Abstract
Hepatic sinusoidal obstruction syndrome (SOS/VOD) represents a dramatic complication of hematopoietic stem cell transplantation (HSCT), particularly in children. Recent evidence has suggested a role for the gut microbiome (GM) in the context of HSCT and its related complications, but no data are available on the relationship between GM and SOS/VOD. Here, we conducted a retrospective case–control study in allo-HSCT pediatric patients developing or not SOS/VOD and profiled their GM over time, from before the transplant up to 72 days after. A rich and diverse GM before HSCT was found to be associated with a reduced likelihood of developing SOS/VOD. Furthermore, prior to transplant, patients not developing SOS/VOD showed an enrichment in some typically health-associated commensals, such as Bacteroides, Ruminococcaceae and Lachnospiraceae. Their levels remained overall higher until post-transplant. This high-diversity configuration resembles that described in other studies for other HSCT-related complications, including graft-versus-host disease, potentially representing a common protective GM feature against HSCT complications.
Similar content being viewed by others
Introduction
Hepatic sinusoidal obstruction syndrome (SOS), also known as veno-occlusive disease (VOD) is a potentially life-threatening complication of hematopoietic stem cell transplantation (HSCT). It is caused by the damage generated during the conditioning regimen to sinusoidal endothelial cells and hepatocytes in zone 3 of the hepatic acinus1. Pediatric patients are at higher risk of developing SOS/VOD rather than adults, showing an incidence of approximately 20–30%, compared to 9–14% in adults2,3. In a large pediatric cohort, the survival probability at 1 year for patients with or without severe and very severe SOS/VOD was 61% and 77%, respectively3.
Mounting evidence has suggested that the gut microbiome (GM) is associated with HSCT outcomes4and may play a pivotal role in the pathophysiology of the major HSCT complications in children, namely bloodstream infection (BSI) and graft-vs-host disease (GvHD)5,6. In particular, it is well known that the GM can act as a reservoir for BSI pathogens5 and pre-transplant GM signatures predicting outcomes such as subsequent GvHD occurrence have been consistently identified6,7, These signatures include lower diversity, increased proportions of pathogens or pathobionts, and reduced amounts of beneficial microbes, generally producers of short-chain fatty acids (SCFAs), which are key regulators of metabolic and immunological homeostasis8. To date, no information is available on the relationship between GM and SOS/VOD. However, preclinical studies suggest that microbial products translocated across impaired mucosal barriers may participate in the pathogenesis of endothelial damage, interfering with procoagulant and fibrinolytic endothelial responses9,10. This applies to lipopolysaccharide (LPS), a bacterial endotoxin that activates various signaling mechanisms in endothelial cells, ultimately leading to cellular dysfunction and injury11, as well as to other molecules directly produced or contributed by GM, such as tryptophan metabolites, trimethylamine-N-oxide and SCFAs. While the first two are recognized to have prothrombotic activity12,13, SCFAs are well-known anti-inflammatory metabolites that have also been shown to have hypotensive effects through binding to Gpr41 receptor, thus possibly improving circulation14.
In an attempt to shed some light on the possible relationship between GM and SOS/VOD, here we conducted a retrospective case–control study in allo-HSCT pediatric patients who developed SOS/VOD compared to control patients who did not develop this complication.
Methods
Patients enrollment
We performed a retrospective single-institution case–control study at the pediatric HSCT Unit of the University of Bologna, from 01/01/2015 to 31/12/2019. This study was approved by the Ethics Committee (ref. number 19/2013/U/Tess) and written informed consent was obtained from each enrolled patient or parent/legal guardian. Study inclusion criteria were the availability of a pre-HSCT fecal sample and of at least two samples collected after HSCT. Allo-HSCT pediatric recipients were selected among previously enrolled patients6,15. Patients in the control group were matched 1:1 by carefully matching for several microbiome-associated and SOS/VOD-related confounding factors16, such as age, sex, type of disease, conditioning regimen, antibiotic prophylaxis and type of nutrition, as well as source of stem cells. SOS/VOD grading was calculated according to the new EBMT (European Society for Blood and Marrow Transplantation) criteria for the diagnosis and grading of pediatric SOS/VOD17. All the patients developing SOS/VOD received defibrotide from the date of diagnosis for at least 21 days until resolution of signs and symptoms of SOS/VOD. When febrile neutropenia occurred, ceftazidime was used as first-line antibiotic therapy. Fecal samples were collected before and after transplant, up to 72 days post-HSCT, and stored at − 80 °C until analysis.
Microbial DNA extraction and sequencing
Microbial DNA was extracted from about 250 mg of stool sample using the repeated bead-beating plus column method, as previously described6,18. The V3–V4 hypervariable region of the 16S rRNA gene was amplified using primers 341F and 785R with Illumina overhang adapter sequences. In addition to being considered among the best options for reliable taxonomic inference19, the V3–V4 region was chosen for comparative purposes, as some sequences had already been generated using primers targeting this region. PCR products were purified using a magnetic bead-based system (Agencourt AMPure XP; Beckman Coulter, Brea, CA) and indexed by limited-cycle PCR using Nextera technology. Indexed libraries were further cleaned up as above and pooled at equimolar concentration. The final library was denatured and diluted to 5 pmol/l with a 20% PhiX control. Sequencing was performed on an Illumina MiSeq platform using the 2 × 250 bp paired-end protocol, per manufacturer's instructions (Illumina, San Diego, CA). Raw sequence reads were deposited in MG-RAST (https://www.mg-rast.org/linkin.cgi?project=mgp97083).
Bioinformatics and statistics
The bioinformatic processing of the sequencing reads was performed using a pipeline combining PANDAseq20 and QIIME 221 as recently described22. Briefly, length and quality-filtered reads were clustered into Amplicon Sequence Variants (ASVs) with DADA223. Singleton ASVs and chimeras were discarded during analysis. The taxonomy was assigned using the vsearch classifier24 against the Greengenes database as a reference (release May 2013). Alpha diversity was calculated using several metrics, such as the number of observed ASVs, the Shannon index, the Chao1 index, the inverse Simpson index and the phylogenetic metrics, Faith’s PD index, phylogenetic entropy (PE) and the abundance weighted evolutionary distinctiveness (AEDt) index. Beta diversity was estimated based on weighted and unweighted UniFrac distances and visualized on a Principal Coordinates Analysis (PCoA) plot.
All statistical analysis was performed in R using the packages vegan25, MADE426, adiv27 and mallorn28. The significance of the separation between study groups in the PCoA space was assessed by a permutation test with pseudo-F ratio using the function adonis in vegan. The bacterial genera most contributing to the ordination space were identified using the function envfit of vegan. Differences between groups for both taxonomic profile and alpha diversity were evaluated by Wilcoxon test. P values were corrected for multiple comparisons using the Benjamini–Hochberg method when appropriate. A P value ≤ 0.05 was considered statistically significant, while a P value ≤ 0.1 a tendency. Samples were grouped according to the sampling date relative to HSCT as previously reported6,18, i.e., “PRE” (samples taken before HSCT within a maximum of 30 days before HSCT), “HSCT” (samples taken up to 30 days after HSCT), and “POST” (samples taken more than 30 days after HSCT).
Quantitative PCR analysis
Quantitative PCR (qPCR) was used to confirm trends in taxa of interest, as emerged from the UniFrac-based PCoA analysis and the reconstruction of the longitudinal GM profiles. Specifically, we used a primer set targeting the 16S rDNA of Bacteroides29, and two primer sets for the 16S rDNA of Lachnospiraceae and Ruminococcaceae genera30,31. For the qPCR assays, genomic DNA was diluted with PCR-grade water (Hoffmann-La Roche, Basel, Switzerland) to a final concentration of 5 ng/µl. Reaction mixtures were prepared in a total volume of 20 μl using SYBR Select PCR master mix (Thermo Fisher Scientific, Waltham, MA), 0.2 μmol/l of each primer and 2 µl of diluted DNA. Amplification was performed on a StepOne Real-Time PCR System instrument (Thermo Fisher Scientific) with an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 55–61 °C for 30–60 s and elongation at 72 °C for 45 s, with a final elongation step at 72 °C for 5 min. For the annealing temperature and time for each primer set, see the original papers30,31. Melting curves for PCR product identification were obtained immediately after amplification. Standard curves were obtained by amplification of DNA from Bacteroides eggerthii DSM 20697 and Coprococcus comes ATCC 27758. Results were expressed as log10 16S rRNA gene copies/ng of DNA. Differences between and within groups were evaluated by Wilcoxon test.
Ethics declarations
This study was approved by the IRCCS Azienda Ospedaliero-Universitaria di Bologna ethics committee (ref. number 19/2013/U/Tess). Written informed consent was obtained from each enrolled patient or parent/legal guardian. The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines.
Results
Patients characteristics
Eighteen allo-HSCT pediatric recipients were selected among previously enrolled patients (Table 1). Nine patients developed severe or very severe SOS/VOD and nine patients were matched as controls (see Supplemental Tables S1–S4 for the distribution of microbiome-associated and SOS/VOD-related confounding factors, see Table 1 footnotes for exact matching). The cumulative incidence of SOS/VOD in our center was 15.38%. Three patients received haplo-HSCT with post-transplant cyclophosphamide. None of the patients received either Gemtuzumab or Inotuzumab prior to HSCT, or total body irradiation-based therapy in the conditioning regimen. A detailed description of antibiotic exposure is given in Table 2.
GM profiling of SOS/VOD patients vs controls from before HSCT up to more than 2 months later
A total of 74 fecal samples were available, collected before and after transplant, up to 72 days post-HSCT (Fig. 1). 16S rRNA gene sequencing of these samples yielded a total of 3,805,437 reads, ranging from 5246 to 233,603 per sample (mean 51,425, SD 39,961).
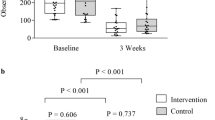
Pre-transplant samples (i.e., PRE samples) from patients who developed SOS/VOD showed significantly lower alpha diversity than controls, as measured by different metrics (i.e., the Shannon index, the number of observed ASVs, the Chao1 index, the Faith’s PD index, PE and AEDt index) (P ≤ 0.05, Wilcoxon test, false discovery rate (FDR) corrected). Consistent with the available literature4,6, during the first 30 days after HSCT, all subjects showed comparable low levels of GM diversity (HSCT samples). The diversity of post-transplant samples (POST samples) was again significantly lower in SOS/VOD-diagnosed patients than in controls (P ≤ 0.05), with the latter showing overall diversity levels comparable to those calculated for the PRE samples (Fig. 2 and Supplemental Figs. S1 and S2). A combined index of evenness and richness (inverse Simpson index) showed a similar trend, albeit in the absence of significance, with decreased average values in the PRE samples for patients who developed SOS/VOD compared to controls (Fig. 2 and Supplemental Fig. S1).
The gut microbiota of SOS/VOD patients before HSCT shows less diversity compared to control patients. Alpha diversity of fecal microbiota in samples taken before transplant (PRE), up to 30 days after transplant (HSCT) and more than 30 days after transplant (POST), calculated for SOS/VOD patients (on the left in each graph) and controls (on the right) using the following metrics: the Shannon index, the Faith’s PD index, the number of observed amplicon sequence variants (ASVs), and the Chao1 index. *P ≤ 0.05; **P ≤ 0.01; Wilcoxon test. See also Supplemental Figure s S1 and S2.
Weighted and unweighted UniFrac distances between GM profiles were then computed and used to construct PCoA plots for each of the three defined time intervals, i.e., PRE, HSCT and POST. As shown in Fig. 3, PRE samples from SOS/VOD patients and controls significantly separated in the weighted UniFrac-based PCoA (P = 0.005, permutation test with pseudo-F ratio), confirming that early compositional differences might characterize the GM of pediatric patients who subsequently develop SOS/VOD. An additional analysis with PRE samples revealed no difference in the GM structure in relation to the administration of antibiotic prophylaxis (Supplemental Fig. S3).
GM structure of SOS/VOD patients vs controls from before HSCT up to more than 2 months later. PCoA are based on weighted (left) and unweighted (right) UniFrac distances of microbiota profiles of samples collected (from top to bottom) before transplant (PRE), during the first 30 days after transplant (HSCT) and > 30 days after transplant (POST) from patients who subsequently developed SOS/VOD and controls (see the color legend on the left). First and second principal components (MDS1 and MDS2) are plotted for each analysis. Percentages of variance in the dataset accounted for by MDS1 and MDS2 are reported. Permutation tests with pseudo-F ratio (Adonis function in vegan package of R software) were carried on for each analysis to test the significance of separation between patients and controls; P values ≤ 0.05 are reported (top left in each plot).
Similarly to what is described above for alpha diversity, no significant difference was found within the first 30 days after HSCT, with the samples from SOS/VOD patients and controls overlapping in the PCoA space (Fig. 3). On the other hand, in the POST, i.e., beyond 30 days from HSCT, the two groups were again significantly separated in the PCoA using the unweighted UniFrac metric (P = 0.05), likely suggesting the persistence of some structural changes in low-abundance taxa.
In order to identify discriminating taxa between patients who did or did not develop SOS/VOD, we plotted the bacterial genera contributing most to the ordination space, using the function envfit in the vegan package of R (P ≤ 0.05). It was therefore possible to ascertain that the PRE samples from control subjects were characterized by the Bacteroides genus and unclassified bacteria belonging to the Clostridiales, Ruminococcaceae and [Mogibacteriaceae] groups (Fig. 4A). The comparison of the relative abundances of these taxa in the two subject groups allowed to confirm the observed trends (P ≤ 0.1, Wilcoxon test) (Fig. 4B). Furthermore, the trend of Bacteroides was confirmed by qPCR (Supplemental Fig. S4).
The gut microbiota of SOS/VOD patients before HSCT shows a different compositional structure compared to control patients. (A) PCoA based on weighted UniFrac distances between microbiota profiles of samples collected before transplant from patients who subsequently developed SOS/VOD (dark grey circles) and controls (empty white circles). First and second principal components (MDS1 and MDS2) are plotted, accounting for 38.61% and 15.55% of variance in the dataset, respectively. The separation between the two groups of samples (Adonis P = 0.008) is highlighted by plotting the SEM-based ellipse around the centroid. The biplot of the average bacterial coordinates weighted by the corresponding relative abundance per sample was superimposed on the PCoA plot for the 10 genera significantly contributing to the ordination space, as calculated using the function envfit in the vegan package of R (black arrows). The number associated with each arrow identifies the bacterial group contributing to the ordination space; the legend is shown at the bottom. (B) Boxplots showing the relative abundance distribution of taxa identified with envfit function in SOS/VOD and control groups. *P ≤ 0.05; #0.05 < P ≤ 0.1; Wilcoxon test. Uncl. unclassified. (C) Heatmap showing Ward-linkage clustering based on the Pearson’s correlation coefficients of the relative abundance of genus-level taxa from SOS/VOD patients (see the grey bar at the top) and controls (white bar) before HSCT. Only taxa with relative abundance > 0.1% in at least 1 sample are shown. Unclassified genera are reported at higher taxonomic level. See also Fig. 5 and Supplemental Figure S4.
In contrast, no clearly defined groups of bacteria appeared to characterize patients who subsequently developed SOS/VOD, whose samples were actually more dispersed in the PCoA plot, indicating greater inter-sample variability. However, it is worth noting that Granulicatella (P = 0.03) and Bifidobacterium (P = 0.1) were significantly more represented or tended to be, in the PRE samples belonging to SOS/VOD patients (Fig. 4). This was likely related to the presence in this group of two patients aged less than 2 years, in line with the available literature on the infant-type GM32,33. Furthermore, some patients shared relatively high proportions of Actinomyces, as well as Erysipelotrichaceae and Peptostreptococcaceae genera (Fig. 4C), which are commonly overrepresented in inflammation-related disorders34,35. A subgroup analysis by removing the samples corresponding to the infants from both SOS/VOD patients and controls, confirmed the main findings of this study (Supplemental Fig. S5), thus suggesting that previously identified microbiota signatures may be age independent.
Finally, we evaluated the GM compositional changes in the whole cohort, i.e., within and between groups over time (Fig. 5). Notably, we found that the PRE samples from SOS/VOD patients were also characterized by depletion in other typically health-associated taxa, such as Lachnospiraceae genera (i.e., Coprococcus and Dorea) and Akkermansia, as well as in unclassified members of Coriobacteriaceae (P ≤ 0.1). Furthermore, the opportunistic pathogen Eggerthella was overrepresented in the POST samples of the SOS/VOD patients compared to the controls, while the opposite was observed for [Eubacterium] (P ≤ 0.05). Although the decrease over time in beneficial taxa, such as Blautia, Faecalibacterium and Ruminococcus, was shared by both groups of subjects, SOS/VOD patients showed generally lower proportions or faster decreases (e.g. already in the HSCT samples for Blautia and Faecalibacterium) (P ≤ 0.1). The data on the differential trends of members of the Lachnospiraceae and Ruminococcaceae families were confirmed by qPCR (Supplemental Fig. S4).
Gut microbiota compositional changes in SOS/VOD patients vs controls from before HSCT up to more than 2 months later. Boxplots showing the relative abundance distribution of genera differentially represented between and within groups. Dynamics of each genus were reconstructed from before transplant (PRE), during the first 30 days after transplant (HSCT) up to > 30 days after transplant (POST), for patients who subsequently developed SOS/VOD (on the left in each graph) and controls (on the right). *P ≤ 0.05; #0.05 < P ≤ 0.1; Wilcoxon test. Uncl. unclassified.
Discussion
Potential protective GM signature against SOS/VOD
This pilot study reports for the first time a possible association between GM and the onset of SOS/VOD. Well aware of the impossibility of postulating any causal relationship, here we advance the hypothesis that a rich and diverse GM before HSCT could be associated with a decreased probability of developing SOS/VOD. This observation is consistent with the recent scenario in which a high level of GM diversity, a cornerstone of a so-called “healthy” GM, has been associated with decreased mortality after HSCT4. Greater alpha diversity is linked as well to a reduced likelihood of developing another post-HSCT complication, aGvHD in pediatric patients6. The protective signature identified, rich in Bacteroides, Lachnospiraceae genera and unclassified members of Clostridiales and Ruminococcaceae, also shares some similarity with the protective configuration observed in studies regarding aGvHD. In particular, other authors have previously shown that higher relative abundances of Lachnospiraceae and Ruminococcus were associated with better clinical outcomes, mainly reduced aGvHD incidence and severity36,37. As briefly discussed above, the presence of Bifidobacterium in two patients who went on developing SOS/VOD could be partially explained by their age, these being children under the age of two and breastfed38. Similarly, Granulicatella, which was more represented in SOS/VOD patients, has been identified as a signature of the infant gut microbiota, probably due to its involvement in lactate metabolism33. However, some species have been recognized as possible agents of bloodstream and endovascular infections, especially when predisposing conditions such as mucositis and neutropenia are present39,40, thus potentially representing a “red flag” for clinicians. The protective effect exerted by the peculiar pre-HSCT GM composition in both SOS/VOD and aGvHD could be related to the common endothelial damage that characterizes both diseases. As for the other complications, the mechanistic explanation for this effect remains poorly understood. Based on previous preclinical studies9,10, we hypothesize that an altered intestinal ecosystem, depleted of health-associated taxa and with low production of SCFAs, could lead to greater translocation of microbial molecules through the damaged intestinal mucosa. Consistent with this, the proportions of the most common SCFA producers (from Lachnospiraceae and Ruminococcaceae families) remained overall lower in SOS/VOD patients than controls, even over time. Translocated bacterial products, and in particular LPS, could reach the liver sinusoid through the portal vein and participate in endothelial damage9,10. It is also worth mentioning that Bacteroides, found to be enriched in the control group, is one of the major producers of propionate, which has been shown to induce vasodilatation ex vivo and in animal models14, and appears to lack the potential to produce trimethylamine41, precursor of TMAO, which is well known to be involved in vasculo-occlusive events12. However, since metabolic profiles were not evaluated in this study, future investigations will be mandatory to test our hypotheses and clarify which functional players are actually involved.
Analysis of confounding variables
This study has some limitations, mainly due to its retrospective design. The number of patients is relatively small because of the low incidence of this complication. The GM sequences obtained in this study were combined with sequences from previous studies, but all samples were collected by the authors, processed in the same laboratory and thus subjected to the same wet and in silico analysis steps. Moreover, the timing of fecal sample collection, which depends on patients’ bowel movements, that in children undergoing HSCT are particularly irregular, is not fully matched between the two groups. To address this possible confounding variable, we performed a diversity analysis using only one sample per subject in each time window and the results were very similar to those found considering all samples, i.e., the patients who developed SOS/VOD had a less diverse GM in the pre and post-HSCT samples compared to the control patients (Supplemental Fig. S2). As for the relative imbalance in the administration of antibiotics between the two study groups, no difference was found in the total antibiotic exposure and in the administration of specific antimicrobial molecules (Table 2). Moreover, a subgroup analysis to better address the imbalance of levofloxacin prophylaxis between the two groups, revealed no separation in the PRE GM samples between patients who did or did not receive levofloxacin prophylaxis (Supplemental Fig. S3).
In conclusion, our pilot study’s findings stress the relevance of having a GM characterized by high diversity and richness of beneficial microorganisms in the pre-transplant period, as it could be associated with a reduced occurrence of SOS/VOD, as well as other HSCT-related complications and overall survival. The present results should prompt additional studies to better investigate the possible interplay between GM dysbiosis, endothelial cell injury and alloreactivity occurring in the pathogenesis of SOS/VOD, also using other techniques, including metabolomics. Studies involving larger cohorts of patients, possibly of various age groups to account for age-related specificities of the GM, are warranted to explore whether GM structure and functionality could serve as a predictive biomarker for clinicians to assess the risk, onset and progression of SOS/VOD. Such studies should include, as far as possible, a finer sampling even in the post-HSCT period, to define healthy/unhealthy trajectories of the gut microbiota across HSCT. This study also corroborates to ensure scientific bases for future GM-based interventions, such as fecal microbiota transplantation, to assess whether GM manipulation may impact on the incidence of HSCT complications.
Data availability
Sequencing reads from samples of patients 5, 8, 9 and 16 are available at http://www.ebi.ac.uk/ena/data/view/PRJEB23820 (corresponding to subjects B2, B1, B3 and B5, respectively; Biagi et al.6). Sequencing reads from samples of patients 1, 2, 11–15 are available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA592853 (corresponding to patients BP9, BE4, BP10, BE3, BP4, BE6 and BP6; D’Amico et al.18). Sequencing reads from newly sequenced samples (from patients 3, 4, 6, 7, 10, 17 and 18) are available at https://www.mg-rast.org/linkin.cgi?project=mgp97083, labelled as patients BE19, BP11, BE14, BE15, BE13, BE12 and BE16, respectively.
References
Mohty, M. et al. Sinusoidal obstruction syndrome/veno-occlusive disease: Current situation and perspectives—a position statement from the European Society for Blood and Marrow Transplantation (EBMT). Bone Marrow Transplant. 50, 781–789 (2015).
Corbacioglu, S., Jabbour, E. J. & Mohty, M. Risk factors for development of and progression of hepatic veno-occlusive disease/sinusoidal obstruction syndrome. Biol. Blood Marrow Transplant. 25, 1271–1280 (2019).
Faraci, M. et al. Sinusoidal obstruction syndrome/veno-occlusive disease after autologous or allogeneic hematopoietic stem cell transplantation in children: A retrospective study of the Italian Hematology-Oncology Association-Hematopoietic Stem Cell Transplantation Group. Biol. Blood Marrow Transplant. 25, 313–320 (2019).
Peled, J. U. et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N. Engl. J. Med. 382, 822–834 (2020).
Kelly, M. S. et al. Gut colonization preceding mucosal barrier injury bloodstream infection in pediatric hematopoietic stem cell transplantation recipients. Biol. Blood Marrow Transplant. 25, 2274–2280 (2019).
Biagi, E. et al. Early gut microbiota signature of aGvHD in children given allogeneic hematopoietic cell transplantation for hematological disorders. BMC Med. Genomics 12, 49 (2019).
Doki, N. et al. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann. Hematol. 96, 1517–1523 (2017).
Zama, D., Biagi, E., Masetti, R. et al. Gut microbiota and hematopoietic stem cell transplantation: where do we stand?. Bone Marrow Transplant 52, 7–14. https://doi.org/10.1038/bmt.2016.173 (2017).
Eissner, G., Multhoff, G. & Holler, E. Influence of bacterial endotoxin on the allogenicity of human endothelial cells. Bone Marrow Transplant. 21, 1286–1288 (1998).
Falanga, A., Vignoli, A., Marchetti, M. & Barbui, T. Defibrotide reduces procoagulant activity and increases fibrinolytic properties of endothelial cells. Leukemia 17, 1636–1642 (2003).
Dayang, E.-Z. et al. Identification of LPS-activated endothelial subpopulations with distinct inflammatory phenotypes and regulatory signaling mechanisms. Front. Immunol. 10, 1169 (2019).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 (2016).
Gondouin, B. et al. Indolic uremic solutes increase tissue factor production in endothelial cells by the aryl hydrocarbon receptor pathway. Kidney Int. 84, 733–744 (2013).
Pluznick, J. L. et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 4410–4415 (2013).
D’Amico, F. et al. Gut resistome plasticity in pediatric patients undergoing hematopoietic stem cell transplantation. Sci. Rep. 9, 5649 (2019).
Vujkovic-Cvijin, I. et al. Host variables confound gut microbiota studies of human disease. Nature 587, 448–454 (2020).
Corbacioglu, S. et al. Diagnosis and severity criteria for sinusoidal obstruction syndrome/veno-occlusive disease in pediatric patients: A new classification from the European society for blood and marrow transplantation. Bone Marrow Transplant. 53, 138–145 (2018).
D’Amico, F. et al. Enteral nutrition in pediatric patients undergoing hematopoietic SCT promotes the recovery of gut microbiome homeostasis. Nutrients 11, 2958 (2019).
Almeida, A., Mitchell, A. L., Tarkowska, A. & Finn, R. D. Benchmarking taxonomic assignments based on 16S rRNA gene profiling of the microbiota from commonly sampled environments. Gigascience 7, 1–10 (2018).
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G. & Neufeld, J. D. PANDAseq: Paired-end assembler for Illumina sequences. BMC Bioinform. 13, 31 (2012).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Saresella, M. et al. Alterations in circulating fatty acid are associated with gut microbiota dysbiosis and inflammation in multiple sclerosis. Front. Immunol. 11, 1390 (2020).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46 (2001).
Culhane, A. C., Thioulouse, J., Perrière, G. & Higgins, D. G. MADE4: An R package for multivariate analysis of gene expression data. Bioinformatics 21, 2789–2790 (2005).
Pavoine, S. adiv: An R package to analyse biodiversity in ecology. Methods Ecol. Evol. 11, 1106–1112 (2020).
Davis, M., Faurby, S. & Svenning, J.-C. Mammal diversity will take millions of years to recover from the current biodiversity crisis. Proc. Natl. Acad. Sci. U.S.A. 115, 11262–11267 (2018).
Tanaka, Y., Ito, S. & Isobe, K. Vancomycin-sensitive bacteria trigger development of colitis-associated colon cancer by attracting neutrophils. Sci. Rep. 6, 23920 (2016).
Van Dyke, M. I. & McCarthy, A. J. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl. Environ. Microbiol. 68, 2049–2053 (2002).
Nava, G. M., Friedrichsen, H. J. & Stappenbeck, T. S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638 (2011).
Milani, C. et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 81, 1–67 (2017).
Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015).
Kaakoush, N. O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 5, 84 (2015).
Cheng, W. Y., Wu, C.-Y. & Yu, J. The role of gut microbiota in cancer treatment: Friend or foe?. Gut 69, 1867–1876 (2020).
Ingham, A. C. et al. Specific gut microbiome members are associated with distinct immune markers in pediatric allogeneic hematopoietic stem cell transplantation. Microbiome 7, 131 (2019).
Payen, M. et al. Functional and phylogenetic alterations in gut microbiome are linked to graft-versus-host disease severity. Blood Adv. 4, 1824–1832 (2020).
Masetti, R. et al. The gut microbiome in pediatric patients undergoing allogeneic hematopoietic stem cell transplantation. Pediatr. Blood Cancer 67, e28711 (2020).
Senn, L. et al. Bloodstream and endovascular infections due to Abiotrophia defectiva and Granulicatella species. BMC Infect. Dis. 6, 9 (2006).
Senn, L., Entenza, J. M. & Prod’hom, G. Adherence of Abiotrophia defectiva and Granulicatella species to fibronectin: Is there a link with endovascular infections?. FEMS Immunol. Med. Microbiol. 48, 215–217 (2006).
Falony, G., Vieira-Silva, S. & Raes, J. Microbiology meets big data: The case of gut microbiota-derived trimethylamine. Annu. Rev. Microbiol. 69, 305–321 (2015).
Acknowledgements
Authors thank Alessio Cardilli and Sara Roggiani, students of the Department of Pharmacy and Biotechnology, for the contribution in DNA extraction and data analysis processes, as well as Dr. Simone Rampelli for bioinformatics support.
Funding
This study was supported by research funding from the Italian Ministero della Salute (Bando Ricerca Finalizzata 2013, Giovani Ricercatori section, code GR-2013-02357136) to R.M.
Author information
Authors and Affiliations
Contributions
R.M., D.Z. and E.B. contributed to the study design. E.M. and D.L. collected clinical data and fecal samples and wrote the manuscript. F.D. and E.B. analyzed the data, performed statistical analysis and wrote the manuscript. S.T., A.Pr., P.B. and A.Pe. critically analyzed the manuscript and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Masetti, R., Biagi, E., Zama, D. et al. Early modifications of the gut microbiome in children with hepatic sinusoidal obstruction syndrome after hematopoietic stem cell transplantation. Sci Rep 11, 14307 (2021). https://doi.org/10.1038/s41598-021-93571-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-93571-4
This article is cited by
-
Levofloxacin prophylaxis and parenteral nutrition have a detrimental effect on intestinal microbial networks in pediatric patients undergoing HSCT
Communications Biology (2023)
-
Linking gastrointestinal microbiota and metabolome dynamics to clinical outcomes in paediatric haematopoietic stem cell transplantation
Microbiome (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.