Abstract
Early bacterial infection (BI) identification in resource-limiting Emergency Departments (ED) is challenging, especially in low- and middle-income counties (LMIC). Misdiagnosis predisposes to antibiotic overuse and propagates antimicrobial resistance. This study evaluates new emerging biomarkers, secretory phospholipase A2 group IIA (sPLA2-IIA) and compares with other biomarkers on their performance characteristic of BI detection in Malaysia, an LMIC. A prospective cohort study was conducted involving 151 consecutive patients admitted to the ED. A single measurement was taken upon patient arrival in ED and was analysed for serum levels of sPLA2-IIA, high-sensitive C-reactive protein (CRP), procalcitonin (PCT), neutrophil percentage (N%), and lactate. All biomarkers’ performance was compared for the outcomes using area under the receiver operating characteristic curve (AUROC), sensitivity, and specificity. The performance of sPLA2-IIA (AUROC 0.93 [95% CI: 0.89–0.97]; Sn 80% [95% CI: 72–87]; Sp 94% [95% CI: 81–89]) was the highest among all. It was comparable with high-sensitive CRP (AUROC 0.93 [95% CI: 0.88–0.97]; Sn 75% [95% CI: 66–83]; Sp 91 [95% CI: 77–98]) but had a higher Sn and Sp. The sPLA2-IIA was also found superior to N%, PCT, and lactate. This finding suggested sPLA2-IIA was recommended biomarkers for BI detection in LMIC.
Similar content being viewed by others
Introduction
The Global Burden of Disease Study reported that infection caused more than 10 million lives lost per year1. Various studies show a marked difference in mortality rate from high-income to low- and middle-income countries (LMIC), with the highest death, observed in Sub-Saharan Africa and Southeast Asia (SEA)2,3,4. In Malaysia, an LMIC in SEA, pneumonia remains amongst the leading cause of death right after ischemic heart disease since 2014 across all age groups, gender, ethnicities, and stratum5.
Infection, not limited to pneumonia, can happen elsewhere caused by a diversity of microbial pathogens. Hence, identification of the aetiological agent is the key in determining patient recovery. It is clinically essential to differentiate bacterial infection (BI), and non-bacterial infection (NBI) as the treatment protocol differs significantly. Nevertheless, considering the similar clinical presentation between bacterial and viral infections, it could be challenging in distinguishing the two based on both history taking and examinations6.
Theoretically, by isolating out the causative agents, blood culture is always recognised as the gold standard to diagnose BI. The biggest drawback is the significant turnaround time of approximately 24–48 h to isolate the causative agents. This time-intensive limitation had become the disadvantages for blood culture and rendering it impracticable at ED for its triage responsibility7,8. Hence, the delay in diagnosis would render the initiation of empirical antibiotics. Saleh et al. (2019) illustrated that up to 30% of the clinicians proceed to continue the antibiotic, although the results were yet to confirm bacterial infection7. Frequent abuse and misuse of the antibiotic attribute to antimicrobial resistance becoming one of the biggest threats to global public health. Moreover, individual patient health is at stake with prolonged hospitalisation, radical treatment, and soaring healthcare expenses6.
Therefore, there is an ongoing effort to pursue a simple and accurate diagnostic tool. Biomarkers appear as a promising point-of-care test in the triage system. Culture and sensitivity would play a supportive role to further assist in proper clinical judgement. Among the biomarkers, CRP and PCT are the most extensively studied. However, most of them being studied in high-income countries with limited data generated from LMIC9. Nevertheless, the performances were somewhat inconsistent and fluctuated10,11. For the new emerging biomarkers, secretory phospholipase A2 group IIA (sPLA2-IIA) was hypothesised able to distinguish BI, but few studies were available, hence requiring further validation12,13. This prospective study aimed to investigate the performance of new emerging biomarker sPLA2-IIA with other biomarkers inclusive of high-sensitive CRP, PCT, N%, and lactate in their diagnostic value to identify BI from NBI.
Methods
Study design, population and setting
The ED UKM Medical Centre, with 72,000 ED visits annually, served as an urban, academic teaching hospital with 1000-beds. The target population includes ≥ 18 years old who presented to the ED suspicious of infection throughout the study. Patients enrolled had given their written informed consent to participate in this study. The diagnosis of infection was determined by attending ED physicians based on the criteria stated in Horan et al. (2008)14. The patients were then be grouped into either BI or NBI. The bacterial infection is defined as a clinical bacterial infection or positive bacterial cultures (sputum, body fluids, blood, et cetera). However, the exclusion criteria were immunosuppressed patients, in either case, oncologic patients15,16,17,18,19, partially treated with antibiotics before ED presentation, preexisting liver cirrhosis20,21,22,23 and end-stage renal failure who required regular dialysis24,25,26, or sterile inflammatory disease. The research was approved by the Universiti Kebangsaan Malaysia (UKM) ethical review board (ethic code FF-2015-322) and carried out according to Good Clinical Practice guideline. This single-centre prospective cohort study was conducted over 30 months (May 2015–October 2017) in ED UKM Medical Centre, Malaysia.
Data collection and quality control
The data collections were done by two well-trained clinicians who work in the Emergency Department. Once patients with suspected infections fulfil the study criterion upon registration in the ED, single-time blood (either venous or arterial) sampling was performed for each interest biomarker (high-sensitive CRP, sPLA2-IIA, N%, PCT, and Lactate) before the patients received any form of medical treatment. The patients recruited were monitored continuously during the ED stay until they were discharged or passed away for their current admission. For each patient, a total of 5 mL of whole blood was collected with 2.5 mL dispensed into an EDTA tube for a full blood count. Another 3 mL whole blood was withdrawn to the serum tube for high-sensitive CRP, sPLA2-IIA, and PCT. The samples were centrifuged immediately and stored at − 85 °C until the moment of analysis. The serum lactate level was detected from the site lab arterial blood gas results, whereas N% was traced from full blood count results. Relevant culture and serology tests were ordered as determined by the treating physician based on a case-to-case basis.
Method of determination of biomarkers
The high-sensitive latex immunoassay MULTIGENT CRP Vario with a measurement range of 0.10–160.00 mg/L was used to measure high-sensitive CRP levels. For sPLA2-IIA serum activity, the samples were tested in triplicates on sPLA2-IIA (human type IIA) Enzyme Immunometric Assay Kit (Cayman Chemical, USA) as per the manufacturer’s instructions. N% was determined by flow cytometry analysis on an SYSMEX XN-3000 Analyzer. In vitro quantitative determination of PCT value was done with Elecsys BRAHMS PCT, which was utilised as an immunoassay. Lactate levels were measured with ABL 800 BASIC analyser.
Statistical analysis
All statistical analysis was accomplished using SPSS software, version 24. The recruited patients’ demographic data and causative microorganisms were summarised as frequency (%), mean and standard deviation. Parametric variables with normal distribution were presented as mean ± standard deviation. Otherwise, the median and interquartile values were reported instead. Subsequently, non-parametric variables were tested with the Mann–Whitney U test for two-group comparisons and Kruskal–Wallis tests for multi-group comparisons. A 2-sided P value of < 0.05 was used in simple comparisons to indicate statistical significance. A 2-sided P value of < 0.01 was adjusted for multiple comparisons to indicate the statistical significance based on Bonferroni’s correction27. Pearson chi-square, χ2 test was used to compare the association of categorical variables. Area under receiver operating characteristic curve (AUROC) was used to assess each biomarker’s performance in discriminating between BI and non-BI. The cut-off point of each biomarker was then determined and used as the reference. The MedCalc online calculator was used to determine Sn, Sp, positive predictive values (PPV), negative predictive values (NPV), and accuracy with a 95% confidence interval (95%CI) of each biomarker. The accuracy of each parameter was verified by Cohen’s kappa (κ) agreement test. An online sample calculator, “easyROC”, was used to calculate the sample size using PCT as a standard biomarker based on past literature. With a type 1 error of 0.05, a power of 0.8, an AUROC PCT of 0.93 from the Luzzani et al. (2003), and an AUROC sPLA2-IIA of 0.93 from Tan et al. (2016) at the lower difference of 0.01, we calculated a total sample size of 11212,28.
Results
Characteristic of study cohort
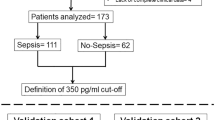
From March 2015–October 2017, a total of 154 patients admitted to the Emergency Department were who fulfilled the inclusion criteria, of which 151 consecutive patients were eligible and selected for this study after exclusion of three missing blood samples (Fig. 1). The demographic data of the patient characteristics were shown in Table 1, and causative pathogen data was presented in Table 2. The mean age was around 57.8 years old with equal gender distribution. The age for BI appeared older as compared with the NBI group (p = 0.008). Among the 151 patients, 115 were diagnosed to have BI. All biomarker results were compared at a cut-off level determined via AUROC analysis.
Study on-site recruitment and workflow (May 2015–October 2017). High-sensitive CRP = High-Sensitive C-Reactive Protein; sPLA2-IIA = Secretory Phospholipase 2- IIA; N%, = Neutrophil Percentage; PCT = Procalcitonin; HIV = Human Immunodeficiency Virus; ESRF = End-Stage Renal Failure; BI = Bacterial Infection; NBI = Non-Bacterial Infection; HL = High Level*; LL = Low Level†. *High level refers to biomarker’s level that equal to or higher than the cut-off point. † Low level refers to biomarker’s level that equal to or lower than the cut-off point.
Detection of bacterial infection
According to BI and NBI groups, the median and interquartile range for all biomarker levels were summarised in Table 1 and Fig. 2. All biomarker median levels were significantly higher in the BI group than the NBI group (P < 0.001). The AUROC values, Sn, Sp, PPV, NPV, accuracy, and κ for all biomarkers for detecting sepsis and BI were shown in Table 3 and Fig. 3. Table 1 showed that high-sensitive CRP and sPLA2-IIA were found to have the highest AUROC values, both 0.93 (95% CI, 0.88–0.97). Figure 3 presents the AUROC for all biomarkers in differentiating BI. Interestingly, sPLA2-IIA was found to have the best Sp (94%, 95% CI, 0.81–0.99) with a cut-off point of 7.04 μg/l. N% had the highest Sn (84%, 95% CI, 0.77–0.91) among all the biomarkers, with a cut-off of 75%. High-sensitive CRP, PCT, and sPLA2-IIA had higher cut-off points in BI detection than in sepsis detection. In further analyses of AUROC among biomarkers, high-sensitive CRP, sPLA2-IIA, and N% predictability of BI were found equally good (P > 0.05). These three biomarkers also had significantly higher AUROC than PCT and lactate (P < 0.05) (Table 4).
Box plots for high-sensitive CRP, sPLA2-IIA, N%, PCT, and lactate levels in detecting bacterial infection. hsCRP = High-Sensitive C-Reactive Protein; sPLA2-IIA = Secretory Phospholipase 2-IIA; N% = Neutrophil Percentage; PCT = Procalcitonin. Boxes show the 25th–75th centiles, while whiskers indicate the 10th and 90th centiles. Horizontal lines within the boxes indicate the median. Outliers are shown as circles and stars.
AUROCs of five tested biomarkers in differentiating bacterial infection in ED. This figure showed that all biomarkers able to predict sepsis and bacterial infection (BI). High-sensitive CRP, sPLA2-IIA, and N% have the highest AUROC among all for both sepsis and BI detection in ED. AUROC = Area Under Receiver Operating Characteristic Curve; hsCRP = High-Sensitive C-Reactive Protein; sPLA2-IIA = Secretory Phospholipase 2-IIA; N% = Neutrophil Percentage; PCT = Procalcitonin.
Discussion
In our study, high-sensitive CRP and sPLA2-IIA had better AUROCs followed by N%, PCT, and lactate in discriminating sepsis and differentiating BI.
Despite limited literature available, the sPLA2-IIA performance in our study parallel with the findings from Rintala et al. (1993), showing that sPLA2-IIA was significantly higher in BI patients29. The same study also highlighted a strong correlation of sPLA2-IIA level to high-sensitive CRP and PCT. In our current study, we found sPLA2-IIA has the equal performance to high-sensitive CRP and outweigh PCT. The primitive role of sPLA2-IIA coined as a bactericidal enzyme, catalysed bacterial membranes’ hydrolysis13. This acute-phase protein engages the body host in response to inflammation and generates pro-inflammatory metabolites12. Hence, its activity level is reliable to measure the degree of systemic inflammation in various bacteremic and non-bacteremic infections and differentiate between bacterial and viral infections12,30.
Several studies comparing CRP and PCT in the detection of BI showed that PCT was superior to CRP;31,32,33,34,35 however, our results showed otherwise. Since CRP and PCT are mostly evaluated among the high-income countries, there was a lack of reference tests for comparative analysis in a geographical speicfic region. Therefore, Escadafal et al. (2017) highlighted the necessity to appraise biomarkers in LMIC populations for its intended application settings36. Our CRP cut-off point falls within the cut-off range above 60–80 mg/dL that bacterial infection may be present8. We hope the biomarkers value and their cut-off point in our study could serve as a reference and guidance for future result interpretation among the LMIC countries. Being the leading most extensive study on CRP and PCT’s efficacy for predicting bacterial infection in tropical, malarial endemic settings in the Southeast Asian context, Lubell et al. (2015, 2016) concluded that CRP outperforms PCT in its accuracy37,38. Although the role of CRP was debatable as a biomarker of either inflammation or infection7,8, our result affirmed the stand of Escadafal et al. (2020) whereby CRP levels correlated with the presence of bacterial infection and was consistent across various studies39. Our study showed better performance of CRP than PCT which is comparable to other LMIC context studies in contrary to the observations in high-income countries38,40,41.
PCT played an essential role in antibiotic stewardship (AMS) and was recognised internationally for its diagnostic and prognostic properties42,43. The efficacy was validated in several trials showing a decreased antibiotic prescription rate and improvement in patients’ clinical outcomes. The Berlin 2018 expert consensus had developed a PCT-guided AMS concerning illness severity and likelihood for BI. Subsequently, algorithm adaptations were made to harmonise the PCT usage across the Asia–Pacific region, given the differences in LMIC background. Nevertheless, the modified consensus also agreed that the PCT-guided AMS was not applicable in patients suspected of tropical disease43. From the Berlin 2018 consensus, the PCT cut-off was fixed at 0.5 ng/mL at the intensive care unit (ICU) and 0.25 ng/mL non-ICU setting to predict the likelihood of BI42. However, our study reported a higher PCT cut-off at 0.38 ng/mL compared to the non-ICU group. As ED stands a unique setting that harbours a mixture of critical and non-critical ill patients, a higher PCT cut-off is more practical for BI identification. Furthermore, the uprising performance in sPLA2-IIA compared to PCT in our study prompts the consideration of sPLA2-IIA to rise as a potential AMS biomarker. This current study encourages further exploration of sPLA2-IIA on AMS as an approach for more judicious antibiotic usage in the future time.
In previous literature, N% showed weak BI prediction in elderly patients. With an 80% cut-off point, low Sn and Sp were recorded at 35% and 74%, respectively44. Comparatively, our study demonstrated a slightly lower N% cut-off (75%) with a higher Sn (84%) and Sp (69%). Although it is not the best biomarker, N% showed promising performance and outperformed PCT and lactate. Such attribution may relate to the neutrophils’ role as the predominant immune cell population migrated to the affected site regardless of the etiological agents at the early stage of infection. Mainly, the neutrophils were markedly increased during bacterial or fungal infections as compared to viral infections11. Considering N% as an inexpensive and readily available biomarker, it promised to be an excellent parsimonious biomarker in detecting BI in LMICs. Therefore, a high N% should prompt a high BI suspicion and warrant further investigation in our settings.
In contrast, our study found that lactate was poor in differentiating BI, even though it has been used widely in clinical settings as a biomarker to discriminate sepsis from non-sepsis patients. Lactate reflects strained cellular metabolism as it is an end product in anaerobic metabolism from hypoxemia. A wide variety of conditions, such as trauma, endocrine emergency, acute cardiac events, and increased bacterial load, lead to lactate level elevation, impeding its sensitivity.
This study addressed several limitations that merit consideration. However, it was a single-centre ED-based study with all data samples collected solely from an academic medical centre. Hence, selective bias may have occurred, and this study result may not apply to other hospitals’ ED settings. Apart from that, the study excluded the minorities with the inclusion age criterion restricted to those aged ≥ 18 years old. The paediatric population appeared to be an exciting group for further investigation as its application may possess considerable clinical potential and relevance. Undoubtedly, this will potentially support the use of biomarkers in all age groups to identify patients with BI. Lastly, the available clinical algorithm and lab test for microbiological diagnosis may be imperfect. The diagnostic performance may have negatively affected several cases whereby bacterial infection was misclassified as other pathogenic agents at some instances when patients harbour bacterial infections without being microbiologically proven. Regardless, antibiotics would still be administered in these cases as part of a regular clinical routine. Our study’s strength is that the prospective study was carried out as proposed despite limited resources in our setting. Our study had achieved the proposed sample size, which ensured a high-power study.
Conclusion
Taking all together, sPLA2-IIA is comparable to high-sensitive CRP but better than N%, PCT, and lactate in identifying BI at a fast-paced ED setting in LMIC, preliminarily. Combinations of biomarkers can yield better diagnostic performance on bacterial infection management in LMICs. Further studies should be carried out to explore the potency of sPLA2-IIA, particularly validating its implicit roles as a potential AMS biomarker. This would expedite the physicians’ decision-making for proper antimicrobial administration and mitigate the rising threat of antimicrobial resistance globally.
Data availability
The data that support the findings of this study are available from Toh Leong Tan. However, restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are, however, available from the authors upon reasonable request and with permission of Toh Leong Tan.
References
Wang, H. et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544. https://doi.org/10.1016/S0140-6736(16)31012-1 (2016).
Dadonaite, B. and Roser, M. Pneumonia. OurWorldInData.org. https://ourworldindata.org/pneumonia (2018)
Aston, S. J. Pneumonia in the developing world: characteristic features and approach to management. Respirology 22, 1276–1287. https://doi.org/10.1111/resp.13112 (2017).
Zar, H. J., Madhi, S. A., Aston, S. J. & Gordon, S. B. Pneumonia in low and middle income countries: progress and challenges. Thorax 68, 1052–1056. https://doi.org/10.1136/thoraxjnl-2013-204247 (2013).
Ab. Razak, M.Y. Statistic on Causes of Death, Malaysia, 2020. Department of Statistics Malaysia Official Portal. https://www.dosm.gov.my/v1/index.php?r=column/cthemeByCat&cat=401&bul_id=QTU5T0dKQ1g4MHYxd3ZpMzhEMzdRdz09&menu_id=L0pheU43NWJwRWVSZklWdzQ4TlhUUT09 (2020)
Tsao, Y.-T. et al. Differential markers of bacterial and viral infections in children for point-of-care testing. Trends Mol. Med. 26, 1118–1132. https://doi.org/10.1016/j.molmed.2020.09.004 (2020).
Saleh, M. A. A., van de Garde, E. M. W. & van Hasselt, J. G. C. Host-response biomarkers for the diagnosis of bacterial respiratory tract infections. Clin. Chem. Lab. Med. (CCLM) 57, 442–451. https://doi.org/10.1515/cclm-2018-0682 (2019).
Hausfater, P. Biomarkers and infection in the emergency unit. Med. Mal. Infect. 44, 139–145. https://doi.org/10.1016/j.medmal.2014.01.002 (2014).
Kapasi, A. J., Dittrich, S., González, I. J. & Rodwell, T. C. Host biomarkers for distinguishing bacterial from non-bacterial causes of acute febrile illness: a comprehensive review. PLoS ONE 11, e0160278 (2016).
Ljungström, L. et al. Diagnostic accuracy of procalcitonin, neutrophil-lymphocyte count ratio, C-reactive protein, and lactate in patients with suspected bacterial sepsis. PLoS ONE 12, e0181704. https://doi.org/10.1371/journal.pone.0181704 (2017).
Yusa, T., Tateda, K., Ohara, A. & Miyazaki, S. New possible biomarkers for diagnosis of infections and diagnostic distinction between bacterial and viral infections in children. J. Infect. Chemother. 23, 96–100. https://doi.org/10.1016/j.jiac.2016.11.002 (2017).
Tan, T. L. et al. CD64 and group II secretory phospholipase A2 (sPLA2-IIA) as biomarkers for distinguishing adult sepsis and bacterial infections in the emergency department. PLoS ONE 11, e0152065. https://doi.org/10.1371/journal.pone.0152065 (2016).
Tan, T. L. & Goh, Y. Y. The role of group IIA secretory phospholipase A2 (sPLA2-IIA) as a biomarker for the diagnosis of sepsis and bacterial infection in adults—a systematic review. PLoS ONE 12, e0180554. https://doi.org/10.1371/journal.pone.0180554 (2017).
Horan, T. C., Andrus, M. & Dudeck, M. A. CDC/NHSN surveillance definition of health care–associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control 36, 309–332. https://doi.org/10.1016/j.ajic.2008.03.002 (2008).
Allin, K. H. & Nordestgaard, B. G. Elevated C-reactive protein in the diagnosis, prognosis, and cause of cancer. Crit. Rev. Clin. Lab. Sci. 48, 155–170 (2011).
Lu, S. & Dong, Z. Overexpression of secretory phospholipase A2-IIa supports cancer stem cell phenotype via HER/ERBB-elicited signaling in lung and prostate cancer cells. Int. J. Oncol. 50, 2113–2122 (2017).
Schüttrumpf, S. et al. Utility of procalcitonin concentration in the evaluation of patients with malignant diseases and elevated C-reactive protein plasma concentrations. Clin. Infect. Dis. 43, 468–473 (2006).
Vassallo, M. et al. Procalcitonin and C-reactive protein/procalcitonin ratio as markers of infection in patients with solid tumors. Front. Med. 8, 255 (2021).
Ding, S. et al. Diagnostic accuracy of procalcitonin, neutrophil-to-lymphocyte ratio, and C-reactive protein in detection of bacterial infections and prediction of outcome in nonneutropenic febrile patients with lung malignancy. J. Oncol. 2020, 1–13 (2020).
Dong, R. et al. Procalcitonin and liver disease: a literature review. J. Clin. Transl. Hepatol. 7, 51 (2019).
Talvinen, A., Kemppainen, E. & Nevalainen, T. J. K. Expression of group II phospholipase A2 in the liver in acute pancreatitis. Scand. J. Gastroenterol. 36, 1217–1221 (2001).
Vishwanath, B. S., Frey, F. J., Escher, G., Reichen, J. & Frey, B. M. Liver cirrhosis induces renal and liver phospholipase A2 activity in rats. J. Clin. Investig. 98, 365–371 (1996).
Pieri, G., Agarwal, B. & Burroughs, A. K. C-reactive protein and bacterial infection in cirrhosis. Ann. Gastroenterol. Q. Publ. Hell. Soc. Gastroenterol. 27, 113 (2014).
Owen, W. F. C-reactive protein as an outcome predictor for maintenance hemodialysis patients. Kidney Int. 54, 627–636 (1998).
Panichi, V. et al. C reactive protein in patients with chronic renal diseases. Ren. Fail. 23, 551–562 (2001).
Dahaba, A. A., Rehak, P. H. & List, W. F. Procalcitonin and C-reactive protein plasma concentrations in nonseptic uremic patients undergoing hemodialysis. Intensive Care Med. 29, 579–583 (2003).
Bland, J. M. & Altman, D. G. Multiple significance tests: the Bonferroni method. BMJ 310, 170. https://doi.org/10.1136/bmj.310.6973.170 (1995).
Luzzani, A. et al. Comparison of procalcitonin and C-reactive protein as markers of sepsis. Crit. Care Med. 31, 1737–1741. https://doi.org/10.1097/01.CCM.0000063440.19188.ED (2003).
Rintala, E. M. & Nevalainen, T. J. Group II phospholipase A2 in sera of febrile patients with microbiologically or clinically documented infections. Clin. Infect. Dis. 17, 864–870. https://doi.org/10.1093/clinids/17.5.864 (1993).
Nik Mansor, N. N. et al. An amperometric biosensor for the determination of bacterial sepsis biomarker, secretory phospholipase Group 2-IIA using a tri-enzyme system. Sensors 18, 686 (2018).
Simon, L., Gauvin, F., Amre, D. K., Saint-Louis, P. & Lacroix, J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin. Infect. Dis. 39, 206–217. https://doi.org/10.1086/421997 (2004).
Bador, K., Intan, S., Hussin, S. & Gafor, A. H. A. Serum procalcitonin has negative predictive value for bacterial infection in active systemic lupus erythematosus. Lupus 21, 1172–1177. https://doi.org/10.1177/0961203312450085 (2012).
Massaro, K. S., Costa, S. F., Leone, C. & Chamone, D. A. Procalcitonin (PCT) and C-reactive protein (CRP) as severe systemic infection markers in febrile neutropenic adults. BMC Infect. Dis. 7, 137. https://doi.org/10.1186/1471-2334-7-137 (2007).
Gao, L. et al. Early diagnosis of bacterial infection in patients with septicopyemia by laboratory analysis of PCT, CRP and IL-6. Exp. Ther. Med. 13, 3479–3483. https://doi.org/10.3892/etm.2017.4417 (2017).
Cui, N., Zhang, H., Chen, Z. & Yu, Z. Prognostic significance of PCT and CRP evaluation for adult ICU patients with sepsis and septic shock: retrospective analysis of 59 cases. J. Int. Med. Res. 47, 1573–1579. https://doi.org/10.1177/0300060518822404 (2019).
Escadafal, C. et al. New biomarkers and diagnostic tools for the management of fever in low- and middle-income countries: an overview of the challenges. Diagnostics 7, 44 (2017).
Lubell, Y. et al. Modelling the impact and cost-effectiveness of biomarker tests as compared with pathogen-specific diagnostics in the management of undifferentiated fever in remote tropical settings. PLoS ONE 11, e0152420. https://doi.org/10.1371/journal.pone.0152420 (2016).
Lubell, Y. et al. Performance of C-reactive protein and procalcitonin to distinguish viral from bacterial and malarial causes of fever in Southeast Asia. BMC Infect. Dis. 15, 511. https://doi.org/10.1186/s12879-015-1272-6 (2015).
Escadafal, C., Incardona, S., Fernandez-Carballo, B. L. & Dittrich, S. The good and the bad: using C reactive protein to distinguish bacterial from non-bacterial infection among febrile patients in low-resource settings. BMJ Glob. Health 5, e002396. https://doi.org/10.1136/bmjgh-2020-002396 (2020).
Zakariah, N. A. et al. Is Procalcitonin more superior to hs-CRP in the diagnosis of infection in diabetic foot ulcer?. Malays. J. Pathol. 42, 77–84 (2020).
Prodjosoewojo, S. et al. A novel diagnostic algorithm equipped on an automated hematology analyser to differentiate between common causes of febrile illness in Southeast Asia. PLoS Negl. Trop. Dis. 13, e0007183 (2019).
Schuetz, P. et al. Procalcitonin (PCT)-guided antibiotic stewardship: an international experts consensus on optimised clinical use. Clin. Chem. Lab. Med. (CCLM) 57, 1308–1318. https://doi.org/10.1515/cclm-2018-1181 (2019).
Lee, C.-C. et al. Procalcitonin (PCT)-guided antibiotic stewardship in Asia-Pacific countries: adaptation based on an expert consensus meeting. Clin. Chem. Lab. Med. (CCLM) 58, 1983–1991. https://doi.org/10.1515/cclm-2019-1122 (2020).
Wasserman, M., Levinstein, M., Keller, E., Lee, S. & Yoshikawa, T. T. Utility of fever, white blood cells, and differential count in predicting bacterial infections in the elderly. J. Am. Geriatr. Soc. 37, 537–543. https://doi.org/10.1111/j.1532-5415.1989.tb05686.x (1989).
Acknowledgements
We would also like to thank Universiti Kebangsaan Malaysia and the Ministry of Education, Malaysia, for supporting this research. We thank all the Doctors & EMS personnel, the ED, and all the medical laboratory technicians from the Department of Pathology Universiti Kebangsaan Malaysia Medical Centre.
Funding
This research was funded by Ministry of Education, Malaysia with Grant No. Fundament Research Grant Scheme (FRGS/1/2014/SKK01/UKM/03/3), Prototype Research Grant Scheme (PRGS/1/2017/STG05/UKM/03/1) and Universiti Kebangsaan Malaysia, Malaysia under Geran Galakan Penyelidik Muda with Grant No. GGPM-2013-102 & GGPM-2103-103.
Author information
Authors and Affiliations
Contributions
T.L.T. and C.W.K. initiated the conceptualisation of the study. T.L.T., C.W.K. and N.S.A. wrote the methodology. T.L.T., C.W.K., K.S.O., S.T.T., and N.S.A. handled the software. T.L.T., C.W.K., K.S.O. S.T.T., and D.N.N. validated the data. T.L.T., C.W.K., K.S.O. and S.T.T. did the formal analysis. T.L.T., C.W.K, K.S.O., S.T.T., N.S.A. D.N.N. A.I., and K.T.A.I. performed the investigation. Resources were secure by T.L.T., C.W.K., K.S.O., S.T.T. and N.S.A. Data curation were done by T.L.T., C.W.K., K.S.O., S.T.T., N.S.A., D.N.N. and A.I. All the following: T.L.T. C.W.K., K.S.O., S.T.T. and H.M.N. did the writing and prepared the draft. Write, review & editing were done by T.L.T., C.W.K., K.S.O., S.T.T., N.S.A., D.N.N., A.I., K.T.A., L.Y.H., N.I.H., K.B.G. and H.M.N. Visualisations were prepared by T.L.T., K.S.O. and S.T.T. Project supervision, T.L.T.; Project administration run by T.L.T. and N.S.A. Funding Acquisition by T.L.T., and D.N.N. All tables and figures were prepared by T.L.T. and K.S.O. All authors have reviewed and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tan, T.L., Kang, C.Wl., Ooi, K.S. et al. Comparison of sPLA2IIA performance with high-sensitive CRP neutrophil percentage PCT and lactate to identify bacterial infection. Sci Rep 11, 11369 (2021). https://doi.org/10.1038/s41598-021-90894-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-90894-0
This article is cited by
-
Host-response transcriptional biomarkers accurately discriminate bacterial and viral infections of global relevance
Scientific Reports (2023)
-
Prediction of the Postoperative Fat Volume Retention Rate After Augmentation Mammoplasty with Autologous Fat Grafting: From the Perspective of Preoperative Inflammatory Level
Aesthetic Plastic Surgery (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.