3,16,17,18 were amplified by PCR with gene-specific primers (Supplementary Table S1) and TaKaRa Ex Taq (TAKARA BIO, Shiga, Japan) (IDH1/2, H3F3A, and HIST1H3B) or AmpliTaq Gold 360 (Thermo Fisher Scientific, Waltham, MA) (TERTp) using an Applied Biosystems GeneAmp PCR System 9700 (Thermo Fisher Scientific). PCR products were processed by ExoSAP-IT (Thermo Fisher Scientific), then sequenced with sequencing primer (IDH1) or PCR forward primer as a sequencing primer (IDH2, H3F3A, HIST1H3B, TERTp) and a BigDye Terminator V1.1 Cycle Sequencing Kit (Thermo Fisher Scientific) using the ABI 3130xL Genetic Analyzer (Thermo Fisher Scientific).
MGMT promoter methylation analysis
MGMTp methylation was assessed by quantitative methylation-specific PCR (qMSP), in accordance with previous reports20,38. Genomic DNA samples were processed using an EZ DNA Methylation Gold Kit (Zymo Research Corporation, Irvine, CA). Methylated and unmethylated molecules were quantified by qMSP using a QuantStudio 12 K Flex Real-Time PCR System (Thermo Fisher Scientific) with POWER SYBR Green Master Mix (Thermo Fisher Scientific) and specific primers (Supplementary Table S1)39 according to the standard curve method. The methylation status of samples was determined from the ratio of methylated molecules using the cut-off value at > 1%20.
MLPA
Copy number analyses of 1p/19q, EGFR, PTEN, CDKN2A and PDGFRA were performed with MLPA according to the instructions from the manufacturer (SALSA MLPA KIT probemix P088-C2 for 1p/19q analysis and SALSA MLPA KIT probemix P105-D2 for the others; MRC-Holland, Amsterdam, the Netherlands)19,20. MLPA with probemix P105-D2 can also analyse the alteration of MDM2, CDK4, NFKBIA, and TP53. To determine the copy number status of chromosomes 7p and 10q, in the present study, gain of 7p and loss of 10q were substituted by EGFR gain and PTEN loss, which were determined by MLPA with probemix P105. The statuses of chromosomes 7q and 10p were analysed with SALSA MLPA KIT probemix P370-C1 and P303-A3 (MRC-Holland). The P370-C1 had 13 target probes in chromosome 7q and P303-A3 had 3 target probes in chromosome 10p.
Data on MLPA were collected using an ABI 3130xL Genetic Analyzer (Thermo Fisher Scientific), then analysed using Coffalyzer.Net Software (MRC-Holland). Thresholds of copy number detection were chosen as reported previously19,40, and set at 0.8–1.2. A ratio of 0.4 was set as the threshold between hemizygous and homozygous losses, and ratios > 2.0 were defined as amplifications.
Integrated diagnosis
Tumour grading was performed histologically. Using all molecular pathological information, all cases received integrated diagnoses according to the 2016 WHO classification for central nervous system tumours5.
Clinical outcomes
Clinical data retrospectively collected from electronic records included age at diagnosis, sex, treatment protocol as chemotherapy or radiotherapy, and dates of surgery, last follow-up, and death.
Classification of astrocytomas, IDH-wild type
All IDH-wild-type astrocytomas were analysed by Sanger sequencing and MLPA to reveal the status of TERTp, EGFR gain or amplification, and PTEN loss. Classifications were performed in three steps. The first step was TERTp mutation, the second was EGFR amplification, and the third was + EGFR/− PTEN.
According to the recommendations of a previous study15, “astrocytoma, grade 4” was defined as IDH-wild-type astrocytoma with one or more of TERTp mutation, EGFR amplification, or + 7/− 10. However, showing the whole chromosomal alteration with MLPA (with SALSA MLPA KIT probemix P105-D2) is impossible. Because EGFR and PTEN are located on chromosomes 7 and 10, respectively, we hypothesised that tumours with + 7/− 10 must have + EGFR/− PTEN. In our classification system, + EGFR/− PTEN was therefore used as a criterion in the third step.
Chromosomal microarray
Whole-genome level CNA analysis was performed with Cytoscan 750 K array (Thermo Fisher Scientific) according to the protocol provided by the manufacturer. Data were analysed using Chromosome Analysis Suite (ChAS) software (Thermo Fisher Scientific). The reference database included the Database of Genomic Variants (GRCh38/hg38).
Statistical analysis
All statistical analyses were performed using JMP Pro version 15.1.0 software (SAS Institute, Cary, NC). Differences in categorical variables were evaluated using Fisher’s exact test or the chi-square test, and Student’s t-test was used for continuous variables. For survival analyses, OS was defined as the interval between the initial operative day and the date of death, or the last follow-up date on which the patient was known to be alive. Survival data were analysed using the Kaplan–Meier curve and the p-value for survival in the present paper was determined by log-rank testing if there was no special comment, while the generalized Wilcoxon test was used when appropriate. Multivariate analysis was performed by Cox proportional hazard modelling. Relationships between OS and the following factors were analysed: age; MGMTp hypermethylation; gain, amplification, and gain/amplification of EGFR, PDGFRA, and MDM2; hemizygous loss, homozygous loss and loss (including both of hemizygous and homozygous loss) of CDKN2A, PTEN, CDK4, MDM2, NFKBIA, and TP53. Differences were considered significant for values of p < 0.05.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AA:
-
Anaplastic astrocytoma, grade 3
- CDK4 :
-
Cyclin-dependent kinase 4
- CDKN2A :
-
Cyclin-dependent kinase inhibitor 2A
- CI:
-
Confidence interval
- CNA:
-
Copy number alteration
- cIMPACT-NOW update 3:
-
The Consortium to Inform Molecular and Practical Approaches to CNS Tumour Taxonomy
- CMA:
-
Chromosomal microarray
- DA:
-
Diffuse astrocytoma, grade 2
- EGFR :
-
Epidermal growth factor receptor
- FFPE:
-
Formalin-fixed paraffine-embedded
- GBM:
-
Glioblastoma
- H3F3A :
-
H3 histone, family 3A
- HIST1H3B :
-
Histone cluster 1 H3 family member b
- IDH :
-
Isocitrate dehydrogenase
- MDM2 :
-
Murine double minute 2
- MGMTp:
-
O6-Methylguanine-DNA methyltransferase promoter
- MLPA:
-
Multiplex ligation-dependent probe amplification
- NFKBIA :
-
Nuclear factor kappa-B inhibitor alpha
- OS:
-
Overall survival
- PDGFRA :
-
Platelet-derived growth factor receptor A
- PTEN :
-
Phosphatase and tensin homolog deleted from chromosome 10
- qMSP:
-
Quantitative methylation-specific PCR
- TCGA:
-
The Cancer Genome Atlas
- TERT :
-
Telomerase reverse transcriptase
- TERTp:
-
Telomerase reverse transcriptase promoter
- TP53 :
-
Tumour protein p53
- WHO:
-
World Health Organization
References
Ostrom, Q. T. et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 16, iv1–iv63. https://doi.org/10.1093/neuonc/nou223 (2014).
Louis, D. N. et al. The 2007 WHO Classification of Tumours Of The Central Nervous System. Acta Neuropathol. 114, 97–109. https://doi.org/10.1007/s00401-007-0243-4 (2007).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773. https://doi.org/10.1056/NEJMoa0808710 (2009).
Cairncross, J. G. et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. JNCI J. Natl. Cancer Inst. 90, 1473–1479. https://doi.org/10.1093/jnci/90.19.1473 (1998).
Cavenee, W. K., Louis, D. N., Ohgaki, H., Wiestler, O. D. & International Agency for Research on Cancer. WHO Classification of Tumours of the Central Nervous System 4th edn. (International Agency for Research on Cancer, 2016).
Ceccarelli, M. et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563. https://doi.org/10.1016/j.cell.2015.12.028 (2016).
Eckel-Passow, J. E. et al. Glioma groups based on 1p/19q, IDH, and TERTPromoter mutations in tumors. N. Engl. J. Med. 372, 2499–2508. https://doi.org/10.1056/nejmoa1407279 (2015).
Cancer Genome Atlas Research, N et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372, 2481–2498. https://doi.org/10.1056/NEJMoa1402121 (2015).
Reuss, D. E. et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 130, 407–417. https://doi.org/10.1007/s00401-015-1454-8 (2015).
Aibaidula, A. et al. Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol. 19, 1327–1337. https://doi.org/10.1093/neuonc/nox078 (2017).
Aoki, K. et al. Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol. 20, 66–77. https://doi.org/10.1093/neuonc/nox132 (2018).
Wijnenga, M. M. J. et al. Molecular and clinical heterogeneity of adult diffuse low-grade IDH wild-type gliomas: Assessment of TERT promoter mutation and chromosome 7 and 10 copy number status allows superior prognostic stratification. Acta Neuropathol. 134, 957–959. https://doi.org/10.1007/s00401-017-1781-z (2017).
Stichel, D. et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 136, 793–803. https://doi.org/10.1007/s00401-018-1905-0 (2018).
Tabouret, E. et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 132, 625–634. https://doi.org/10.1007/s00401-016-1611-8 (2016).
Brat, D. J. et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 136, 805–810. https://doi.org/10.1007/s00401-018-1913-0 (2018).
Arita, H. et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 126, 267–276. https://doi.org/10.1007/s00401-013-1141-6 (2013).
Arita, H. et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol. Commun. 4, 79. https://doi.org/10.1186/s40478-016-0351-2 (2016).
Bleeker, F. E. et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum. Mutat. 30, 7–11. https://doi.org/10.1002/humu.20937 (2009).
Jeuken, J., Cornelissen, S., Boots-Sprenger, S., Gijsen, S. & Wesseling, P. Multiplex ligation-dependent probe amplification. J. Mol. Diagn. 8, 433–443. https://doi.org/10.2353/jmoldx.2006.060012 (2006).
Umehara, T. et al. Distribution differences in prognostic copy number alteration profiles in IDH-wild-type glioblastoma cause survival discrepancies across cohorts. Acta Neuropathol. Commun. 7, 99. https://doi.org/10.1186/s40478-019-0749-8 (2019).
Weller, M. et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 129, 679–693. https://doi.org/10.1007/s00401-015-1409-0 (2015).
Schouten, J. P. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30, 57e–557. https://doi.org/10.1093/nar/gnf056 (2002).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996. https://doi.org/10.1056/NEJMoa043330 (2005).
Olar, A. et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II–III diffuse gliomas. Acta Neuropathol. 129, 585–596. https://doi.org/10.1007/s00401-015-1398-z (2015).
Petersen, J. K. et al. Targeted next-generation sequencing of adult gliomas for retrospective prognostic evaluation and up-front diagnostics. Neuropathol. Appl. Neurobiol. https://doi.org/10.1111/nan.12645 (2020).
Akyerli, C. B. et al. Use of telomerase promoter mutations to mark specific molecular subsets with reciprocal clinical behavior in IDH mutant and IDH wild-type diffuse gliomas. J. Neurosurg. 128, 1102–1114. https://doi.org/10.3171/2016.11.jns16973 (2018).
Brito, C. et al. Clinical insights gained by refining the 2016 WHO classification of diffuse gliomas with: EGFR amplification, TERT mutations, PTEN deletion and MGMT methylation. BMC Cancer https://doi.org/10.1186/s12885-019-6177-0 (2019).
Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. https://doi.org/10.1038/nature07385 (2008).
Verhaak, R. G. W. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110. https://doi.org/10.1016/j.ccr.2009.12.020 (2010).
Srividya, M. R. et al. Homozygous 10q23/PTEN deletion and its impact on outcome in glioblastoma: A prospective translational study on a uniformly treated cohort of adult patients. Neuropathology 31, 376–383. https://doi.org/10.1111/j.1440-1789.2010.01178.x (2011).
Carico, C. et al. Loss of PTEN is not associated with poor survival in newly diagnosed glioblastoma patients of the temozolomide era. PLoS ONE 7, e33684. https://doi.org/10.1371/journal.pone.0033684 (2012).
Sabha, N. et al. Analysis of IDH mutation, 1p/19q deletion, and PTEN loss delineates prognosis in clinical low-grade diffuse gliomas. Neuro Oncol. 16, 914–923. https://doi.org/10.1093/neuonc/not299 (2014).
Cristofano, A. D., Pesce, B., Cordon-Cardo, C. & Pandolfi, P. P. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355. https://doi.org/10.1038/1235 (1998).
Knobbe, C. B., Merlo, A. & Reifenberger, G. Pten signaling in gliomas. Neuro Oncol. 4, 196–211 (2002).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437. https://doi.org/10.1016/j.ccr.2012.08.024 (2012).
Martinho, O. et al. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. Br. J. Cancer 101, 973–982. https://doi.org/10.1038/sj.bjc.6605225 (2009).
Puputti, M. et al. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in Gliomas. Mol. Cancer Res. 4, 927–934. https://doi.org/10.1158/1541-7786.mcr-06-0085 (2006).
Okita, Y. et al. (11)C-methinine uptake correlates with MGMT promoter methylation in nonenhancing gliomas. Clin. Neurol. Neurosurg. 125, 212–216. https://doi.org/10.1016/j.clineuro.2014.08.004 (2014).
Esteller, M., Hamilton, S. R., Burger, P. C., Baylin, S. B. & Herman, J. G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Can. Res. 59, 793–797 (1999).
Jeuken, J. et al. Robust detection of EGFR copy number changes and EGFR variant III: Technical aspects and relevance for glioma diagnostics. Brain Pathol. 19, 661–671. https://doi.org/10.1111/j.1750-3639.2009.00320.x (2009).
Acknowledgements
This study included results based upon data generated by The Cancer Genome Atlas (TCGA) Research Network. We would like to acknowledge TCGA and their financial and material support in the development of the TCGA, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of the study authors.
Funding
This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (project nos. 16K10754, 17K19724, 19K09505, and 19K22685).
Author information
Authors and Affiliations
Contributions
Y.A. and Y.K. designed the study. Y.Ma., T.K, E.Y., T.S., and D.K. performed genetic analyses and data analyses. Y.T., M.T. and Y.Mi. collected samples. Y.Ma., Y.A. and Y.K. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Makino, Y., Arakawa, Y., Yoshioka, E. et al. Prognostic stratification for IDH-wild-type lower-grade astrocytoma by Sanger sequencing and copy-number alteration analysis with MLPA. Sci Rep 11, 14408 (2021). https://doi.org/10.1038/s41598-021-93937-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-93937-8
This article is cited by
-
Clinical and imaging characteristics of supratentorial slioma with IDH2 mutation
Neuroradiology (2024)
-
Optimizing the risk stratification of astrocytic tumors by applying the cIMPACT-NOW Update 3 signature: real-word single center experience
Scientific Reports (2023)
-
Easy-to-use machine learning system for the prediction of IDH mutation and 1p/19q codeletion using MRI images of adult-type diffuse gliomas
Brain Tumor Pathology (2023)
-
IDH wild-type lower-grade gliomas with glioblastoma molecular features: a systematic review and meta-analysis
Brain Tumor Pathology (2023)
-
Correlation between brain functional connectivity and neurocognitive function in patients with left frontal glioma
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.