Abstract
Malnutrition affects growth and development in humans and causes socio-economic losses. Normal maize is deficient in essential amino acids, lysine and tryptophan; and vitamin-A. Crop biofortification is a sustainable and economical approach to alleviate micronutrient malnutrition. We combined favorable alleles of crtRB1 and lcyE genes into opaque2 (o2)-based four inbreds viz. QLM11, QLM12, QLM13, and QLM14 using marker-assisted backcross breeding. These are parents of quality protein maize versions of two elite hybrids viz. Buland and PMH1, grown in India. Gene-based SSRs for o2 and InDel markers for crtRB1 and lcyE were successfully employed for foreground selection in BC1F1, BC2F1, and BC2F2 generations. The recurrent parent genome recovery ranged from 88.9 to 96.0% among introgressed progenies. Kernels of pyramided lines possessed a high concentration of proA (7.14–9.63 ppm), compared to 1.05 to 1.41 ppm in the recurrent parents, while lysine and tryptophan ranged from 0.28–0.44% and 0.07–0.09%, respectively. The reconstituted hybrids (RBuland and RPMH1) showed significant enhancement of endosperm proA (6.97–9.82 ppm), tryptophan (0.07–0.09%), and lysine (0.29–0.43%), while grain yield was at par with their original versions. The dissemination of reconstituted hybrids holds significant promise to alleviate vitamin-A deficiency and protein-energy malnutrition in developing countries.
Similar content being viewed by others
Introduction
Maize is treasured as a profitable crop for its productivity and nutritive qualities and is also referred to as “Queen of Cereals”1. It is currently cultivated in more than 150 countries with a total global production of 1147.62 million tons from 193.73 million ha land with an average yield of 5922 kg/ha2. It provides about 30% of the food calories to more than 4.5 billion people in developing countries and also serves as a major component of animal feed3. It is considered as a poor man’s nutria-cereal due to its high protein content, carbohydrates, fats, and few of the vital vitamins and minerals4.
The maize kernel constitutes 16.5% of the protein, 3–5% oil, 3% crude fiber, up to 2% soluble sugars, 15% water, and 65–70% starch5. The major constituent of endosperm is starch, and about 8–10% is grain protein, of which 60% is composed of prolamins known as zeins4. However, zeins are devoid of the essential amino acids, lysine and tryptophan, make maize’s nutritional quality poor. The discovery of classic recessive mutant opaque2 (o2) has improved the grain nutritional value by reducing the synthesis of zein proteins and increases endosperm lysine and tryptophan by about 2-folds6. O2 gene localized on chromosome 7 encodes an endosperm-specific bZIP transcription factor that recognizes O2 box in the promoters of α- and β-zein genes and regulates their expression. As a result, the mutated o2 gene has several pleiotropic effects like soft texture, susceptibility to diseases, shriveled kernels, and decline in yield. Later on, breeding o2 mutants with hard vitreous kernels and retaining the high lysine content has led to the development of Quality Protein Maize (QPM). The nutritional benefits of QPM have been well documented in feeding trials7,8. A number of QPM conversion programs have been successfully accomplished9,10,11,12.
Vitamin-A deficiency (VAD) has emerged as a pressing health issue that affects millions of people from many countries in the world13. Maize possesses both proA (α-carotene, β-carotene, and β-cryptoxanthin) and non-proA (lutein and zeaxanthin) carotenoids. Most of the reports concluded that the maize kernels naturally exhibit an appreciable variation in carotenoid levels14,15. However, commercially grown yellow varieties in many parts of the world contain less than 1.5 ppm of proA compared to the target level of 15 ppm set by Harvest Plus under the biofortification program initiated by CIMMYT16. Optimizing β-carotene accumulation requires enhanced flux to the β-branch of the pathway in combination with limiting hydroxylation of β-carotene to downstream xanthophyll compounds that no longer have provitamin-A activity. The key candidate genes influencing β-carotene concentration are lycopene ε-cyclase (lcyE) located on chromosome 8 and β-carotene hydroxylase1 (crtRB1) present on chromosome 10 that results in enhancement of proA in maize14,17,18. Three functional polymorphisms, 5ˊTE (in the 5ˊ-Untranslated Region), InDel (in the coding region), and 3ˊTE (in the 3ˊ-Untranslated Region) have been identified through association mapping in both genes, and favorable alleles have significantly increased the β-carotene content. Thus, breeding maize for increased levels of proA and essential amino acids will play a crucial role, especially in the developing world to combat vitamin-A deficiency and protein-energy malnutrition (PEM) in achieving nutritional security more holistically14,17,19.
The favorable alleles of crtRB1, lcyE, and o2 genes hold an immense perspective for the development of beta-carotene plus QPM enriched lines in an accelerated and resource proficient manner without progeny testing through marker-assisted selection (MAS)20,21,22. It also considerably reduces the breeding cycles required to retain the recurrent parent genome (RPG)10. Substantial efforts have been put forward to enhance the proA level in maize15,23,24. But a few breeding efforts have been made to combine two traits i.e.QPM and beta-carotene13,21,25. The present scenario necessitates developing maize genotypes with a combination of QPM and proA so that value-added product reaches poor communities. Buland and PMH1 are the popular maize hybrids released for cultivation in the North-Western Plain Zones of India. Our previous investigation has led to the development of QPM versions viz., BulandQ (QLM11 × QLM12) and PMH1Q (QLM13 × QLM14)12. The present study was thus aimed to (i) pyramid the favorable alleles of crtRB1 and lcyE genes into QPM background using marker-assisted backcross breeding (MABB), (ii) evaluate the nutritional quality of the introgressed lines and reconstituted hybrids, and (iii) assess the agronomic performance and yield potential of the MAS-derived genotypes.
Results
Parental polymorphism
The recurrent parents (QLM11, QLM12, QLM13, and QLM14) and donor parent (HP467-15) were authenticated for crtRB1 3ˊTE and lcyE 5ˊTE polymorphism using gene-based markers. All the QLM inbreds revealed 296 bp fragment, while donor inbred showed 543 bp of amplicon for crtRB1 3ˊTE locus (Supplementary Fig. S1a). Likewise, all recurrent parents registered 300 bp unfavorable allele whereas donor parent possessed 650 bp favorable allele at lcyE 5ˊTE locus (Supplementary Fig. S1b). Also, the recessive o2 allele was present in all QPM recurrent parents while the donor parent had dominant O2 allele. The SSR marker phi057 distinguished between the recurrent parents- QLM12, QLM14 (o2: 170 bp) and donor parent- HP467-15 (O2: 140 bp) (Supplementary Fig. S1c). While QLM11, QLM13 (o2: 145 bp), and donor parent (O2: 160 bp) were discriminated for O2 locus with SSR marker umc1066 (Supplementary Fig. S1d). All the gene-based markers were inherited co-dominantly, and thus selection of heterozygotes in backcross generations is simple and straightforward. SSR markers spanning all the 10 maize linkage groups were analyzed for parental polymorphism between QLM inbreds and CIMMYT β-carotene donor inbred for identification of polymorphic markers to follow background selection. Of the 324 genome-wide SSR markers genotyped on parental inbreds, 161 (49.69%), 134 (41.35%), 144 (44.44%), and 120 (37.03%) SSR markers showed polymorphism between QLM11, QLM12, QLM13, QLM14, and HP467-15, respectively (Supplementary Table S1; Supplementary Fig. S2). The polymorphic markers identified were then subsequently used in screening backcross progenies for the selection of genotypes with the highest background recovery.
Marker-assisted selection of crtRB1, lcyE, and o2
A total of 390 BC1F1 progenies of QLM11 × HP467-15/QLM11 were screened with crtRB1 3ˊTE marker and 180 were heterozygous for the crtRB1 allele. These 180 plants were subjected to screening for lcyE 5ˊTE marker. A total of 75 plants were identified as heterozygous for both crtRB1 and lcyE genes. Further, these positive plants were analyzed with o2 specific marker and 35 plants were homozygous for the o2 allele (Table 1). Similarly, a total of 45, 49, 19 plants were heterozygotes for both crtRB1 and lcyE alleles and homozygous for o2 allele in QLM12 × HP467-15/QLM12, QLM13 × HP467-15/QLM13, and QLM14 × HP467-15/QLM14 crosses, respectively (Table 1). The BC1F1 plants were visually assessed for few phenotypic traits (tassel shape, tassel density, anther color, silk color) towards respective recurrent parent type (Supplementary Table S2). Based on visual observations 25, 20, 30, 15 plants of QLM11 × HP467-15/QLM11, QLM12 × HP467-15/QLM12, QLM13 × HP467-15/QLM13, and QLM14 × HP467-15/QLM14 crosses, respectively, were selected for background selection. A total of 100, 90, 121, and 97 SSR markers were employed for background selection in QLM11 × HP467-15/QLM11, QLM12 × HP467-15/QLM12, QLM13 × HP467-15/QLM13, and QLM14 × HP467-15/QLM14 crosses, respectively. The recovery of the recipient genome varied from 70.0 to 92.8% across four crosses in BC1F1 progenies. The best ones were selected and again backcrossed onto respective recurrent parents to generate BC2F1 progenies (Table 2). Three ears from each cross were selected based on kernel color and ear shape towards respective recurrent parent to raise BC2F1 progenies (Supplementary Table S2). Among the BC2F1 progenies 28, 32, 52, and 43 plants from QLM11 × HP467-15//QLM11, QLM12 × HP467-15//QLM12, QLM3 × HP467-15//QLM13, and QLM14 × HP467-15//QLM14, respectively, were heterozygous for crtRB1, lcyE alleles and homozygous for o2 loci. These foreground positive plants were then again assessed phenotypically on the basis of tassel shape, tassel density, anther color, and silk color. The morphologically selected plants 13, 15, 19, and 17 from each cross of QLM11 × HP467-15//QLM11, QLM12 × HP467-15//QLM12, QLM3 × HP467-15//QLM13, and QLM14 × HP467-15//QLM14, respectively, were subsequently subjected to background selection with respect to only that fraction of region which was heterozygous and not recovered to respective recurrent parent in BC1F1. The background recovery varied from 88.9% to 96.0% among selected BC2F1 progenies across the four crosses (Table 2; Fig. 1).
The introgressed progenies with maximum recovery of the recipient genome and phenotypically similar to the recurrent parent for plant architecture, ear, and grain-related traits were chosen from BC2F1 progenies in each cross for developing BC2F2 generation to fix the crtRB1, lcyE, and o2 genes in a homozygous state. A total of 2103 BC2F2 plants across four crosses were raised from three to five ears/cross and subjected to foreground selection. A total of 21, 30, 25, and 17 plants were homozygous at all three loci in question in BC2F2 progenies of QLM11 × HP467-15//QLM11, QLM12 × HP467-15//QLM12, QLM3 × HP467-15//QLM13, and QLM14 × HP467-15//QLM14, respectively (Fig. 2). The selected homozygotes were self-pollinated to generate BC2F3 seeds. The introgressed progenies in the respective backgrounds were grown during the rainy season of 2018 and the presence of all favorable alleles was confirmed (Supplementary Fig. S3).
Foreground screening of BC2F2 progenies using gene specific markers. (a), Multiplex PCR for selection of favorable alleles for crtRB1 and lcyE genes; (b), Amplification profile for selection of o2 gene using umc1066 SSR marker; (c), Amplification profile for selection of o2 gene using phi057 SSR marker. RP: recurrent parent; DP: donor parent; Red star indicates the desirable favorable alleles in homozygous condition; Yellow star indicates homozygous alleles to recurrent parent; Green star indicates both alleles present in heterozygous state.
Agronomic performance of introgressed progenies
The selected 18, 22, 17, and 12 BC2F3 introgressed progenies of QLM11 × HP467-15//QLM11, QLM12 × HP467-15//QLM12, QLM3 × HP467-15//QLM13, and QLM14 × HP467-15//QLM14 crosses, respectively, were evaluated under multi-location trial and revealed high phenotypic similarity with their respective recurrent parents for the various morphological characteristics and grain yield attributing traits (Fig. 3). All the studied traits viz., days to 50% anthesis, days to 50% silking, plant height, cob length, number of kernels per row, and grain yield of the improved versions were at par to original respective inbreds. These introgressed lines exhibited more than 90% similarity to their original inbreds (Table 3; Supplementary Tables S3–S4). The mean yield of QβLM11-1, QβLM12-1, and QβLM13-2 was numerically higher than QLM11, QLM12, and QLM13, respectively. The selection was made on a single plant basis from selected progenies to advance the stable and uniform lines.
Biochemical evaluation of introgressed progenies
All of the introgressed lines showed a significant increase in β-carotene over the original inbreds. Lysine and tryptophan contents varied from 0.28 to 0.44% and 0.07 to 0.09%, with an average of 0.34 and 0.08% across crosses (Table 4). The concentration of β-carotene ranged from 4.95 to 7.46 ppm, while β-cryptoxanthin from 3.62 to 4.47 ppm and proA varied from 7.14 to 9.63 ppm. The higher accumulation of proA content may be due to both favorable alleles of crtRB1 and lcyE were introgressed in the background of QPM that has contributed additively. QβLM11-A, QβLM12-C, QβLM13-B, and QβLM14-C ascertained high proA content of 9.28, 9.63, 9.01, and 8.36 ppm, respectively (Table 4). The biochemical parameters were also confirmed in the advanced generations (data not given).
Agronomic performance of reconstituted hybrids
The improved lines with a similar degree of kernel texture, shape, and color owing to their respective recurrent parents were crossed in original combination to reconstitute QPM + β-carotene enriched maize hybrids. Numerically the reconstituted hybrid versions were at par to their original respective hybrids as well as within same reconstituted hybrid versions with respect to traits like plant height, ear height, cob girt, cob length, and the number of kernels per row (Table 5 and Supplementary Tables S5, S6). The reconstituted hybrids also exhibited a high degree of resemblance for various morphological characters with the respective original versions like late-maturing behavior and medium long ears. The grain characteristics like orange and yellow flint grains of Buland and PMH1, respectively, were retained among the newly reconstituted RBuland and RPMH1 hybrids (Fig. 4).
Biochemical analysis of reconstituted hybrids
Tryptophan content ranged from 0.06 to 0.09% in RBuland versions whereas it varied from 0.07 to 0.09% in RPMH1 versions (Fig. 5a). The proA concentration among the RBuland hybrids ranged from 6.97 to 9.82 ppm whereas it varied from 7.28 to 9.39 ppm in RPMH1 hybrids with an increase of 4–6-folds over their original Buland and PMH1 checks (Fig. 5b). The reconstituted hybrid versions had significantly higher β-carotene, β-cryptoxanthin, lysine and tryptophan as compared to their original hybrids (Supplementary Table S7). The reconstituted hybrids of RBuland had Zn content from 34.52 to 36.53 ppm and Fe from 32.13 to 35.94 ppm while Buland possessed 34.82 ppm Zn and 32.55 ppm Fe. Zn and Fe content among RPMH1 hybrid versions varied from 47.87–49.41 ppm and 35.27–38.75 ppm, respectively, whereas PMH1 had 48.30 and 37.10 ppm Zn and Fe, respectively (Supplementary Table S8).
Quality parameters of reconstituted hybrids versions vis‐à‐vis original hybrids. (a), Lysine and tryptophan concentration in reconstituted and original hybrids; (b), BC, BCX and proA concentration in reconstituted and original hybrids. BC, β-carotene; BCX, β-cryptoxanthin; proA, ProvitaminA; bar indicates standard error.
Chapatti making performance and sensory quality of reconstituted hybrids
The physico-chemical characteristics are an important group as it determines the quality of maize chapattis. Statistically significant variations were observed relative to water absorption of flour, roll-ability, and puffing in all reconstituted hybrids versus checks (Table 6). Sensory parameters of chapatti prepared from RBuland and RPMH1 hybrids and original hybrids as well as control flour from the market were assigned score based on the Hedonic scale. Sensory scores for appearance varied from 6.80 to 9.73, with higher scores for RBuland-2 chapattis as the latter showed an appealing color with light brown spots spread evenly over the surface. Overall acceptability of chapatti quality was higher for RPMH1-4 (9.45) and RBuland-8 (8.55) hybrids. Further, the grains of these hybrids were processed to form ready-to-eat finished Massa products using the nixtamalization method, transforming the raw material into food products that are more suitable for commercial trade as they provide convenience and extended shelf life. These shelf-stable products were then used in the production of homemade tortillas which are the main source of carbohydrate and calcium with reduced meal preparation time and convenience.
Discussion
Globally, QPM varieties were developed to address protein-energy malnutrition problems and significantly benefited the people in underdeveloped nations, particularly in Africa5. It has been reported that the QPM diet greatly improved the health of children prone to severe malnutrition26,27. However, normal maize cultivars commonly grown by farmers and QPM varieties contain less than 2 mg g−1 of proA15 which is below the recommended value to meet the daily requirements in a diet28. Henceforth, the development and deployment of nutritious biofortified maize with higher protein quality and proA content is one of the most sustainable strategies to tackle “hidden hunger,” more effectively for millions of people who depend upon maize for sustenance. Previous studies have shown that down regulation of lcyE reduces the ratio of the α-carotene branch to the β-carotene branch and crtRB1 favors the accumulation of β-carotene over that of β-cryptoxanthin in the carotenoid biosynthetic pathway, leading to an increase in levels of provitamin-A14,17. Four natural lcyE polymorphisms (lcyE 5ˊTE, lcyE SNP216, lcyE SNP2238 and lcyE 3ˊInDel) and three crtRB1 polymorphisms (crtRB1 5ˊTE, crtRB1InDel4 and crtRB1 3ˊTE) were identified. The favorable allele of lcyE 5ˊTE and crtRB1 3ˊTE causes a significant increase in provitamin-A in the endosperm14,16,17,18. Therefore these two loci (lcy 5ˊTE and crtRB1 3ˊTE) were targeted to introgress into QPM version of elite maize inbreds using MABB.
We employed co-dominant gene specific markers that clearly distinguished all the four recurrent parents (QLM11, QLM12, QLM13, and QLM14) from the donor line HP467-15. The results were consistent with other studies regarding allele size for each marker10,13,16,21,25. The foreground selection of target genes in each backcross and selfed generation facilitated the identification of desirable genotypes at the early stage of plant growth. In the BC1F1 generation, we selected plants that were homozygous for o2 allele but heterozygous for crtRB1 and lcyE alleles. Therefore, the selected progenies were fixed for o2 allele and further no segregation occurred for o2 allele in the subsequent generations. These results are in accordance with earlier reports13,18,21,29,30,31.
It has been observed that during foreground selection in different generations crtRB1 gene showed segregation distortion across the crosses. Significant segregation distortion (SD) for crtRB1 was also documented in other studies13,16,21. On the contrary, lcyE and o2 genes segregated as per Mendelian inheritance10,11,12,25. We performed simultaneously background selection for recovery of the parental genome using 80–100 genome-wide SSR markers for the selection of desirable genotype in each cross. Background analysis revealed 88.9–96.0% recovery of recurrent parent genome. This enabled us in precise selection of the foreground positive progenies possessing high RPG10,11,13,16,21,32. The main emphasis of background selection was to recover the maximum proportion of recurrent parent genome at non-target loci using DNA markers distributed evenly on the genome in an accelerated manner12. The identified individuals with a range from low to high RPG content in BC1 progenies revealed unbiased sampling and marker data points. Further high RPG recovery in BC2 progenies demonstrated that two rounds of background selection resulted in selections of desirable genotypes. Servin and Hospital33 illustrated that the use of either a few optimally placed markers or more sub-optimally placed markers can efficiently control large chromosomal regions and leads to better control of the return to the recipient genome. Thus, the criteria of selecting optimal positioning of markers in the present study resulted in maximization of the expected proportion.
Phenotypic evaluation and selection was done in the genotypically identified BC2F3 improved progenies having crtRB1, lcyE, and o2 genes in homozygous condition. The MAS-derived pyramided lines with high RPG also exhibited a high degree of resemblance with their corresponding recurrent parent for plant architecture, ear type, and grain characteristics as evident from our results and previous studies10,16,34. Similarly, reconstituted hybrid (RBuland and RPMH1) versions were at par to checks and original hybrids in grain yield and other attributing traits. This high degree of phenotypic similarity among the reconstituted hybrids is also ascribed to high RPG recovery of the introgressed progenies10,11,12,16,25.
The proA content of the improved lines was increased from 4–8-folds when compared to their original QPM parents while lysine and tryptophan content showed an increase of 1.38–2.50 and 1.68–2.86-folds, respectively across the crosses over the normal parental inbreds. The reconstituted hybrids had 4–6-folds higher proA while 1.40–2.50 and 1.54–2.83-folds increase in lysine and tryptophan over their original versions. The variation for lysine and tryptophan among improved progenies could be attributed to the presence of amino acid modifies which varied with different genetic backgrounds11,12. Similar results were obtained by previous studies10,25,35,36,37,38. It was observed that the QPM introgressed inbreds used in the present study possessed comparatively more tryptophan content that might be due to the fact of genetic interaction of QPM inbred with non-QPM β-carotene donor inbred. Also, differences for β-carotene content among the improved lines and reconstituted hybrid versions were due to other genetic loci like crtRB3, CCD1, and ZEP1 in β-carotene synthesis pathway apart from favorable alleles of crtRB1 and lcyE genes that might have contributed to the increase of proA39,40,41,42,43. The retention of high proA even after four months of storage was observed. It might be due to the presence of favorable alleles for higher retention of proA44. Studies on retention of proA during different storage periods showed that degradation of proA occurs at the first three months of storage and gradually stabilizes after six months (4–6 months)44. It suggests that crtRB1 and lcyE -based genotypes would still hold immense significance nutritionally over normal maize. The advantage of stacking favorable alleles of crtRB1 and lcyE genes for proA over as single effects has been reported in a number of studies45,46,47. The micronutrient concentrations in the reconstituted hybrids were at par to original hybrids that could be due to high background recovery of introgressed progenies. Our study revealed variability for Fe and Zn between RBuland and RPMH1 indicating the accessibility of wider genetic variation to be utilized for the genetic improvement of kernel micronutrient traits in maize. Previous studies also documented considerable variations for the kernel Fe and Zn concentrations among maize genotypes48,49,50,51.
It was observed that combined quality parameters of QPM and proA affected flour color, flavor, the texture of chapatti but all the values were in acceptable range among versions of reconstituted hybrids as compared to original hybrids. Thus, chapattis from reconstituted hybrids have QPM addition with β-carotene and can be considered acceptable to avail nutritional, phytochemical, and health benefits of maize. These reconstituted hybrids when subjected to nixtamalization resulted in Massa with higher nutritional value with value addition of QPM and proA compared to the original hybrids. Similar studies reported that nixtamalized maize has a higher nutritional value (increased bioavailability of niacin, improved protein quality, increased calcium) with reduced mycotoxins content52,53,54. Nutritionally RBuland-4 and RPMH1-4 were better as compared to other versions while overall acceptability as chapatti quality was for RBuland-8 and RPMH1-4 versions. Also, the yield of RPMH1-4 was higher whereas yields of both RBuland-4 and RBuland-11 were comparable to each other as well as both outperformed at all locations as compared to the original checks. Overall, RBuland-4 and RPMH1-4 are selected for further seed multiplication and commercialization.
Conclusions
We report here the successful stacking of o2, crtRB1, and lcyE genes into the background of four QPM inbreds and developed nutritionally enriched hybrids using MABB. The dual (MAS and phenotypic) selection approach provided an opportunity to unite the desirable agronomic traits with enriched nutritional traits in an accelerated manner. Thus, in the near future, the reconstituted hybrids could likely be grown in India without any difference in grain yield and to greater gain in terms of grain nutrition compared with their originals. The newly developed β-carotene enriched lines in the QPM background and their reconstituted hybrids are value-added products that could serve as pre-breeding material for elite line conversion. The clinical trial of nutritionally enriched hybrids in collaboration with a hospital could also be tested on under-nutrition people at a small scale.
Materials and methods
The present study was conducted at Punjab Agricultural University (PAU), Ludhiana and ICAR-Indian Agricultural Research Institute (IARI), New Delhi, India during the years from 2015 to 2019 with relevant institutional guidelines and legislation. Necessary permission was obtained from the institute for the collection of plant material.
Plant materials
The investigational material consisted of four elite QPM maize inbreds viz. QLM11, QLM12, QLM13, and QLM14. These QPM inbreds were developed by introgression of o2 gene from different QPM donors to locally adapted inbreds, LM11, LM12, LM13, and LM14 at PAU, Ludhiana12. These are the parental inbreds of single cross QPM version hybrids of BulandQ (QLM11 × QLM12) and PMH1Q (QLM13 × QLM14). QLM11 had cylindrical ears with orange-red round and bold kernels, whereas QLM12 possessed small ears with pointed dull orange kernels. Similarly, QLM13 exhibited medium conical-cylindrical ears with bold round dull yellow kernels, while QLM14 also had conical-cylindrical ears having yellow kernels. Likewise, leaf attitude was semi-dropping and dropping in QLM11 and QLM12; and semi-erect in QLM13 and QLM14. The other phenotypic descriptors (tassel shape, tassel density, anther glume color, anther color, and silk color) of QLM inbreds are enlisted in Table S2. However, β-carotene in the kernels of QPM inbreds and their hybrids is low; hence were targeted for β-carotene enrichment. HP467-15 inbred (CIMMYT, Mexico) was used as a pollen contributor for the introgression of high β-carotene content into the QPM genetic background of each recurrent parent. The pedigree information of recurrent parents and their tryptophan content is given in Supplementary Table S9.
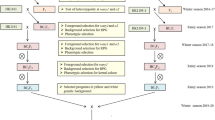
Marker-assisted backcross breeding for development of improved progenies and hybrids
The crossing scheme followed for the reconstitution of nutritionally enriched hybrids and details of population development is represented in Fig. 6 and Supplementary Table S10. The population development involved two parallel crossing schemes. Each QLM inbred was crossed to HP467-15 at ICAR- IARI, New Delhi during the rainy season (July–October) 2015. The F1s were raised at Punjab Agricultural University (PAU), Ludhiana during the spring season (February-May 2016), and the hybridity of F1s was confirmed by using target gene-specific markers. The F1s of each cross were backcrossed twice to develop BC2F1 progenies during rainy 2016 and spring season of 2017. The selected BC2F1 plants were selfed and advanced to BC2F2, progenies that were grown during rainy 2017. The homozygous progenies for target loci were selected and selfed to generate BC2F3 and BC2F4 progenies during rainy 2018 and spring 2019. Line conversion for stacking of genes in recurrent background involved different selection indices- foreground, background, phenotypic and biochemical at the appropriate step as specified in the crossing scheme. The promising converted lines were crossed in original hybrid combination to reconstitute biofortified maize hybrids, Buland and PMH1 during spring season 2019 and were evaluated during rainy season 2019.
Marker-assisted breeding scheme for the development of crtRB1, lcyE introgressed lines in background of four QPM inbreds and reconstitution of original hybrids versions. RP: recurrent parent; DP: donor parent; A: homozygous allele to donor parent; H: heterozygous allele; FS: foreground selection; BS: background selection.
Genomic DNA isolation and PCR analysis
Genomic DNA was extracted following modified CTAB protocol from two-week old seedlings55. The DNA quality and quantity was checked by 0.8% agarose gel. Polymerase chain reaction was performed using functional markers for both the crtRB1 3ˊTE and lcyE 5ˊTE favorable alleles16,47. The PCR amplification was performed in a Veriti 96 well (Applied Biosciences, Invitrogen, UK) and GeneAmp PCR system 9700 (Applied Biosystems by Thermo Fisher Scientific) thermal cyclers. The amplification of lcyE 5ˊTE was executed by a “touch-down” profile of initial denaturation at 95 °C for 5 min, followed by 16 cycles of denaturation at 95 °C for 45 s, annealing at 58 °C for 45 s and extension at 72 °C for 45 s with each cycle decrease in annealing temperature by 0.5 °C, then 20 cycles were profiled at 95 °C for 45 s, annealing at 56 °C for 45 s and extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. The amplification for crtRB1 3ˊTE was achieved using a thermal profile of initial denaturation at 94 °C for 4 min, followed by 25 cycles of denaturation at 94 °C for 1 min, annealing at 53.5ºC for 1.30 min and extension at 72 °C for 1.30 min, and a final extension at 72 °C for 7 min. Multiplex amplification for crtRB1 3ˊTE and lcyE 5ˊTE was also standardized using a thermal profile of initial denaturation at 95 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 50 s, annealing at 59 °C for 50 s and extension at 72 °C for 55 s, and a final extension at 72 °C for 8 min. The amplified product was kept on hold at 4 °C after the completion of the thermal profile. SSR gene-based markers (umc 1066 and phi 057) were deployed for the selection of o2 gene. The PCR profile for amplification of o2 gene and genome-wide SSR markers was performed using the protocol of Kaur et al.12. A negative control (without template DNA) was included in each plate during every amplification reaction. Amplicons were resolved and analyzed using 3.5% Metaphor agarose gel electrophoresis at 120 V for 3-5 h.
Foreground selection for target genes
Selection for target alleles was performed in both backcrossed populations (BC1F1 and BC2F1) using crtRB1 3ˊTE, lcyE 5ˊTE, and o2 gene specific markers. Plants having crtRB1 3ˊTE and lcyE 5ˊTE alleles in heterozygous form were further directed for the selection of o2 allele with phi057 and umc1066 markers in QLM12, QLM14, and QLM11, QLM13 crosses, respectively. The plants having all three genes were subsequently selfed to generate the BC2F2 progenies. The plants with all three genes in a homozygous state were selected in BC2F2 progenies across four crosses and subsequently advanced to BC2F3 progenies. The segregation pattern of each marker locus in each generation was checked by using standard Chi-square analysis for the goodness of fit.
Background selection for recurrent parent genome
A total of 324 SSR markers spanning all the 10 chromosomes of maize genome were surveyed for polymorphism between each recurrent and donor parent. The coverage of SSR markers per chromosome from each chromosomal bin varied from 14 to 55 markers (Supplementary Table S11). The sequences of the SSR primers were retrieved from maize genome database (www.maizegdb.org) and custom synthesized (IDT, USA). The polymorphic markers were surveyed in BC1F1 progenies of each cross involving QLM11, QLM12, QLM13, and QLM14 as recurrent parents respectively, for selecting the individuals having high background coverage. The background selection was again preceded in BC2F1 progenies across crosses for those genomic regions which were not recovered in selected BC1F1 plants. The amplicon of each marker was registered as ‘A’ for the recurrent parent allele and ‘B’ for the donor allele while ‘H’ for both the alleles. Graphical genotyping (GGT) analysis was carried out to analyze the introgression from donor parent on the individual chromosome using GGT software56. The % recurrent parent genome (RPG) recovery was calculated as the ratio of the number of SSRs showing recurrent parent allele in a homozygous state to the total number of polymorphic SSRs applied for background selection.
Morphological characterization of introgressed progenies
Improved BC2F3 progenies of each cross along with their original parents were evaluated for phenotypic characteristics and grain yield attributing traits during rainy season 2018 at three locations viz. (i) Punjab Agricultural University (PAU), Ludhiana, (ii) Regional Research Station (PAU) Gurdaspur and (iii) IARI, New Delhi. Each progeny was raised in two replications in a randomized complete block design (RBCD) and two rows/replication of 3 m row length was grown with a plant-to-plant distance of 20 cm and row to row distance of 60 cm. Standard agronomic practices were followed for raising the crop at each experimental site. The data was recorded for various agronomic characters such as plant height (cm), days to anthesis (days), days to silking (days), cob length (cm), cob girth (cm), number of kernel rows/cob, number of kernels/row and grain yield (Kg/ha). Data were recorded on daily visual observations during the flowering period. Anther glume color, anther color, silk color, tassel shape, tassel density (low to high), and kernel color was documented on a visual basis. Data was subjected to analysis of variance (ANOVA) to determine the significant differences among treatments using SAS (9.4 version) computer software.
Estimation of proA, lysine, and tryptophan contents
The quality parameters were estimated from BC2F4 kernels of selected lines across the crosses after 4 months of storage. The procedures specified by Vignesh et al.29 were followed for extraction of β-carotene (BC) and β-cryptoxanthin (BCX). BC and BCX were estimated using Dionex Ultimate 3000 UHPLC System (Ultra High-Performance Liquid Chromatography; Thermo Scientific, Massachusetts, USA). The proA concentration (ppm on a dry weight basis) was calculated as the sum of BC plus half the BCX concentration18. Lysine and tryptophan content in the endosperm was estimated using Dionex Ultimate 3000 UHPLC system according to the procedure described by Sarika et al.38. Each parameter from each sample was estimated from two technical replications. The data generated were analyzed to calculate the standard error of the mean and CD (critical difference) value for comparing the averages among sets.
Reconstitution of hybrids and their evaluation
The selected BC2F4 (QβLM11, QβLM13 as a seed parent and QβLM12, QβLM14 as a male parent) progenies of the improved inbreds were crossed in different combinations to reconstitute hybrids (RBuland and RPMH1) during 2019 (spring season) at Punjab Agricultural University, Ludhiana. The nutritionally enriched reconstituted hybrids with their corresponding original hybrids were raised in two rows of 3 m row length with three replications in a randomized complete block design (RBCD) and were evaluated for their agronomic performance at three locations viz. (i) PAU, Ludhiana, (ii) Regional Research Station (PAU) Gurdaspur and (iii) IARI, New Delhi during 2019 (rainy season). To enhance the efficiency of selection various morphological characteristics were recorded as per DUS (Distinctness, Uniformity, and Stability) guidelines. Lysine, tryptophan, β-carotene, β-cryptoxanthin, and proA content were estimated from selfed seeds of reconstituted hybrid versions with the combination of three genes i.e. crtRB1, lcyE, and o2, along with their corresponding original hybrids. For each entry the grains were divided into three parts and analyzed as three replicates for estimation of different mineral nutrients viz., Fe, Zn, Cu, Se, Mn, S, K, P, and Mg using simultaneous multi-element inductively coupled plasma–optical emission spectrometer (ICP-OES, Perkin Elmer) as per the methodology described by Arora et al.57. The value obtained was then multiplied by initial sample volume, divided by initial weight of grains, and expressed as μg element g−1 dry grain material (ppm)58.
Assessment of reconstituted hybrids for chapatti (flatbread) making quality
Nutritionally enriched reconstituted hybrids with their corresponding checks along-with a local market sample were used for chapatti formulation. Whole grain flour was obtained by grinding the grains in domestic attachakki (burr mill) using the mesh sieves of 60 µm particle size. 100 gm of flour was mixed with optimum water to make dough and was then allowed to rest for 20–30 min. The optimum level of water for dough development was recorded in all the reconstituted hybrids. Dough ball of 60 g each was rolled and baked on a hot skillet; until brown spots appeared on it. The chapattis dough was evaluated for dough handling, roll-ability, and puffing. Further, the maize grains were subjected to a nixtamalization process53 to make soft dough known as “Massa”. This Massa was then used for the production of tortillas which are the main source of carbohydrates and calcium54. Sensory properties like appearance, color, flavor, texture, and overall acceptability were assessed using ten points Hedonic scale (9 for maximum and 0 for minimum) by the panel of sixteen semi-trained judges59. The panelists (8 males and 8 females) provided signed written informed consent for assessment of sensory parameters. This study was approved by the local ethics committee of Department of Food Science and Technology, PAU, Ludhiana. All methods were carried out in accordance with relevant guidelines and regulations.
References
Murdia, L. K., Wadhwani, R., Wadhawan, N., Bajpai, P. & Shekhawat, S. Maize utilization in India: an overview. Am. J. Food Nutr. 4(6), 169–176 (2016).
FAOSTAT. Faostat statistical database. Food and Agriculture Organization of the United Nations. Preprint at http://www.fao.org/faostat/en/#home (2020).
Shiferaw, B., Prasanna, B. M., Hellin, J. & Banziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 3, 307–327 (2011).
Prasanna, B. M., Vasal, S. K. & Kassahun, B. Quality protein maize. Curr. Sci. 81, 1308–1319 (2001).
Nuss, E. T. & Tanumihardjo, S. A. Maize: a paramount staple crop in the context of global nutrition. Compr. Rev. Food Sci. Food Saf. 9, 417–336 (2010).
Mertz, E. T., Bates, L. S. & Nelson, O. E. Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145, 279–280 (1964).
Panda, A. K. et al. Replacement of normal maize with quality protein maize on performance, immune response and carcass characteristics of broiler chickens. Asian Aust. J. Anim. Sci. 23, 1626–1631 (2010).
Bidi, T. N., Gasura, E., Ncube, S., Saidi, P. T. & Maphosa, M. Prospects of quality protein maize as feed for indigenous chickens in Zimbabwe: a review. Afr. Crop Sci. J. 27, 709–720 (2019).
Babu, R. et al. Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor. Appl. Genet. 111, 888–897 (2005).
Gupta, H. S. et al. Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 132, 77–82 (2013).
Hossain, F. et al. Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J. Genet. 97, 287–298 (2018).
Kaur, R. et al. Genetic enhancement of essential amino acids for nutritional enrichment of maize protein quality through marker assisted selection. Physiol. Mol. Biol. Plants 26(11), 2243–2254 (2020).
Chandran, S. et al. Marker-assisted selection to pyramid the Opaque-2 (O2) and β-Carotene (crtRB1) genes in maize. Front. Genet. 10, 859. https://doi.org/10.3389/fgene00859 (2019).
Harjes, C. E. et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333 (2008).
Pixley, K. et al. “Biofortification of maize with provitamin A carotenoids,” in Carotenoids and Human Health, ed S. Tanumihardjo (New York, NY: Springer Science+ Business Media), 271–292 (2013).
Muthusamy, V. et al. Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9(12), e11583 (2014).
Yan, J. B. et al. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42, 322–327 (2010).
Babu, R., Rojas, N. P., Gao, S., Yan, J. & Pixley, K. Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor. Appl. Genet. 126, 389–399 (2013).
Zhang, X., Pfeiffer, W. H., Palacios-Rojas, N., Babu, R. & Bouis, H. Probability of success of breeding strategies for improving provitamin A content in maize. Theor. Appl. Genet. 125, 235–246 (2012).
Das, G., Patra, J. K. & Baek, K. H. Insight into MAS: a molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 8, 985. https://doi.org/10.3389/fpls00985 (2017).
Liu, L. et al. Introgression of the crtRB1 gene into quality protein maize inbred lines using molecular markers. Mol. Breeding 35, 154. https://doi.org/10.1007/s11032-015-0349-7 (2015).
Babu, R. Palacios, N. & Prasanna, B. M. Biofortified Maize- A genetic avenue for nutritional security. Editor(s): Varshney, R. K. & Tuberosa, R. Translational Genomics for Crop Breeding: Abiotic Stress, Yield and Quality, 2, 61-76 (2013)
Andersson, M. S., Saltzman, A., Virk, P. S. & Pfeiffer, W. H. Progress update: crop development of biofortified staple food crops under Harvestplus. Afr. J. Food Agric. Nutr. Dev. 17, 11905–11935 (2017).
Natesan, S. et al. Characterization of crtRB1 gene polymorphism and β-carotene content in maize landraces originated from north eastern himalayan region (NEHR) of India. Front. Sustain. Food Syst. 4, 78. https://doi.org/10.3389/fsufs00078 (2020).
Zunjare, R. U. et al. Development of biofortified maize hybrids through marker-assisted stacking of β-Carotene Hydroxylase, Lycopene-ε-Cyclase, and Opaque2 genes. Front. Plant Sci. 9, 17. https://doi.org/10.3389/fpls00178 (2018).
Akalu, G., Taffesse, S., Gunaratna, N. & De Groote, H. The Effectiveness of quality protein maize in improving the nutritional status of young children in the Ethiopian highlands. Food Nutr. Bull. 31(3), 418–430 (2010).
De Gunaratna, N. S., Groote, H., Nestel, P., Pixley, K. V. & McCabe, G. P. A meta-analysis of community-based studies on quality protein maize. Food Policy 35, 202–210 (2010).
Institute of Medicine, Dietary reference intake (DRIs): Estimated average requirements (Washington, DC: Food and Nutrition Board, Institute of Medicine). Preprint at http://www.iom.edu/Activities/Nutrition/summaryDRIs/media (2012).
Vignesh, M. et al. Genetic variability for kernel β-carotene and utilization of crtRB1-3′TE gene for biofortification in maize (Zea mays L.). Indian J. Genet. 72, 189–194 (2012).
Selvi, D. T. et al. Assesment of crtRB1 polymorphism associated with increased β-carotene content in maize (Zea mays L.) seeds. Food Biotech. 28, 41–49 (2014).
Goswami, R. et al. Marker-assisted introgression of rare allele of β-carotene hydroxylase (crtRB1) gene into elite quality protein maize inbred for combining high lysine, tryptophan and provitamin A in maize. Plant Breed. 138, 174–183 (2019).
Feng, F., Wang, Q., Liang, C., Yang, R. & Li, X. Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 206, 513–521 (2015).
Servin, B. & Hospital, F. Optimal positioning of markers to control genetic background in marker-assisted backcrossing. J. Hered. 93, 214–217 (2002).
Choudhary, M. et al. Characterization of β-carotene rich MAS derived maize inbreds possessing rare genetic variation in β-carotene hydroxylase gene. Indian J. Genet. 74, 620–623 (2014).
Wu, R. et al. An improved genetic model generates high-resolution mapping of QTL for protein quality in maize endosperm. Proc. Natl. Acad. Sci. U.S.A. 99, 11281–11286 (2002).
Yang, W., Zheng, Y., Zheng, W. & Feng, R. Molecular genetic mapping of a high-lysine mutant gene (opaque-16) and the double recessive effect with opaque-2 in maize. Mol. Breed. 15, 257–269 (2005).
Pandey, N. et al. Molecular characterization of endosperm and amino acids modifications among quality protein maize inbreds. Plant Breed. 135, 47–54 (2015).
Sarika, K. et al. Exploration of novel opaque16 mutation as a source for high lysine and tryptophan in maize endosperm. Indian J. Genet. 77, 59–64 (2017).
Wong, J. C., Lambert, R. J., Wurtzel, E. T. & Rocheford, T. R. QTL and candidate genes phytoene synthase and ζ-carotene desaturase associated with the accumulation of carotenoids in maize. Theor. Appl. Genet. 108, 349–359 (2004).
Chander, S. et al. Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor. Appl. Genet. 116, 223–233 (2008).
Zhou, Y. et al. ZmcrtRB3 encodes a carotenoid hydroxylase that affects the accumulation of α-carotene in maize. J. Integr. Plant Biol. 54, 260–269 (2012).
Kandianis, C. B. et al. Genetic architecture controlling variation in grain carotenoid composition and concentrations in two maize populations. Theor. Appl. Genet. 126, 2879–2895 (2013).
Suwarno, W. B., Pixley, K. V., Palacios-Rojas, N., Kaeppler, S. M. & Babu, R. Genome-wide association analysis reveals new targets for carotenoid biofortification in maize. Theor. Appl. Genet. 128, 851–864 (2015).
Dutta, S. et al. Effect of storage period on provitamin-A carotenoids retention in biofortified maize hybrids. Int. J. Food Sci. Technol. https://doi.org/10.1111/ijfs.14785 (2020).
Azmach, G., Gedil, M., Menkir, A. & Spillane, C. Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol. 13, 227. https://doi.org/10.1186/1471-2229-13-227 (2013).
Gebremeskel, S., Garcia-oliveira, A. L., Menkir, A., Adetimirin, V. & Gedil, M. Effectiveness of predictive markers for marker assisted selection of pro-vitamin A carotenoids in medium-late maturing maize (Zea mays L.) inbred lines. J. Cereal Sci. 79, 27–34 (2017).
Zunjare, R. U. et al. Influence of rare alleles of β-carotene hydroxylase and lycopene epsilon cyclase genes on accumulation of provitamin A carotenoids in maize kernels. Plant Breed. 136, 872–880 (2017).
Banziger, M. & Long, J. The potential for increasing the iron and zinc density of maize through plant-breeding. Food Nutr. Bull. 21, 397–400 (2000).
Dixon, B. M., Kling, J. G., Menkir, A. & Dixon, A. Genetic variation in total carotene, iron and zinc contents of maize and cassava genotypes. Food Nutr. Bull. 21, 419–422 (2000).
Oikeh, S. O., Menkir, A., Dixon, B. M., Welch, R. M. & Glahn, R. P. Genotypic differences in concentration and bioavailabilty of kernel-iron in tropical maize varieties grown under field conditions. J. Plant Nutr. 26, 2307–2319 (2003).
Prasanna, B. M. et al. Genetic variability and genotype × environment interactions for kernel iron and zinc concentrations in maize (Zea mays) genotypes. Indian J. Agric. Sci. 81, 704–711 (2011).
Afoakwa, E., Seaf-Dedah, S. & Simpson, A. Influence of spontenous fermentation on same quality characteristics of maize based cowpea-fortified nixtamalized foods. Afr. J. Food Agric. Nutr. Dev. 7, 1–18 (2007).
Gwirtz, J. A. & Garcia-Casal, M. N. Processing maize flour and corn meal food products. Ann. N. Y. Acad. Sci. 1312, 66–75 (2014).
Perez, L. A. et al. Chemical and physiochemical properties of dried wet masa and dry masa flour. J. Sci. Food Agric. 83, 408–412 (2003).
Saghai-Maroof, H. A., Soliman, K. M., Jorgensen, A. R. & Allard, R. W. Ribosomal DNA spacer length polymorphism in barley: mendelian inheritance, chromosomal location and population dynamics. Proc. Natl. Acad. Sci. U. S. A. 81, 8014–8018 (1984).
Van Berloo, R. GGT: software for display of graphical genotypes. J. Hered. 90, 328–329 (1999).
Arora, S., Cheema, J., Poland, J., Uauy, C. & Chhuneja, P. Genome-wide association mapping of grain micronutrients concentration in Aegilops tauschii. Front. Plant Sci. 10, 54. https://doi.org/10.3389/fpls00054 (2020).
Khokhar, J. S. et al. Variation in grain Zn concentration, and the grain ionome in field-grown Indian wheat. PLoS ONE 13(1), e0192026 (2018).
Linda, M. P. Deborah, A. M. Gail, B. & Elizabeth, L. Laboratory methods for sensory analysis of food. (ed. Linda, M. P.) 1–90 Ottawa - Ontario: Agriculture Canada (1991).
Acknowledgements
Financial support from the Department of Biotechnology, Government of India, through the project entitled ‘Enrichment of nutritional quality in maize through molecular breeding’ (BT/PR10922/AGII/106/944/2014/25.3.2015) is dully acknowledged.
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.V., F.H; experiment designing, Y.V., F.H., J.S.C., G.K.G., A.K; methodology, AD.K., A.K.C., B.K.B; phenotypic data collection, J.S., S.S., A.K., P.S; genotyping, data analysis, J.S., S.S., AD.K; A.K.C., B.K.B., N.K; quality analysis, V.M., AJ.K; writing and original draft preparation: J.S., S.S., P.K.M; review and editing: Y.V., G.K.G., F.H. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, J., Sharma, S., Kaur, A. et al. Marker-assisted pyramiding of lycopene-ε-cyclase, β-carotene hydroxylase1 and opaque2 genes for development of biofortified maize hybrids. Sci Rep 11, 12642 (2021). https://doi.org/10.1038/s41598-021-92010-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-92010-8
This article is cited by
-
Enhancement of nutritional quality in maize grain through QTL-based approach
Cereal Research Communications (2024)
-
Heterotic grouping of provitamin A-enriched maize inbred lines for increased provitamin A content in hybrids
BMC Genomic Data (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.