Abstract
Intestinal Behçet’s disease (BD) and Crohn’s disease (CD) present similar manifestations, but there are no specific diagnostic tests to differentiate them. We used a proteomic approach to discover novel diagnostic biomarkers specific to intestinal BD. Colon mucosa tissue samples were obtained from patients with intestinal BD or CD using colonoscopy-guided biopsy of the affected bowel. Peptides from seven intestinal BD and seven CD patients were extracted and labeled using tandem mass tag (TMT) reagents. The labeled peptides were identified and quantified using liquid chromatography-tandem mass spectrometry (LC–MS/MS). The proteins were further validated using immunohistochemical (IHC) analysis with tissue samples and an ELISA test with serum samples from 20 intestinal BD and 20 CD patients. Using TMT/LC–MS/MS-based proteomic quantification, we identified 39 proteins differentially expressed between intestinal BD and CD. Beta-2 glycoprotein 1 (APOH) and maltase-glucoamylase (MGAM) showed higher intensity in the IHC staining of intestinal BD tissues than in CD tissues. The serum MGAM level was higher in intestinal BD patients. Proteomic analysis revealed that some proteins were differentially expressed in patients with intestinal BD compared with those with CD. Differential MGAM expression in intestinal BD suggests its role as a potential novel diagnostic biomarker.
Similar content being viewed by others
Introduction
Behçet’s disease (BD) is an idiopathic, chronic, relapsing, multi-systemic vasculitis characterized by recurrent oral or genital ulcers, arthritis, and ocular, dermal, neurovascular, and gastrointestinal manifestations1,2. Intestinal BD is diagnosed when a patient with BD has clinical gastrointestinal symptoms and typical endoscopic findings3. It is a very rare disease, but the prevalence of BD is the highest in countries located along the Silk Road stretching from Asia to the Mediterranean countries4. Crohn’s disease (CD) is a chronic, relapsing, inflammatory, bowel disease (IBD) that may affect the gastrointestinal tract from the mouth to the anus. Once considered to be a Western disease, the incidence of CD has rapidly increased in East Asian countries, such as Japan, Korea, and Hong Kong, while plateauing in the West5,6.
Both intestinal BD and CD often present similar gastrointestinal and extraintestinal manifestations, as well as endoscopic findings. Therefore, it is sometimes difficult to differentiate between these two diseases. Clinically, oral and genital ulcerations are more common in intestinal BD patients, and perianal lesions are more common in CD patients7. However, both diseases develop nonspecific, waxing and waning, life-long gastrointestinal symptoms8. Moreover, ocular and skin manifestations, and arthropathy, can occur in both diseases9. Endoscopically, a typical intestinal BD ulcer is characterized by being single or few in number, round or oval in shape, deep, possessing a sharp margin, being larger than 10 mm, and showing localization to the ileocecal valve10. The “atypical” intestinal BD ulcer without the systemic manifestations could be confused with CD. There have been several attempts to develop a diagnostic biomarker to differentiate between the two diseases, but there are practically no available tests because of several limitations.
Proteomic analysis is a promising technology that is a powerful tool for identifying biomarkers to help diagnose and choose personalized treatment. It allows high-throughput study of protein expression with high accuracy, sensitivity, and repeatability, and it enables the identification of molecular mechanisms that are responsible for the development of a specific disease11,12. The tandem mass tag (TMT)-liquid chromatography-tandem mass spectrometric (LC–MS/MS) method has been recently developed to identify and quantify proteins. Although several studies have investigated colon mucosal biopsies using gel-based proteomic approaches to identify protein biomarkers that differentiate intestinal BD from IBD13,14, no applicable biomarkers are currently available. Since gel-free proteomic approaches have several advantages compared to gel-based approaches, particularly for identifying membrane-bound and/or glycosylated large proteins (e.g., mucins)15, we used gel-free approaches to analyze intestinal BD proteomes in comparison with CD proteomes.
Here, we aimed to develop diagnostic biomarkers to differentiate between intestinal BD and CD. We employed a coupled TMT/LC–MS/MS-based method to identify proteins that were differentially expressed between patients with intestinal BD and CD.
Methods
Study population and sample collection
In total, 47 patients with intestinal BD and 47 patients with CD were recruited from the IBD Clinic of Severance Hospital, Seoul, Korea. Intestinal BD and CD were diagnosed according to clinical, histological, endoscopic, and radiological criteria16,17. Exclusion criteria included indeterminate colitis, ulcerative colitis, intestinal tuberculosis, a history of malignancy, or insufficient available medical records. The intestinal BD ulcer type was defined according to the Korean Inflammatory Bowel Disease study group, and the CD phenotype was defined according to the Montreal Classification18,19. The severity of intestinal BD was determined using the Disease Activity Index for Intestinal Behçet’s Disease (DAIBD) score, and CD severity was determined using the Crohn’s Disease Activity Index (CDAI) score20,21.
The intestinal mucosa tissue samples were obtained from patients with intestinal BD or CD using colonoscopy-guided biopsy of the affected bowel. The tissue and serum samples were preserved at −80 °C.
Informed consent was obtained from all individuals enrolled in this study. This study was approved by the Institutional Review Board of Yonsei University College of Medicine (IRB No: 2012-0039-030) and was conducted in accordance with the Declaration of Helsinki.
TMT sample processing and protein quantitation
For proteomic analysis, the colon mucosa tissue samples were lysed and labeled with tandem mass tag (TMT, Thermo Scientific, San Jose, CA, USA) according to the manufacturer’s instructions. The labeled peptide samples were pooled into a new vial and dried using SpeedVac (Thermo Scientific). The following processes, including strong cation exchange fraction and liquid chromatography (LC)-mass spectrometry (MS) and database searching, were performed by Poochon Scientific (Frederick, MD) as described previously22. Briefly, TMT-multiplex labeled peptide mixture (100 µg protein/each plex) fractionation was performed using an Agilent AdvanceBio Column and Agilent UHPLC 1290 system (Agilent, Santa Clara, CA). LC/MS/MS analysis was performed using a Thermo Scientific Q-Exactive hybrid Quadrupole-Orbitrap Mass Spectrometer and Thermo Dionex UltiMate 3000 RSLCnano System (Thermo Scientific). Raw MS data files were searched against the human protein sequence databases obtained from the NCBI website using Proteome Discoverer 1.4 software (Thermo Scientific) based on the SEQUEST and percolator algorithms. The false positive discovery rate (FDR) was set at 5%. The resulting Proteome Discoverer Report from Poochon Scientific contained all assembled proteins with peptide sequences and peptide spectrum match counts (PSM#) and TMT-tag-based quantification ratios.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded colonic biopsy sections from patients with intestinal BD (n = 20) and those with CD (n = 20) were stained with 39 antibodies and counterstained with hematoxylin–eosin. Antibodies against APOH (HPA003732) and MGAM (HPA002270) were purchased from Atlas Antibodies AB (Bromma, Sweden). Detailed antibody information, antibody dilution factors, and antigen retrieval methods are provided in Supplementary Table I. IHC analysis was performed as previously described23. Primary antibodies (1:1000 dilution for anti-APOH and 1:2000 dilution for anti-MGAM) were applied overnight at 4 °C. Staining was visualized using an Olympus BX43 microscope with the Olympus CellSens Entry software (Hamburg, Germany).
We used a semi-quantitative grading method to assess protein expression, as described previously24. Briefly, 100 × magnification was used to grade all fields for each sample, and the staining intensity was scored from 1 to 3. Staining extent was scored from 1 to 4. The scores were multiplied together, and the final scores were classified as follows: 1–3, weak; 4–8, moderate; and 9–12, strong staining. Fisher’s exact test was used to assess the immunochemical scores for protein expression. In this particular test, values of p less than 0.2 were determined as a threshold for moving to the next validation test.
Enzyme-linked immunosorbent assay (ELISA)
Serum samples from patients with BD (n = 20) and those with CD (n = 20)—constituting an independent cohort of patients distinct from those providing samples for IHC staining—were analyzed using ELISA. The serum was diluted 1:50,000 for APOH and 1:200 for MGAM. ELISA was performed according to the manufacturer’s instructions using the APOH ELISA kit (KA0982, Abnova, Taipei, Taiwan) and the MGA ELISA kit (MBS2021345, MyBioSource, San Diego, CA).
Data analysis and statistical methods
A heat map was generated using the “limma” package for R25. The fold changes of proteins and p values were calculated using linear regression in “limma.” The heat map was drawn using the heatmap.2 function in the “gplots” package for R (https://CRAN.R-project.org/package=gplots), and colors were changed using the “RColorBrewer” package for R (1.1-2. https://CRAN.R-project.org/package=RColorBrewer). Volcano plots were plotted using GraphPad Prism 8 software. Data were expressed as the mean ± SEM. Student’s t-test was used for the statistical analysis of serum ELISA data. Statistical significance was set at p < 0.05.
Result
Descriptive patient characteristics
The colon mucosa tissue samples of 27 patients with intestinal BD and 27 patients with CD, and serum samples of 20 patients with intestinal BD and 20 patients with CD, were obtained. Detailed patient characteristics of the 94 patients are shown in Table 1. The mean age of the patients with intestinal BD and CD was 51. 0 ± 11.4 years and 36. 1 ± 16.0 years (p < 0.001), respectively, which were comparable to those obtained in a previous study7. A total of 42.6% of the patients with intestinal BD and 74.5% of the patients with CD were men. The mean disease duration of intestinal BD and CD was 7.1 ± 6.0 years and 8.0 ± 5.5 years, respectively. Oral ulcers, genital ulcers, skin manifestations, and joint manifestations were more common in patients with intestinal BD and perianal lesions were more common in those with CD. Intestinal BD ulcers were located in the ileocecal valve (74.5%), ascending colon (10.6%), and postoperative anastomosis site (14.6%). The intestinal BD ulcers occurred with the following frequency: solitary (48.9%), two (19.1%), and multiple (31.9%). The CD showed localization in the ileal (14.9%), colonic (2.1%), and ileocolic (83.0%) regions and could be characterized as non-stricturing and non-penetrating (63.0%), stricturing (15.2%), and penetrating (21.7%). The number of patients with a DAIBD score ≥ 40 was 35 (74.5%) and that of patients with a CDAI score ≥ 150 was 24 (51.1%).
Discovery of biomarker candidates using TMT/LC–MS/MS-based proteomic approach
The colon mucosa tissue samples of patients with intestinal BD (n = 7) and those with CD (n = 7) were analyzed using a TMT/LC–MS/MS-based proteomic approach to identify intestinal BD-specific markers (Fig. 1). A total of 3,266 proteins were quantitatively identified, and at least two identified peptides were detected for each protein (Fig. 2A). Logistic regression analysis showed that 39 proteins were significantly different between intestinal BD and CD patients (p < 0.05) (Fig. 2B). Among them, 34 proteins were overexpressed, and 5 proteins were under-expressed in patients with intestinal BD compared to patients with CD (Table 2). The proteins were classified by cellular compartment (CC) based on gene ontology (GO) enrichment analysis using a functional annotation tool (DAVID Bioinformatics Resources, version 6.8) (Supplementary Table II). Among the top ten CC categories, 72.31% of GO items were associated with membrane-bounded vesicle (GO:0031988), extracellular region (GO:0005576), extracellular region (GO:0044421), extracellular exosome (GO:0070062), extracellular vesicle (GO:1903561), and extracellular organelle (GO:0043230), indicating that although there were a certain number of proteins from other cellular compartments such as the cytosol, nucleoplasm, mitochondrion, or cell junction, a large portion of colon mucosal proteomes were composed of secretion-related proteins (Fig. 2C). Notably, APOH and MGAM were included in all six secretion-related CC categories in the GO analysis.
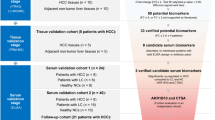
The colon mucosa tissue samples were obtained and analyzed using a TMT/LC–MS/MS-based approach for the discovery cohort (intestinal BD, n = 7; CD, n = 7). Candidate protein biomarker validation was performed using tissue IHC staining with the validation cohort 1 (intestinal BD, n = 20; CD, n = 20) and serum ELISA testing with the validation cohort 2 (intestinal BD, n = 20; CD, n = 20).
Proteomic characterization of 14 samples (seven from patients with intestinal BD and seven from patients with CD) using TMT-10plex labeling-based quantitative proteomics. (A) A heat map showing the relative abundance of 3266 proteins identified across two groups of human intestinal samples. The color key indicates the relative abundance of each protein (− 2 to 2) across 14 samples. (B) Volcano plot demonstrating fold changes in protein abundance between intestinal BD and CD. The x-axis represents the log2 ratio, and the y-axis represents significant differences (-log10 of p value). Proteins showing significantly altered expression (p < 0.05) are colored in magenta, and include the top ten up- or down-regulated proteins (fold change > 1.5). (C) A pie diagram showing cellular localization of human colon mucosa proteomes classified by the top ten cellular compartments in functional annotation with gene ontology (GO) with secretion-related proteins highlighted in green.
IHC validation of biomarker candidates
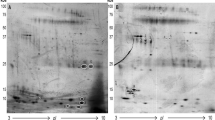
IHC analysis was performed to validate 39 candidate biomarkers. A total of 40 colonic biopsy sections from patients with intestinal BD (n = 20) and CD (n = 20) were stained with each antibody and analyzed using a semi-quantitative grading method as described in the Methods section. Seven candidate proteins were selected based on their differential expression between intestinal BD and CD: maltase-glucoamylase (MGAM), beta-2 glycoprotein 1 (APOH), plasminogen (PLG), pro-interleukin 16 (IL16), serine/arginine-rich splicing factor 3 (SRSF3), clusterin (CLU), and serine/threonine-protein phosphatase 4 catalytic subunit (PPP4C). Fisher’s exact test showed that APOH (p = 0.039) and MGAM (p = 0.192) levels were consistently higher in intestinal BD than in CD, suggesting that these two proteins might be distinctive biomarkers of intestinal BD distinguishing it from CD. There were no significant differences in the expression of PLG (p = 0.480), IL16 (p = 0.563), SRSF3 (p = 0.591), CLU (p = 1.000), or PPP4C (p = 0.450) between intestinal BD and CD. IHC analysis of the colon mucosa tissue obtained from patients with intestinal BD showed that stronger APOH immunoreactivity was detected in a portion of the lamina propria (Fig. 3A, lower panels), and the MGAM immunoreactive signal was exclusively present in the brush border membrane of the epithelium (Fig. 3B, lower panels), consistent with previous findings26.
Serum ELISA validation of biomarker candidates
Since APOH and MGAM were included in all six secretion-related CC categories of GO analysis (Fig. 2C), we hypothesized that these proteins could also be detected in the blood. This notion was supported by several biochemical studies that identified APOH as a component of circulating plasma lipoproteins27, and the fact that MGAM is found in brush border membrane vesicles28. Both proteins were also recently detected in exosomes using in-depth proteomic analyses29. Although there was no difference in APOH concentration between the two groups (Fig. 4A), serum concentrations of MGAM were statistically higher in patients with intestinal BD compared to those with CD (p < 0.05), when the two candidate biomarkers were further tested with serum ELISA in an independent validation cohort (intestinal BD, n = 20; CD, n = 20) (Fig. 4B). These results suggest that MGAM can be a specific, diagnostic biomarker of intestinal BD.
Analysis of the MGAM-related pathway in the colon mucosa proteome
The DAVID functional annotation tool was used to identify the biological pathways of the colon mucosa proteome of intestinal BD and CD patients, in which MGAM could be involved. The Kyoto Encyclopedia of Genes and Genomes (KEGG) demonstrated that 85 pathways were significantly enriched (p < 0.05) (Fig. 5A). Among them, MGAM was involved in three enriched pathways: metabolic pathways (hsa01100, 467 proteins), galactose metabolism (hsa00052, 19 proteins), and starch and sucrose metabolism (hsa00500, 17 proteins). Unexpectedly, APOH was not related to any of the 85 enriched pathways.
MGAM and APOH characterization in human colon mucosal proteomes. (A) Significant biological pathways enriched among 3,266 proteins identified by TMT are represented as bar graphs (p < 0.05) with the MGAM-containing categories highlighted in red. Venn diagrams showing numbers of MGAM-and APOH-containing categories classified by cellular compartment (B), molecular function (C), and biological process (D) based on functional annotation with gene ontology (GO) analysis.
To determine whether MGAM and APOH share biological functions, 3,266 proteins were divided into three GO annotation categories: cellular compartment (CC), molecular function (MF), and biological process (BP). Next, the categories containing MGAM or APOH were selected and compared. There were six CC categories, but no MF or BP categories, correlating with both MGAM and APOH as represented in Figs. 2C and 5B–D. This suggests that the physiological roles of MGAM and APOH might differ in the colon mucosa, but both are assumed to be secreted.
Predicted upstream regulators and disease-related functions of MGAM in intestinal BD and CD
Ingenuity pathway analysis (IPA) was performed to predict canonical pathways, upstream regulators, and disease-related functions to gain more insight into the pathophysiological role of MGAM in intestinal BD and CD. As shown in Supplementary Fig. 1A, the only canonical pathway related to MGAM was enriched in “Glycogen degradation III.” Although absolute z scores (CD, − 0.302; BD, 0.302) were lower than the cut-off (z-score > 2), the pathway significantly differed between intestinal BD and CD (p < 0.000001). The pathway was predicted to be inhibited in CD and activated in intestinal BD (Supplementary Fig. 1B). Upstream regulator analysis revealed that the dexamethasone-related pathway was inactivated in CD compared to its status in intestinal BD (Fig. 6A), but the medication history of corticosteroids (31.9%) was similar between the two patient groups participating in this study (Table 1). This result is consistent with previous observations that patients with CD require corticosteroid therapy more often than patients with intestinal BD7. Comparison of the diseases and functional analyses of the two patient groups revealed that the activation of degranulation and the inactivation of genitourinary carcinogenesis were conserved, and the former is predicted to be more upregulated in intestinal BD than in CD, and the latter is more downregulated in CD than in intestinal BD (Fig. 6B).
Ingenuity pathway analysis (IPA) of the MGAM-related upstream regulator (A) and disease or functional annotations (B) in colon mucosa proteomes of internal BD and CD. IPA core analyses were performed for a proteomic comparison between internal BD and CD using 3,614 UniProt accession numbers. Significant MGAM-related functional annotations are shown (z-score > 2, p < 0.05). Heat maps illustrating the predicted activation z-score. The color range indicates its predicted activation state: increasing (orange) or decreasing (blue).
Discussion
Accurate diagnosis of intestinal BD and CD is important for establishing proper treatment plans and predicting disease prognosis30. Clinical, laboratory, and endoscopic approaches have limitations in differentiating between these two diseases. Gastrointestinal and systemic symptoms, elevated inflammatory markers, and endoscopic findings, such as asymmetric deep ulcers in the ileocecal valve, are often shared between the two diseases in clinical practice31.
We quantified 3,266 proteins from the colon mucosa tissue samples, identified 39 novel diagnostic biomarkers, and validated the MGAM protein as a novel diagnostic biomarker using patient serum samples. To the best of our knowledge, this study is the first to use proteomics to identify a diagnostic marker that can differentiate intestinal BD from CD.
Quantitative proteomic analysis using isobaric chemical labeling, including super-stable isotope labeling with amino acids in cell culture, isobaric tags for relative and absolute quantitation, or TMT, is emerging as a highly effective approach with good quantification performance and reproducibility for profiling new biomarkers in numerous diseases32. These relatively new proteomic techniques enable the discovery of diagnostic biomarkers by providing methods not only for peptide identification, but also for the quantification of biological samples. Thus far, they have been applied to IBD33 or intestinal TB24, but not to intestinal BD. We identified 39 potential novel diagnostic biomarkers using quantitative proteomic analysis, and validated MGAM as a biomarker for differentiating intestinal BD from CD.
MGAM is involved in carbohydrate digestion in the small intestine. MGAM deficiency has been reported in congenital diarrheal diseases34. MGAM and sucrase-isomaltase (SI) have identical exon structures. They are anchored in the small intestinal mucosal brush border and hydrolyze substrates to glucose35. Romach et al. reported a trinitrobenzene sulfonic acid-induced colitis rat model that showed a loss of SI expression and activity36. Lackeyram et al. observed that a dextran sodium sulfate-induced colitis piglet model revealed decreased maximal specific activities of MGAM and SI37. Here, the tissue expression and serum concentrations of MGAM were lower in patients with CD patients than in those with intestinal BD. Our IPA analysis also predicted that the MGAM-related “glycogen degradation III” pathway was inactivated in CD (Supplementary Fig. 1A,B). Consistent with our findings, the involvement of the small intestine is relatively common in CD than in intestinal BD, resulting in digestive problems and nutrient malabsorption. Thus, differential MGAM expression may be related to different clinical manifestations between intestinal BD and CD, which should be further validated.
MGAM has been shown to be important in neutrophil biology but not in lymphocytes38,39. We identified MGAM in the colon mucosa tissue samples, but MGAM was also detected in the serum samples. The detected serum MGAM may be secreted from the gastrointestinal tract or contained in neutrophils. All five degranulation-related pathways (degranulation, degranulation of cells, degranulation of phagocytes, degranulation of neutrophils, and degranulation of granulocytes) enriched in the disease or functional annotations with high activation z-score indicated that MGAM might be involved in the degranulation process in which mast cell activation is essential. Although little is known regarding the degranulation in intestinal BD, degranulation signaling in IBD has been implicated in the regulation of inflammatory responses in the gastrointestinal tract, where the largest population of mast cells in the body resides40,41,42,43. Higher MGAM levels were detected in intestinal BD patients than in CD patients, and this was related to innate immunity pathogenesis. Further studies are required to understand how MGAM influences immune cell degranulation and the resulting gut inflammation. Moreover, it would be valuable to determine whether MGAM levels correlate with the disease activity of intestinal BD.
Here, MGAM was shown to differentiate BD from CD with an area under the curve of 0.805 (95% confidence interval, 0.665–0.945), 85% sensitivity, and 70% specificity at a cut-off of 150 ng/mL (Fig. 7). However, further studies with a larger sample size are warranted to validate our data on the role of MGAM in chronic enterocolitis and to understand its molecular mechanism in depth. In addition, a significant proportion of our CD cohort population had perianal involvement, which distinguishes CD from intestinal BD. Perianal lesions in CD occur more frequently in Korea than in Western countries, but the ideal cohort population for the development of diagnostic biomarkers should present common or resembling clinical, endoscopic, radiologic, and histologic features of the two diseases. Future studies using such a cohort will be needed to validate MGAM as a distinguishing diagnostic biomarker. Finally, the measurement of MGAM levels in a variety of inflammatory diseases such as ulcerative colitis and intestinal tuberculosis could be worthwhile.
In summary, we used TMT-based proteomic quantification to identify 39 candidate proteins that were differentially expressed between intestinal BD and CD. Then, we selected APOH and MGAM proteins as possible biomarkers for intestinal BD based on the results of IHC staining and semi-quantitative grading. Finally, we suggest that the levels of MGAM protein in patient serum can potentially be used to differentially diagnose intestinal BD from CD.
Abbreviations
- BD:
-
Behçet’s disease
- CD:
-
Crohn’s disease
- IBD:
-
Inflammatory bowel disease
- TMT:
-
Tandem mass tag
- LC–MS:
-
Liquid chromatography-tandem mass spectrometry
- DAIBD:
-
Disease Activity Index for Intestinal Behçet’s Disease
- CDAI:
-
Crohn’s Disease Activity Index
- IHC:
-
Immunohistochemistry
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- MGAM:
-
Maltase-glucoamylase
- APOH:
-
Beta-2 glycoprotein 1
- PLG:
-
Plasminogen
- IL16:
-
Interleukin 16
- SRSF3:
-
Serine/arginine-rich splicing factor 3
- CLU:
-
Clusterin
- PPP4C:
-
Serine/threonine-protein phosphatase 4 catalytic subunit
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- IPA:
-
Ingenuity Pathway Analysis
- HLA:
-
Human leukocyte antigen
- MICA:
-
MHC class I related gene A
- CRP:
-
C-reactive protein
- ESR:
-
Erythrocyte sedimentation rate
- ASCA:
-
Anti-Saccharomyces cerevisiae antibodies
- AAEA:
-
Anti-alpha-enolase antibodies
- UC:
-
Ulcerative colitis
- TB:
-
Tuberculosis
- ACE:
-
Angiotensin-converting enzyme
- MT-2:
-
Metallothionein-2
References
Sakane, T., Takeno, M., Suzuki, N. & Inaba, G. Behcet’s disease. N. Engl. J. Med. 341, 1284–1291. https://doi.org/10.1056/NEJM199910213411707 (1999).
James, D. G. Behcet’s syndrome. N. Engl. J. Med. 301, 431–432. https://doi.org/10.1056/NEJM197908233010811 (1979).
Lee, H. J. & Cheon, J. H. Optimal diagnosis and disease activity monitoring of intestinal Behcet’s disease. Intest. Res. 15, 311–317. https://doi.org/10.5217/ir.2017.15.3.311 (2017).
Bayraktar, Y., Ozaslan, E. & Van Thiel, D. H. Gastrointestinal manifestations of Behcet’s disease. J. Clin. Gastroenterol. 30, 144–154. https://doi.org/10.1097/00004836-200003000-00006 (2000).
Mak, W. Y., Zhao, M., Ng, S. C. & Burisch, J. The epidemiology of inflammatory bowel disease: East meets west. J. Gastroenterol. Hepatol. https://doi.org/10.1111/jgh.14872 (2019).
Park, S. H. et al. A 30-year trend analysis in the epidemiology of inflammatory bowel disease in the Songpa-Kangdong District of Seoul, Korea in 1986–2015. J. Crohns Colitis 13, 1410–1417. https://doi.org/10.1093/ecco-jcc/jjz081 (2019).
Jung, Y. S. et al. Long-term clinical outcomes of Crohn’s disease and intestinal Behcet’s disease. Inflamm. Bowel. Dis. 19, 99–105. https://doi.org/10.1002/ibd.22991 (2013).
Yazici, Y., Yurdakul, S. & Yazici, H. Behcet’s syndrome. Curr. Rheumatol. Rep. 12, 429–435. https://doi.org/10.1007/s11926-010-0132-z (2010).
Valenti, S., Gallizzi, R., De Vivo, D. & Romano, C. Intestinal Behcet and Crohn’s disease: Two sides of the same coin. Pediatr. Rheumatol. Online J. 15, 33. https://doi.org/10.1186/s12969-017-0162-4 (2017).
Lee, C. R. et al. Colonoscopic findings in intestinal Behcet’s disease. Inflamm. Bowel. Dis. 7, 243–249. https://doi.org/10.1097/00054725-200108000-00010 (2001).
Gisbert, J. P. & Chaparro, M. Clinical usefulness of proteomics in inflammatory bowel disease: A comprehensive review. J. Crohns Colitis 13, 374–384. https://doi.org/10.1093/ecco-jcc/jjy158 (2019).
Titz, B. et al. Proteomics and lipidomics in inflammatory bowel disease research: From mechanistic insights to biomarker identification. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19092775 (2018).
Lee, H. J. et al. Proteomic analysis of serum amyloid a as a potential marker in intestinal Behcet’s disease. Dig. Dis. Sci. 62, 1953–1962. https://doi.org/10.1007/s10620-017-4606-y (2017).
Lee, H. J. et al. Proteomics-based functional studies reveal that galectin-3 plays a protective role in the pathogenesis of intestinal Behcet’s disease. Sci. Rep. 9, 11716. https://doi.org/10.1038/s41598-019-48291-1 (2019).
Rabilloud, T. Membrane proteins and proteomics: Love is possible, but so difficult. Electrophoresis 30(Suppl 1), S174-180. https://doi.org/10.1002/elps.200900050 (2009).
Skef, W., Hamilton, M. J. & Arayssi, T. Gastrointestinal Behcet’s disease: A review. World J. Gastroenterol. 21, 3801–3812. https://doi.org/10.3748/wjg.v21.i13.3801 (2015).
Maaser, C. et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J. Crohns Colitis 13, 144–164. https://doi.org/10.1093/ecco-jcc/jjy113 (2019).
Satsangi, J., Silverberg, M. S., Vermeire, S. & Colombel, J. F. The Montreal classification of inflammatory bowel disease: Controversies, consensus, and implications. Gut 55, 749–753. https://doi.org/10.1136/gut.2005.082909 (2006).
Cheon, J. H. et al. Development and validation of novel diagnostic criteria for intestinal Behcet’s disease in Korean patients with ileocolonic ulcers. Am. J. Gastroenterol. 104, 2492–2499. https://doi.org/10.1038/ajg.2009.331 (2009).
Cheon, J. H. et al. Development, validation, and responsiveness of a novel disease activity index for intestinal Behcet’s disease. Inflamm. Bowel. Dis. 17, 605–613. https://doi.org/10.1002/ibd.21313 (2011).
Best, W. R., Becktel, J. M., Singleton, J. W. & Kern, F. Jr. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology 70, 439–444 (1976).
Chung, Y. W. et al. Targeted disruption of PDE3B, but not PDE3A, protects murine heart from ischemia/reperfusion injury. Proc. Natl. Acad. Sci. USA 112, E2253-2262. https://doi.org/10.1073/pnas.1416230112 (2015).
Chung, Y. W. et al. Apolipoprotein E and periostin are potential biomarkers of nasal mucosal inflammation a parallel approach of in vitro and in vivo secretomes. Am. J. Respir. Cell Mol. Biol. 62, 23–34. https://doi.org/10.1165/rcmb.2018-0248OC (2020).
Rukmangadachar, L. A. et al. Proteome analysis of the macroscopically affected colonic mucosa of Crohn’s disease and intestinal tuberculosis. Sci. Rep. 6, 23162. https://doi.org/10.1038/srep23162 (2016).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Sukumaran, S. K. et al. Taste cell-expressed alpha-glucosidase enzymes contribute to gustatory responses to disaccharides. Proc. Natl. Acad. Sci. USA 113, 6035–6040. https://doi.org/10.1073/pnas.1520843113 (2016).
Lee, N. S., Brewer, H. B. Jr. & Osborne, J. C. Jr. beta 2-glycoprotein I. Molecular properties of an unusual apolipoprotein, apolipoprotein H. J. Biol. Chem. 258, 4765–4770 (1983).
Naim, H. Y., Sterchi, E. E. & Lentze, M. J. Structure, biosynthesis, and glycosylation of human small intestinal maltase-glucoamylase. J. Biol. Chem. 263, 19709–19717 (1988).
Principe, S. et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics 13, 1667–1671. https://doi.org/10.1002/pmic.201200561 (2013).
Limsrivilai, J. & Pausawasdi, N. Intestinal tuberculosis or Crohn’s disease: A review of the diagnostic models designed to differentiate between these two gastrointestinal diseases. Intest. Res. https://doi.org/10.5217/ir.2019.09142 (2020).
Kim, J. M. & Cheon, J. H. Pathogenesis and clinical perspectives of extraintestinal manifestations in inflammatory bowel diseases. Intest. Res. https://doi.org/10.5217/ir.2019.00128 (2020).
Li, Z. et al. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J. Proteome Res. 11, 1582–1590. https://doi.org/10.1021/pr200748h (2012).
Starr, A. E. et al. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn’s disease from UC. Gut 66, 1573–1583. https://doi.org/10.1136/gutjnl-2015-310705 (2017).
Canani, R. B. & Terrin, G. Recent progress in congenital diarrheal disorders. Curr. Gastroenterol. Rep. 13, 257–264. https://doi.org/10.1007/s11894-011-0188-6 (2011).
Nichols, B. L. et al. The maltase-glucoamylase gene: Common ancestry to sucrase-isomaltase with complementary starch digestion activities. Proc. Natl. Acad. Sci. USA 100, 1432–1437. https://doi.org/10.1073/pnas.0237170100 (2003).
Amit-Romach, E., Reifen, R. & Uni, Z. Mucosal function in rat jejunum and ileum is altered by induction of colitis. Int. J. Mol. Med. 18, 721–727 (2006).
Lackeyram, D., Mine, Y., Archbold, T. & Fan, M. Z. The small intestinal apical hydrolase activities are decreased in the piglet with bowel inflammation induced by dextran sodium sulfate. J. Anim. Sci. 90(Suppl 4), 287–289. https://doi.org/10.2527/jas.54010 (2012).
Ericson, J. A. et al. Gene expression during the generation and activation of mouse neutrophils: Implication of novel functional and regulatory pathways. PLoS ONE 9, e108553. https://doi.org/10.1371/journal.pone.0108553 (2014).
Dreyfus, J. C. & Poenaru, L. Alpha glucosidases in white blood cells, with reference to the detection of acid alpha 1–4 glucosidase deficiency. Biochem. Biophys. Res. Commun. 85, 615–622. https://doi.org/10.1016/0006-291x(78)91207-x (1978).
Crowe, S. E., Luthra, G. K. & Perdue, M. H. Mast cell mediated ion transport in intestine from patients with and without inflammatory bowel disease. Gut 41, 785–792. https://doi.org/10.1136/gut.41.6.785 (1997).
He, S. H. Key role of mast cells and their major secretory products in inflammatory bowel disease. World J. Gastroenterol. 10, 309–318. https://doi.org/10.3748/wjg.v10.i3.309 (2004).
Hamilton, M. J., Frei, S. M. & Stevens, R. L. The multifaceted mast cell in inflammatory bowel disease. Inflamm. Bowel Dis. 20, 2364–2378. https://doi.org/10.1097/MIB.0000000000000142 (2014).
Albert-Bayo, M. et al. Intestinal mucosal mast cells: Key modulators of barrier function and homeostasis. Cells https://doi.org/10.3390/cells8020135 (2019).
Acknowledgements
This work was supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2017M3A9F3041229 to J.-H.R.) and by a Grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI18C0094 to J.H.C.).
Author information
Authors and Affiliations
Contributions
Guarantor of the article: J.H.C. and J.-H.R. Development of study concept and design: Y.W.C. and J.H.C. Study supervision: J.H.C. and J.-H.R. Acquisition data: J.P., D.J., Y.W.C., S.H., D.H.K., and J.Y.. Data analysis and interpretation of data: J.P., D.J., S.H., and Y.W.C.. Drafting of the manuscript: J.P., D.J., and Y.W.C. Critical revision of the manuscript for important intellectual content: J.H.C. and J.-H.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, J., Jeong, D., Chung, Y.W. et al. Proteomic analysis-based discovery of a novel biomarker that differentiates intestinal Behçet’s disease from Crohn’s disease. Sci Rep 11, 11019 (2021). https://doi.org/10.1038/s41598-021-90250-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-90250-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.