Abstract
Autologous blood-derived products such as platelet-rich plasma (PRP) are widely used to treat musculoskeletal conditions, including knee osteoarthritis (OA). However, the clinical outcomes after PRP administration are often variable, and there is limited information about the specific characteristics of PRP that impact bioactivity and clinical responses. In this study, we aimed to develop an integrative workflow to evaluate responses to PRP in vitro, and to assess if the in vitro responses to PRP are associated with the PRP composition and clinical outcomes in patients with knee OA. To do this, we used a coculture system of macrophages and fibroblasts paired with transcriptomic analyses to comprehensively characterize the modulation of inflammatory responses by PRP in vitro. Relying on patient-reported outcomes and achievement of minimal clinically important differences in OA patients receiving PRP injections, we identified responders and non-responders to the treatment. Comparisons of PRP from these patient groups allowed us to identify differences in the composition and in vitro activity of PRP. We believe that our integrative workflow may enable the development of targeted approaches that rely on PRP and other orthobiologics to treat musculoskeletal pathologies.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a whole joint disorder and a major cause of pain and disability in adults. Inflammation is a known contributing factor to OA, and both local and peripheral inflammation are associated with disease activity, pain and knee stiffness. The complex pathogenesis of OA has resulted in significant challenges for the development of therapeutic strategies and currently there are few viable alternatives to total joint replacement. The available non-surgical interventions aim to delay disease progression, improve function, and treat pain by decreasing inflammation1,2.
The current standard methods available to clinicians to treat joint inflammation include oral non-steroidal anti-inflammatory medications and corticosteroid injection. However, these treatments are associated with variable outcomes, and as result clinicians have begun to use “orthobiologics” due to their potential to diminish inflammation and promote anabolism and repair, and because its “minimally manipulated” and autologous nature is not subjected to pre-market regulatory clearance from the Food and Drug Administration3.
Platelet-rich-plasma (PRP) is an autologous blood product generated by centrifugation of whole venous blood to isolate and concentrate platelets. It is believed that platelets, which contain and release numerous growth factors, promote healing, dampen inflammation, and reduce pain3,4,5. These blood-derived products have been used to enhance chondrogenesis6,7, treat bone8,9, tendon10,11 and ligament12,13 injuries, and have recently emerged as a potential therapeutic for knee OA14.
The available studies comparing intra-articular PRP injections to other means of non-surgical intervention for knee OA often report benefits for patients receiving PRP. The current view is that positive clinical results in OA patients are mainly related to the anti-inflammatory effects of PRP rather than to increased anabolic and reparative effects in articular cartilage4,14. However, these reports are inconclusive because of the high variability in the available PRP preparations, the limited available information about the relevant characteristics in these blood products that impact clinical responses in patients with knee OA, the lack of consensus and standardization of treatment protocols and collection of outcomes, and the poor understanding of disease candidates or specific cell/tissue targets for a given therapy15.
In the current study, we aimed to develop an integrative workflow to assess if the PRP composition and bioactivity in vitro is associated with clinical outcomes in patients with knee OA. Because of the well-characterized cellular crosstalk between synovial fibroblasts and macrophages16 and the contribution of these cells to OA disease17,18, we evaluated the anti-inflammatory actions of PRP in cocultures that reproduce the cellular crosstalk that occurs in inflamed synovial tissues. We characterized the responses to PRP in macrophages and fibroblasts using transcriptomics analyses. Relying on clinical outcomes, we identified knee OA patients with variable pain responses to intra-articular PRP injection. Using our bioassay and PRP from these patients we were able to detect differences in the PRP bioactivity that were associated with patient-reported outcomes.
Patients and methods
Subjects
The study protocol was approved by the Institutional Review Board (IRB) of the Hospital for Special Surgery (HSS). Written informed consent was obtained from all participants before entering the study, and the study and all methods were performed in accordance with the relevant guidelines and regulations. After IRB approval (HSS IRB#2016-0267) and patient consent, 51 knee OA patients were enrolled in our study upon presentation to clinic and eligibility for platelet-rich-plasma (PRP) injection, following the inclusion and exclusion criteria indicated in Supplementary Table S1, including Kellgren-Lawrence OA grade 1–3 based on plain radiographs. Knee OA patients received 1 intra-articular injection of leukocyte-reduced PRP. See Table 1 for patient demographics and PRP details. We collected patient-reported outcome measures (Knee injury and Osteoarthritis Outcome (KOOS-JR) and Numeric Pain Rating Scale (NPRS))19,20 at baseline, 6 weeks, and 6 months after PRP injection. For selection of PRP and downstream analyses, patients were categorized as responders (resp) or non-responders (n-resp) based on achievement of Minimal Clinically Important Difference (MCID) values (10-point increase for KOOS JR, and 20% decrease for NPRS)21,22 on the outcome measures reported at 6 months23. Additionally, we prepared PRP from healthy donors to obtain enough PRP volume to conduct in vitro experiments. Participants were enrolled in the study between 10/28/2017 and 11/26/2018.
Platelet-rich-plasma (PRP) preparation
We used the Arthrex Autologous Conditioned Plasma kit series 1 (Arthrex, Naples, FL) for sample preparation. The characteristics of the PRP preparations, volume collected and injected, and time from collection to injection are summarized in Table 1 and reported following the MIBO guidelines24. Immediately after PRP preparation, patients received an intra-articular injection of PRP following standard procedures. The remaining PRP was immediately transported to the laboratory, treated with an additional 10% acid citrate dextrose solution A, aliquoted, and used for downstream analyses. One aliquot of PRP (200 µl) and one aliquot of whole blood collected at the time of the PRP preparation were used to determine the complete blood count (CBC) with differential in the Department of Pathology and Laboratory Medicine at HSS (see Table 1). Exogenous activation of PRP for in vitro experiments or ELISA assays was done using calcium chloride (CaCl2, final concentration 22.8 mM), as described25.
Coculture system of macrophages and synovial fibroblasts
To evaluate the in vitro biologic activity of PRP, we used a previously described coculture system of primary human synovial fibroblasts and macrophages16. Fibroblasts were isolated as described26 from deidentified synovial tissues retrieved from patients undergoing total knee arthroplasty for rheumatoid arthritis (RA) or osteoarthritis (OA), after IRB approval and patient consent, and used for coculture experiments between passages 4 and 7, also as described16. Human CD14 + monocytes were freshly isolated from whole blood obtained from the New York Blood Bank and treated with 10 ng/ml of monocyte colony stimulating factor (M-CSF, PeproTech) for 24 h to promote a pro-macrophage lineage27. At 24 h after M-CSF treatment, macrophages and fibroblasts were cocultured in a 0.4 µm polyester trans-well system (Corning), as described16. The cocultured cells were left untreated (control, medium) or incubated with 10% v:v of activated PRP, 20 ng/ml of recombinant human TNFα (PeproTech), or PRP + TNFα for an additional 24 h. At 24 h, RNA was isolated for RNA-seq, NanoString, and RTqPCR analyses. For dose–response experiments, cells were left untreated or treated with 1%, 10%, and 25% of PRP (v:v) alone or combined with 20 ng/ml of TNFα for 24 h. Time-course experiments were performed using cells treated with 10% of PRP, alone or combined with TNFα, for 1 h, 6 h, and 24 h. For selected experiments, human recombinant IL-1β (R&D Systems, 10 ng/ml) was used instead of TNFα.
RNA sequencing (RNA-seq)
Total RNA was extracted using the RNeasy mini kit (Qiagen) from cocultures of macrophages and fibroblasts, treated with the indicated conditions (control, PRP, TNFα, or PRP + TNFα) for 24 h. RNA-seq analyses were done in 4 independent experiments, using fibroblasts and macrophages obtained from 4 different tissue donors, and PRP samples prepared from 4 different subjects. A total of 100 ng of high-quality RNA (RIN > 8 and A260/280 > 1.8) were used to construct libraries. Sequencing was performed using an Illumina HiSeq 2500 at the Epigenomics Core Facility of Weill Cornell Medicine (50-bp single-end protocol) at a depth of 25 million mappable reads per sample. Read quality evaluation and adapter trimming was performed using fastp28. Read mapping to human genome (hg38) and counting against Gencode annotation was performed with STAR. Differential gene expression analysis was performed with edgeR using quasilikelyhood framework. Batch correction to account for the day of the experiment was performed by including a relevant batch term into a general-linear model. Genes with FDR-corrected p-values less than 0.05 and log2 fold change > 1 were considered differentially expressed. Downstream analyses were performed in R using a Shiny-driven visualization platform (RNAseq DRaMA) developed at the HSS David Z. Rosensweig Genomics Research Center.
Quantitative set analysis of gene expression (QuSAGE) pathway analyses
To determine differentially regulated pathways we performed QuSAGE pathway analysis29 using MsigDB 6.2 C2 set (curated gene sets) as a reference30. QuSAGE approach relies on comparisons of differences between pathway-level probability density functions (PDF) for control and treatment samples. For a detailed description of the method, see30. Briefly, first, the PDF for the difference between control and treatment for each gene in the pathway is constructed by sampling from a standardized t-distribution with the degrees of freedom calculated from the data. Then, individual gene PDFs for all genes in a gene set are combined using numeric convolution to yield a pathway PDF. To account for the differences in pathway sizes, the pathways PDF is scaled by 1/N where N is the number of genes in a set. To account for correlations between genes, a pathway PDF is further scaled using variance inflation factor (VIF) that is estimated from the data. All gene sets with FDR-corrected p values < 0.05 were visualized using RNAseq DRaMA.
Magnetic resonance imaging (MRI)
For patients with MRI obtained at HSS, MRI was performed using a cartilage-sensitive pulse sequence with an intermediate-weighted moderate echo time (TE) protocol that has already been assessed for accuracy based on an arthroscopic standard, with almost perfect agreement for reproducibility31. MRI readings of the affected knees were performed independently by two trained radiologists blinded to all other data using the Whole-Organ Magnetic Resonance Imaging Score (WORMS) method, as described32.
nCounter NanoString gene expression analyses
For NanoString gene expression analyses, we used 100 ng of total RNA (RIN > 8, 260/280 > 1.8) isolated from 3 independent co-culture experiments, using macrophages and fibroblasts from 3 different blood and tissue donors, and PRP prepared from 3 different subjects. Analyses were performed using the nCounter PanCancer Human Pathways Panel (NanoString Technologies, Seattle, WA), following the manufacturer’s instructions. Data analysis was performed using the accompanying software (nSolver 4.0, NanoString Technologies, Seattle, WA).
RTqPCR analyses
Reverse transcriptase quantitative polymerase chain reaction (RTqPCR) analyses of selected target genes were carried out using SYBR Green I-based real-time PCR, as described33, and specific primers against CCL5, CXCL1, CXCL3, CYLD, IL1B, IL23A, MMP3, PPBP and GAPDH (Supplementary Table S2). The data was calculated as the ratio of each gene to GAPDH.
ELISA and multiplex ELISA
We performed magnetic bead-based sandwich immunoassays using the MILLIPLEX MAP multiplex Human Cytokine Panel 1 (cat #HCYTOMAG-60 k; EMD-Millipore Corporation, St. Charles, MO) following the manufacturer’s instructions. Duplicate wells of activated PRP samples were analyzed using a Luminex MagPix (Luminex). Cytokine concentrations were determined using the Luminex Xponent 4.2 and EMD-Millipore Milliplex Analyst v5.1. A human TGFb1 Quantikine ELISA Kit (R&D Systems) was used to evaluate TGFβ1 in PRP, using a Tecan Infinite M200 PRO plate reader, with the accompanying i-control 1.12 software (Tecan).
Statistical analysis
Analyses of the patient-reported outcomes and multiplex ELISA assays were done at the Biostatistics Core of the Hospital for Special Surgery. Data were assessed for approximation to the Gaussian distribution using the D’Agostino-Pearson omnibus test of normality. Distributions were considered to be Gaussian if the P-value for the null hypothesis was greater than 0.05. Unpaired student t-test was used to compare two groups. Analyses of non-normally distributed data was performed using Mann–Whitney test. Statistical analyses of the qPCR results were performed using GraphPad Prism 8 (GraphPad Software, San Diego, CA). For data involving multiple comparisons, one-way analysis of variance (ANOVA) was performed followed by Tukey’s post-hoc test. Unless otherwise indicated, data are reported as means ± S.D. of at least 3 independent experiments. P < 0.05 was considered significant.
Results
PRP modulates TNFα-induced responses in macrophages
We aimed to develop an integrative approach that permits the evaluation of PRP bioactivity in vitro, using PRP from patients with different responses to PRP (see Fig. 1 for study design). First, we established a coculture system of human macrophages and synovial fibroblasts and compared their transcriptional profiles in response to TNFα, alone or in combination with PRP. We selected this coculture system because of the well-known contribution of synovial fibroblasts and macrophages to OA17,18, and because this coculture system and the crosstalk of these cells in response to TNFα is also well-established16. We performed RNA-seq analyses in 4 independent cocultures (prepared with macrophages and fibroblasts from different tissue donors and using PRP samples from 4 independent donors) treated with 10 ng/ml of TNFα and 10% (vol:vol) of PRP, alone or combined. Macrophages and fibroblasts were co-treated for 24 h and then RNA was isolated from each cell type separately for RNA-seq analyses.
Study workflow. (A) We evaluated the anti-inflammatory activity of PRP using cocultures of macrophages and fibroblasts treated with PRP without or with TNFα for 24 h. After treatment, RNA was isolated from macrophages and fibroblasts for RNA-seq. (B) We collected PRP and patient reported outcomes measures (PROMs, Knee injury and Osteoarthritis Outcome and Numeric Pain Rating Scale) in knee OA patients that received one intra-articular injection of PRP. We defined two patient subtypes based on achievement of minimal clinically important differences at 6 weeks (6 wks) and 6 months (6 mo) after injection, relative to baseline. Based on these criteria, patients that report positive responses (reduced pain) were considered responders (resp), and patients that did not respond to the treatment were considered non-responders (n-resp). (C) We compared the composition of PRP selected from resp and n-resp using ELISA and multiplex ELISA. We compared the bioactivity of PRP from resp and n-resp using the coculture system and changes in the expression of specific gene targets. Created with BioRender.com.
Analyses of RNA isolated from macrophages (Fig. 2A) after PRP, TNFα, and TNFα + PRP treatment uncovered unique transcriptional profiles relative to non-treated controls, as shown by the principal component analyses (PCA, Fig. 2B). Relative to non-treated controls, macrophages treated with PRP showed decreased expression of several canonical NF-κB targets and stress-response genes, including S100A12 and TNFα, and pronounced upregulation of MAP2K6 mRNA. As expected, TNFα-treated cells (Fig. 2C) showed increased expression of canonical NF-κB targets (including CCL5 and NFKB1) and the previously reported TNFα-induced IFN responses16, with increased CXCL10, NKG7 and IL1B mRNA. Comparisons between PRP + TNFα vs. TNFα-treated macrophages uncovered 559 differentially expressed genes (DEGs, Fig. 2D), including decreased expression of known modulators of inflammation (JUN, IRF4 and SMAD3; Supplementary Table S3 for the complete list). NanoString multiplex gene expression analyses confirmed changes in expression of genes identified by RNA-seq analyses in macrophages (Fig. 2E, F, and Supplementary Table S4).
Transcriptomic analyses of the modulation of TNFα-driven responses by PRP in macrophages. (A) Schematic representation of the experimental workflow. Created with BioRender.com (B) Principal component analysis (PCA, using top 1,000 genes) of RNA-seq data from macrophages left untreated, or treated with TNFα and PRP, alone or combined, for 24 h. RNA-seq analysis is based on 4 independent coculture experiments, with macrophages isolated from 4 different tissue donors and PRP prepared from 4 different subjects. (C) Volcano plot of differentially expressed genes comparing TNFα-treated macrophages to untreated controls. Colored dots correspond to genes with significant changes greater than 1 log2-fold expression change (FDR < 0.05, LogFC > 1). Red indicates increased expression and blue indicates decreased expression after TNFα treatment. (D) Volcano plot representing PRP-induced changes in macrophages co-treated with TNFα + PRP. Red dots represent differentially expressed genes (DEGs) induced by TNFα relative to untreated controls, and blue dots represent PRP-induced changes in PRP + TNFα treated cells relative to TNFα-treated macrophages (FDR < 0.05, LogFC > 1). (E) Heatmap representation of selected DEGs by RNA-seq, and (F) NanoString analyses showing the normalized counts of these same selected genes, confirming changes in the control (Ct), PRP- (P), TNFα- (T) and PRP + TNFα-treated (P + T) macrophages. Dotted lines indicate background signal. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by t-test, calculated using ratio data and the nSolver analysis software. NanoString data is represented as the mean ± S.D (error bars) normalized counts of 3 independent coculture experiments, using macrophages isolated from 3 different tissue donors and PRP prepared from 3 different subjects. Graphs were created with GraphPad Prism 8. (G) Representation of the QuSAGE pathway analyses in PRP-treated macrophages relative to untreated controls, showing functional pathways differentially expressed in PRP-treated cells (FDR < 0.05). Heatmap representation of representative genes associated with the (H) NF-κB/MAPK, (I) TNFR1, (J) NLRP3 inflammasome, and (K) PPAR signaling pathways. Data was z-score normalized.
Next, we performed QuSAGE pathway analyses29 using RNA-seq data. Comparison of PRP-treated macrophages relative to non-treated controls (Fig. 2G) uncovered changes in NF-κB/MAPK (primarily driven by the reduced expression of S100A12 and MAP2K6, Fig. 2H), decreased expression of the TNFR1 (Fig. 2I) and NLRP3 inflammasome (Fig. 2J) pathways, and increased expression of the PPAR signaling pathway (Fig. 2K, and Supplementary Table S5). TNFα-treated macrophages showed decreased WNT signaling and the expected increased expression of genes in the TNFα pathway relative to non-treated controls (Supplementary Table S6). The comparison between the TNFα + PRP and TNFα groups uncovered seven differentially expressed pathways, including decreased expression of the cholesterol biosynthesis pathway and increased expression of the pyruvate dehydrogenase PDH complex signaling in macrophages (Supplementary Table S7).
PRP modulates TNFα-induced responses in synovial fibroblasts
Analyses of RNA isolated from cocultured fibroblasts (Fig. 3A) after PRP, TNFα, and TNFα + PRP treatment also uncovered unique transcriptional profiles, with robust and pronounced differences in TNFα-treated fibroblasts vs. non-treated controls, and remarkable differences between TNFα + PRP and TNFα-treated cells (Fig. 3B). Comparison of fibroblasts treated with PRP relative to non-treated controls uncovered 776 differentially expressed genes, including IGF1, VEGFA, CDK1, IL1A, and other genes involved in replication, proliferation, DNA repair, and cell survival. Comparison of TNFα-treated fibroblasts with control cells (Fig. 3C) showed a robust induction of classic inflammatory and NF-κB-responsive genes (including CCL5, NFKBIA, or RELA) as well as up-regulation of IFN signature genes (including MX1, MX2, IRF1, IRF7 or SOCS). Fibroblasts co-treated with PRP and TNFα showed very pronounced differences relative to the TNFα-treated cells, with 2,738 DEGs in PRP + TNFα-treated fibroblasts relative to the TNFα-treated group (Fig. 3D). In agreement with its proposed anti-inflammatory actions, PRP co-treatment modulated and, in many cases, abolished TNFα-driven induction of NF-κB-responsive genes (including CCL5 or RELA) and IFN signatures (with decreased expression of IRF1, MX1 or MX2, Supplementary Table S8). NanoString analyses confirmed changes in expression of genes identified by RNA-seq analyses in fibroblasts. See Fig. 3E, F for selected genes and Supplementary Table S9 for a summary of the NanoString normalized counts in fibroblasts.
Transcriptomic analyses of the modulation of TNFα-driven responses by PRP in fibroblasts. (A) Schematic representation of the experimental workflow. Created with BioRender.com (B) Principal component analysis (PCA, using top 1,000 genes) of RNA-seq data from fibroblasts left untreated, or treated with TNFα and PRP, alone or combined, for 24 h. RNA-seq analysis is based on 4 independent coculture experiments, with fibroblasts isolated from 4 different tissue donors and PRP prepared from 4 different subjects. (C) Volcano plot of differentially expressed genes comparing TNFα-treated fibroblasts to untreated controls. Colored dots correspond to genes with significant changes greater than 1 log2-fold expression change (FDR < 0.05, LogFC > 1). Red indicates increased expression and blue indicates decreased expression after TNFα treatment. (D) Volcano plot representing PRP-induced changes in fibroblasts co-treated with TNFα + PRP. Red dots represent differentially expressed genes (DEGs) induced by TNFα relative to untreated controls, and blue dots represent PRP-induced changes in PRP + TNFα treated cells relative to TNF-treated cells (FDR < 0.05, LogFC > 1). (E) Heatmap representation of selected DEGs by RNA-seq, and (F) NanoString analyses showing the normalized counts of these same selected genes, confirming changes in the control (Ct), PRP- (P), TNFα- (T) and PRP + TNFα-treated (P + T) fibroblasts. Dotted lines indicate background signal. **p < 0.01, ***p < 0.001, ****p < 0.0001 by t-test, calculated using ratio data and the nSolver analysis software. NanoString data is represented as the mean ± S.D (error bars) normalized counts of 3 independent coculture experiments, using fibroblasts isolated from 3 different tissue donors and PRP prepared from 3 different subjects. Graphs were created with GraphPad Prism 8. (G) Representation of the QuSAGE pathway analyses in TNFα + PRP-treated fibroblasts relative to TNFα-treated cells, showing functional pathways differentially expressed in TNFα + PRP-treated cells (FDR < 0.05). Heatmap representation of representative genes associated with the (H) interferon gamma, (I) interferon alpha beta, (J) IL23, and (K) canonical NF-κB signaling pathways. Data was z-score normalized.
QuSAGE pathway analyses in PRP-treated fibroblasts relative to untreated controls confirmed the differential modulation of functional pathways relevant to cell cycle, DNA synthesis and cell survival, and also showed repression of ATF4, ATF6, and PI3K/AKT signaling relative to non-treated controls (Supplementary Table S10). Pathway analyses in TNFα-treated cells relative to non-treated controls confirmed the potent upregulation of IFN and NF-κB signatures, as well increased expression of the extrinsic pathway for apoptosis and repression of DNA repair (Supplementary Table S11). Comparison of TNFα + PRP- versus TNFα-treated fibroblasts showed pronounced changes in different functional pathways (Fig. 3G and Supplementary Table S12), including decreased expression of the interferon gamma (Fig. 3H), interferon alpha beta (Fig. 3I), IL23 (Fig. 3J), and NF-κB (Fig. 3K) signaling pathways.
These analyses in macrophages and fibroblasts demonstrated a profound anti-inflammatory effect of PRP, both at baseline and in response to TNFα-induced inflammation. In additional experiments, we treated macrophages and fibroblasts with IL-1β alone or combined with PRP for 24 h. As shown is Supplementary Figure S1, PRP co-treatment also down-modulated responses to IL-1β in our coculture system, indicating that the immunomodulatory and anti-inflammatory actions of PRP are not cytokine-specific. Experiments using macrophage monocultures treated with TNFα and PRP suggested that the PRP actions on macrophages did not depend upon the fibroblast-macrophage crosstalk (Supplementary Figure S2).
The PRP modulation of TNFα-driven responses is concentration- and time-dependent
We next aimed to identify specific gene targets that can be used as readouts for the cellular responses to PRP in macrophages and fibroblasts. Our transcriptomic analyses showed that PRP strongly modulated the expression of canonical NF-κB targets in macrophages and fibroblasts, in agreement with studies that reported that the modulation of NF-κB by PRP is cell- and PRP composition-dependent34,35,36. We identified CCL5 (RANTES) as one of the direct canonical NF-κB targets37,38 with the most robust (logFC > 6 and FDR < 0.0001) increased expression upon TNFα treatment in both macrophages and fibroblasts (Fig. 4A, E). CCL5 also displayed PRP-dependent inhibition in these cells, particularly evident in TNFα-treated fibroblasts (Supplementary Tables S3 and S8). RTqPCR analyses in cocultures confirmed the TNFα-induced expression of CCL5 in macrophages, with reduced expression in TNFα + PRP-treated cells (Fig. 4B). Cocultures of macrophages and fibroblasts with TNFα, alone or combined with different concentrations (% vol:vol) of PRP for 24 h showed that CCL5 displayed a robust dose–response to PRP in macrophages (Fig. 4C). RTqPCR analyses of time-course experiments in cocultures treated with TNFα, alone or combined with 10% PRP for 1, 6 and 24 h showed time-dependent induction of CCL5 mRNA upon TNFα treatment in macrophages, and inhibition by PRP co-treatment at 6 and 24 h after treatment (Fig. 4D). Similarly, qPCR analyses in cocultures confirmed the TNFα-dependent induction of CCL5 and reduced expression after co-treatment with PRP in fibroblasts (Fig. 4F). In agreement with the results in the cocultured macrophages, the responses to PRP were also concentration- (Fig. 4G) and time-dependent (Fig. 4H) in fibroblasts. Other tested genes in macrophages and fibroblasts displayed less robust dose- or time-dependent responses or were only reliable in one of the cell types (Supplementary Figure S3).
Dose- and time-dependent PRP modulation of the TNFα-induced CCL5 expression in macrophages and fibroblasts. (A) Volcano plot of differentially expressed genes comparing TNFα-treated macrophages to untreated controls. Genes with significant changes (FDR < 0.0001) and logFC > 6 are in red. CCL5 is highlighted in blue. (B) RTqPCR analyses in macrophages left untreated (Ct) or treated with TNFα (T) and 10% v:v PRP (P) alone or combined (T + P) for 24 h. ****p < 0.001 by ANOVA followed by Tukey’s post-hoc test. (C) RTqPCR analyses in macrophages left untreated (Ct) or treated with TNFα alone (T) or combined (T + P) with different concentrations (%, vol:vol) of PRP for 24 h. ***p < 0.001, ****p < 0.0001 versus TNFα by ANOVA followed by Tukey’s post-hoc test. (D) RTqPCR analyses in macrophages left untreated (Ct), or treated with TNFα (T), 10% (v:v) PRP (P), or TNFα + PRP (T + P) for 1 h, 6 h and 24 h. *p < 0.05, ***p < 0.001, ****p < 0.0001 by ANOVA followed by Tukey’s post-hoc test. (E) Volcano plot of differentially expressed genes comparing TNFα-treated fibroblasts to untreated controls. Genes with significant changes (FDR < 0.0001) and logFC > 6 are in red. CCL5 is highlighted in blue. (F) RTqPCR analyses in fibroblasts left untreated (Ct) or treated with TNFα (T) and 10% v:v PRP (P) alone or combined (T + P) for 24 h. *p < 0.05 by ANOVA followed by Tukey’s post-hoc test. (G) RTqPCR analyses in fibroblasts left untreated (Ct) or treated with TNFα alone (T) or combined (T + P) with different concentrations (%, vol:vol) of PRP for 24 h. ns = not significant, *p < 0.05 versus TNFα by ANOVA followed by Tukey’s post-hoc test. (H) RTqPCR analyses in fibroblasts left untreated (Ct), or treated with TNFα (T), 10% (v:v) PRP (P), or TNFα + PRP (T + P) for 1 h, 6 h and 24 h. *p < 0.05, **p < 0.01, by ANOVA followed by Tukey’s post-hoc test. RTqPCR data is represented as mean ± S.D (error bars) of 3 independent coculture experiments, using macrophages and fibroblasts isolated from 3 different tissue donors and PRP prepared from 3 different subjects. RNA-seq is based on 4 independent coculture experiments, with macrophages and fibroblasts isolated from 4 different tissue donors and PRP prepared from 4 different subjects. RTqPCR graphs created with GraphPad Prism 8.
Based on these results, we selected CCL5 as a target, and 10% PRP and 24 h stimulation in our coculture conditions for follow-up analyses and evaluation of PRP bioactivity in patient samples.
Differences in bioactivity in PRP from OA patients with poor clinical outcomes
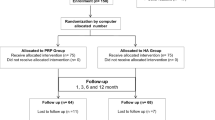
Next, we aimed to assess if the in vitro bioactivity from the PRP obtained from patients reporting improved pain outcomes after PRP treatment is different than the bioactivity of PRP from patients reporting poor outcomes. To do this, we enrolled 51 patients that received one single leukocyte-poor PRP injection (3.7 ± 0.7 ml, prepared using the Arthrex ACP® kit) to treat knee OA. See Table 1 for patient demographics and PRP characteristics. We collected relevant clinical information at baseline, and self-reported Knee injury and Osteoarthritis Outcome (KOOS-JR) and Numeric Pain Rating Scale (NPRS) scores at baseline and at 6 weeks and 6 months after PRP injection. We used these patient-reported outcomes to identify responders (resp) or non-responders (n-resp) to PRP based on achievement of Minimal Clinically Important Difference (MCID) values at 6 months after PRP injection (see Patients and Methods and21,22). We selected 6 months as a time-point to define resp and n-resp to PRP and conduct analyses based on studies that evaluated pain scores in knee OA patients in response to PRP treatment at 6 and 12 months23. Figure 5A and B show the distribution of outcome metrics over time, representing only patients that completed both questionnaires at baseline (d0, immediately before PRP administration) and at 6 weeks and 6 months after PRP treatment (N = 20).
Evaluation of the bioactivity of PRP from patients classified as responders and non-responders using patient-reported outcomes. Responders (resp, green) and non-responders (n-resp, red) to PRP treatment identified using (A) Knee injury and Osteoarthritis Outcome (KOOS JR) Score and (B) Numeric Pain Rating Scale (NPRS) Score in patients with completed questionnaires at baseline (d0) and the 6 weeks (6 wks) and 6 months (6 mo) follow up time-points. For selection of PRP for analyses, patients were categorized as responders or non-responders based on achievement of minimal clinically important difference (MCID) thresholds (dotted lines) of 10-point improvement in KOOS JR and 20% improvement in NPRS at 6 months after PRP injection. Scores are represented as changes relative to self-reported baseline levels (d0) for each patient and each score. RTqPCR analyses of CCL5 mRNA in (C) macrophages and (D) fibroblasts left untreated (Ct) or treated with TNFα (T), PRP (P) or TNFα + PRP (T + P) for 24 h, using PRP from responders or non-responders. RTqPCR data is represented as mean ± S.D (error bars) of 5 (macrophages) and 4 (fibroblasts) independent experiments. **p < 0.01, ****p < 0.0001 by ANOVA followed by Tukey’s post-hoc test. ns = not significant. Graphs were created with GraphPad Prism 8.
Using exclusively outcomes collected at 6 months23 after treatment relative to baseline, we identified 17 responders and 15 non-responders (Supplementary Table S13). We did not observe significant differences in Whole-Organ Magnetic Resonance Imaging Score (WORMS)32, age, sex, BMI, or concentration of platelets in the injected PRP comparing responders and non-responders (Supplementary Table S13). Multiplex ELISA analyses of PRP from responders and non-responders uncovered minor differences in IL-7 protein, but the relative abundance of other analyzed factors that are believed to drive PRP bioactivity4,39 was largely similar between responders and non-responders (Supplementary Table S14), including IL1β (p = 0.264), TNFα (p = 0.602), or VEGF (p = 0.770). Similarly, we did not detect significant differences in TGFβ1 protein by ELISA, comparing PRP from responders and non-responders (Supplementary Figure S4).
Next, we used our in vitro bioassay to evaluate differences in the bioactivity of PRP from responders and non-responders. We selected PRP samples from 5 patients that met the “responder” MCID criteria at 6 months23 for both NPRS and KOOS-JR scores, and we compared them with PRP samples from 5 patients that were categorized as non-responders at 6 months for both scores. We prepared cocultures of macrophages and fibroblasts using cells from 5 independent blood and tissue donors, respectively. We treated the cocultures with TNFα, alone or combined with 10% (v:v) PRP from responders or non-responders. At 24 h after treatment, we analyzed the CCL5 mRNA expression by RTqPCR in macrophages and fibroblasts. The RNA isolated from the cocultured fibroblasts from one experiment was lost during processing. While the co-treatment with PRP from responders and non-responders was able to inhibit the TNFα-induced CCL5 expression in macrophages (Fig. 5C) and fibroblasts (Fig. 5D), we detected cell-specific differences. In macrophages (Fig. 5C), we did not detect differences in the PRP-driven inhibition of TNFα-induced CCL5 expression between responders and non-responders (p = 0.4046). In contrast, in fibroblasts (Fig. 5D) the PRP from responders had a more potent inhibitory effect on the TNFα-induced CCL5 expression (p = 0.0017 vs. non-responders). We also evaluated the effects of PRP from responders and non-responders in the TNFα-induced PPBP and IL23A mRNA expression in macrophages and fibroblasts, respectively (Supplementary Figure S5). In macrophages (Supplementary Figure S5A), the PRP from responders synergized with TNFα to significantly increase PPBP expression (p = 0.0146 vs. TNFα, and p = 0.0071 vs. control), whereas the PRP from non-responders did not (p = 0.7785 vs. TNFα, and p = 0.6000 vs. control). In fibroblasts, co-treatment with PRP from responders and non-responders inhibited the TNFα-induced IL23A expression, and we did not detect differences between cells co-treated with TNFα and PRP from responders and cells co-treated with TNFα and PRP from non-responders (p = 0.5935, Supplementary Figure S5B).
Taken together, our results provide comprehensive molecular-level data on the anti-inflammatory actions of PRP in macrophages and synovial fibroblasts in vitro, and show that the integration of clinical and multi-omics data can help us identify differences in PRP bioactivity that may be associated with clinical outcomes. Our bioassay also highlighted the complex cell- and gene-specific actions of PRP that need to be taken into account for the development of targeted therapeutic approaches.
Discussion
Here we show that integration of clinical data and molecular analyses in a relevant in vitro model that mimics joint inflammation can be effectively used to identify differences in PRP bioactivity and may provide information about its association with clinical outcomes. Using this approach, we characterized the anti-inflammatory actions of PRP in fibroblasts and macrophages treated with TNFα, and we identified differences in bioactivity in PRP samples selected from knee OA patients with different clinical responses to the PRP treatment.
Positive outcomes after PRP treatment for OA are believed to be primarily related to the anti-inflammatory effects of PRP4,14. OA patients receiving leukocyte-poor PRP reported superior Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) scores compared to patients treated with hyaluronic acid40 or placebo41, in agreement with previous reports comparing PRP- and placebo-treated knee OA patients42. Similarly, leukocyte-poor PRP was more effective than conventional therapy (acetaminophen) to treat patients with early knee OA43. However, the functional impact of PRP to treat OA is still not conclusive because of the variable clinical results, which are believed to be related to both the variable PRP composition and to patient-intrinsic factors3,15.
In this study we did not aim to address PRP efficacy, and we relied on patient-reported outcomes exclusively to stratify patients into responders and non-responders and select PRP samples for downstream analyses. We did observe variable responses to PRP that are consistent with the literature, with patients clearly meeting MCID improvement thresholds and patients that did not show a clinically significant improvement following treatment. In our patient cohort, the variation in clinical responses was not associated with disease severity, in agreement with previous reports44. Similarly, we did not detect differences in sex, age, BMI, or MRI scores between patients that we defined as responders and non-responders. It is worth noting that we exclusively analyzed MRI images obtained before PRP administration, and we did not evaluate changes in cartilage morphology in response to PRP treatment with post-treatment MRI. It is conceivable that a longitudinal follow up of patients after PRP treatment could uncover patient-specific changes associated with responses to PRP. Therefore, there is still a need for larger clinical studies that comprehensively evaluate patient-specific features and their association with responses to PRP. These studies should include MRI and other forms of longitudinal evaluation that permit a comprehensive assessment of responses to the treatment over time, to identify good candidates for these emerging therapies.
Technique-45 and patient-dependent46,47 factors are known to contribute to variable PRP composition and are believed to significantly contribute to the variability in clinical outcomes, as also indicated by the differences in efficacy between leukocyte-rich and -poor PRP40. In this study we aimed to minimize technical variability using a single commercially available kit that results in autologous conditioned plasma (ACP, a form of leukocyte-poor PRP with relatively low enrichment in platelets), and standardizing the treatment protocol (single injection, equivalent volume injected). However, we still detected significant variability in the responses to PRP, which were not associated with age, sex, number of platelets in the PRP, or other demographics or technical metrics. Comparing the PRP from responders and non-responders, we did not detect differences in IL-1RA, VEGF, EGF, TGFβ or other cytokines that are classically associated with PRP activity4,39. We did find a minor increase in IL-7 in the PRP from responders relative to non-responders. IL-7 has well-described pleiotropic roles in different cell subsets48,49,50 and has been implicated in pain responses51,52. However, a functional connection between IL-7 and the PRP bioactivity in vitro, or with clinical outcomes in OA patients, was not established. Further, in our study we did not detect significant differences in the presence of platelets or leukocyte subtypes in the whole blood and resulting PRP of responders and non-responders that could explain these differences in IL-7. Follow up studies should evaluate the relative contribution of these molecules to the PRP activity in a cell- and tissue-specific context. These studies should focus on the combinatorial ratios and potential synergistic interactions of different bioactive molecules, including factors classically associated with PRP activity, instead of relative changes in the levels of one single factor. Such studies are required to better understand how the variable composition of PRP impacts clinical outcomes in OA patients and other orthopedic conditions.
Studies in preclinical models and in vitro systems indicated that platelet products can lead to some structural improvement, promote chondrogenesis and facilitate tissue repair53,54. These studies highlighted the immunomodulatory and anti-inflammatory actions of platelet-derived factors in different cell types35,55,56, and studies in knee OA patients have shown that the clinical benefits of PRP are likely related to pain-modifying anti-inflammatory actions14,23. We tested the bioactivity of PRP using a well-established coculture of macrophages and fibroblasts16. These cells contribute to the inflammatory responses in OA17,18 and our coculture reproduces the cellular crosstalk that occurs in inflamed synovial tissues16. PRP treatment or co-treatment with TNFα modulated transcriptional networks that include SMAD3, EGR1, or IRF7 in macrophages and fibroblasts, with a remarkable effect on IFN signatures and canonical NF-κB targets, consistent with other studies35,55,56. Specifically, in fibroblasts, co-treatment with PRP lead to a pronounced inhibition of the TNFα-induced IFNα/β and IFNγ signaling, with well-described roles in tissue homeostasis and inflammatory pathologies (for review, see57). PRP treatment also led to the inhibition of NF-κB-related pathways in macrophages and fibroblasts, including TRAF6-mediated NF-κB activation, TAK1 activation of NF-κB, or NLRP3 inflammasome signaling pathways. NF-κB is known to contribute to OA disease58,59 and to pain responses in different inflammatory contexts60. Thus, these results are consistent with the proposed anti-inflammatory and immunomodulatory actions of PRP. However, the functional impact of the PRP-driven anti-inflammatory actions requires further investigation, particularly because a fine-tuned modulation of these signaling pathways is required for homeostasis, and because the effects of therapeutic blockade in pathological conditions are likely cell/tissue-specific57,59. Taken together, our results further highlighted the complex cell- and gene-dependent actions of PRP in joint-derived cells, reinforcing the notion that studies that address this complexity are required to enable the efficacious targeted application of PRP to knee OA and other musculoskeletal conditions.
Our study has limitations. The targeted evaluation of PRP in vitro uncovered some differences in PRP bioactivity comparing responders and non-responders, but also highlighted the need for using larger group sizes and a wide array of gene targets to capture subtle changes in bioactivity, and to further understand gene- and cell-dependent responses to PRP. The small patient sample size, the lack of a placebo/control group, and the relatively short follow-up time-points used to identify responders and non-responders did not allow us to truly assess the efficacy of PRP as a treatment for OA. The limited set of outcome measures used in this study is also a limitation. Future studies should include placebo controls and longer follow-up time-points, as well as additional metrics for pain, physical function, quality of life, global assessment of the target joint, and adverse effects. Further, using MRI exclusively at baseline may not provide enough information about patient/joint-specific factors associated with clinical responses to PRP, particularly considering that PRP treatment can modulate symptoms independent of the initial disease severity44. Studies that incorporate longitudinal imaging assessment of patients receiving PRP injections could provide additional information about the variable responses to PRP treatment for knee OA. Additionally, while we believe that we have used a relevant and sensitive bioassay for the in vitro evaluation of PRP activity, we are aware that a coculture system with macrophages and synovial fibroblasts does not fully capture the complexity of the joint milieu, that short-term stimulation with cytokines does not capture the more chronic, long-term impact of inflammation in OA, and that in addition to transcriptomics, proteomics analyses should be used to evaluate the functional impact of PRP treatment.
Our results provide novel and relevant insight into the actions of PRP in modulating joint inflammation at a transcriptomics level and establish an integrative workflow that may help us comprehensively evaluate differences in PRP bioactivity associated with clinical outcomes, and therefore identify good candidates for PRP treatment. Larger studies that integrate clinical data with multi-omics analyses are needed to further establish the efficacy of PRP for the targeted treatment of knee OA and other orthopedic conditions, and to identify specific characteristics of PRP that are predictive of optimal response to treatment.
Data availability
The data that supports the findings of this study are available from the corresponding author upon reasonable request. The RNA-seq sequencing data have been deposited at the GEO database with accession code GSE157364.
References
Conaghan, P. G., Cook, A. D., Hamilton, J. A. & Tak, P. P. Therapeutic options for targeting inflammatory osteoarthritis pain. Nat. Rev. Rheumatol. 15, 355–363. https://doi.org/10.1038/s41584-019-0221-y (2019).
Goldring, M. B. & Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23, 471–478. https://doi.org/10.1097/BOR.0b013e328349c2b1 (2011).
Dehghani, B. & Rodeo, S. Cell therapy-a basic science primer for the sports medicine clinician. Curr. Rev. Musculoskelet. Med. 12, 436–445. https://doi.org/10.1007/s12178-019-09578-y (2019).
Andia, I. & Maffulli, N. Platelet-rich plasma for managing pain and inflammation in osteoarthritis. Nat. Rev. Rheumatol. 9, 721–730. https://doi.org/10.1038/nrrheum.2013.141 (2013).
Lopez-Vidriero, E., Goulding, K. A., Simon, D. A., Sanchez, M. & Johnson, D. H. The use of platelet-rich plasma in arthroscopy and sports medicine: optimizing the healing environment. Arthroscopy 26, 269–278. https://doi.org/10.1016/j.arthro.2009.11.015 (2010).
Xu, Z. et al. Comparative evaluation of leukocyte- and platelet-rich plasma and pure platelet-rich plasma for cartilage regeneration. Sci. Rep. 7, 43301. https://doi.org/10.1038/srep43301 (2017).
Barlian, A., Judawisastra, H., Ridwan, A., Wahyuni, A. R. & Lingga, M. E. Chondrogenic differentiation of Wharton’s Jelly mesenchymal stem cells on silk spidroin-fibroin mix scaffold supplemented with L-ascorbic acid and platelet rich plasma. Sci. Rep. 10, 19449. https://doi.org/10.1038/s41598-020-76466-8 (2020).
Roffi, A., Di Matteo, B., Krishnakumar, G. S., Kon, E. & Filardo, G. Platelet-rich plasma for the treatment of bone defects: from pre-clinical rational to evidence in the clinical practice. A systematic review. Int. Orthop. 41, 221–237. https://doi.org/10.1007/s00264-016-3342-9 (2017).
Yin, N. et al. Platelet-rich plasma enhances the repair capacity of muscle-derived mesenchymal stem cells to large humeral bone defect in rabbits. Sci. Rep. 10, 6771. https://doi.org/10.1038/s41598-020-63496-5 (2020).
Filardo, G., Di Matteo, B., Kon, E., Merli, G. & Marcacci, M. Platelet-rich plasma in tendon-related disorders: results and indications. Knee Surg. Sports Traumatol. Arthrosc. 26, 1984–1999. https://doi.org/10.1007/s00167-016-4261-4 (2018).
Zhang, J. et al. Selectively activated PRP exerts differential effects on tendon stem/progenitor cells and tendon healing. J. Tissue Eng. 10, 2041731418820034. https://doi.org/10.1177/2041731418820034 (2019).
Koch, M. et al. Partial anterior cruciate ligament ruptures: advantages by intraligament autologous conditioned plasma injection and healing response technique-midterm outcome evaluation. Biomed. Res. Int. 2018, 3204869. https://doi.org/10.1155/2018/3204869 (2018).
Krismer, A. M. et al. Biologic response of human anterior cruciate ligamentocytes on collagen-patches to platelet-rich plasma formulations with and without leucocytes. J. Orthop. Res. 35, 2733–2739. https://doi.org/10.1002/jor.23599 (2017).
Gato-Calvo, L., Magalhaes, J., Ruiz-Romero, C., Blanco, F. J. & Burguera, E. F. Platelet-rich plasma in osteoarthritis treatment: review of current evidence. Ther. Adv. Chronic Dis. 10, 2040622319825567. https://doi.org/10.1177/2040622319825567 (2019).
Chu, C. R. et al. Optimizing clinical use of biologics in orthopaedic surgery: consensus recommendations from the 2018 AAOS/NIH U-13 conference. J. Am. Acad. Orthop. Surg. 27, e50–e63. https://doi.org/10.5435/JAAOS-D-18-00305 (2019).
Donlin, L. T., Jayatilleke, A., Giannopoulou, E. G., Kalliolias, G. D. & Ivashkiv, L. B. Modulation of TNF-induced macrophage polarization by synovial fibroblasts. J. Immunol. 193, 2373–2383. https://doi.org/10.4049/jimmunol.1400486 (2014).
Griffin, T. M. & Scanzello, C. R. Innate inflammation and synovial macrophages in osteoarthritis pathophysiology. Clin. Exp. Rheumatol. 37(Suppl 120), 57–63 (2019).
Kragstrup, T. W. et al. Fibroblast-like synovial cell production of extra domain A fibronectin associates with inflammation in osteoarthritis. BMC Rheumatol. 3, 46. https://doi.org/10.1186/s41927-019-0093-4 (2019).
Farrar, J. T., Young, J. P. Jr., LaMoreaux, L., Werth, J. L. & Poole, R. M. Clinical importance of changes in chronic pain intensity measured on an 11-point numerical pain rating scale. Pain 94, 149–158. https://doi.org/10.1016/s0304-3959(01)00349-9 (2001).
Lyman, S. et al. Validation of the KOOS, JR: a short-form knee arthroplasty outcomes survey. Clin. Orthop. Relat. Res. 474, 1461–1471. https://doi.org/10.1007/s11999-016-4719-1 (2016).
Hung, M., Bounsanga, J., Voss, M. W. & Saltzman, C. L. Establishing minimum clinically important difference values for the patient-reported outcomes measurement information system physical function, hip disability and osteoarthritis outcome score for joint reconstruction, and knee injury and osteoarthritis outcome score for joint reconstruction in orthopaedics. World J. Orthop. 9, 41–49. https://doi.org/10.5312/wjo.v9.i3.41 (2018).
Salaffi, F., Stancati, A., Silvestri, C. A., Ciapetti, A. & Grassi, W. Minimal clinically important changes in chronic musculoskeletal pain intensity measured on a numerical rating scale. Eur. J. Pain 8, 283–291. https://doi.org/10.1016/j.ejpain.2003.09.004 (2004).
Hohmann, E., Tetsworth, K. & Glatt, V. Is platelet-rich plasma effective for the treatment of knee osteoarthritis? A systematic review and meta-analysis of level 1 and 2 randomized controlled trials. Eur. J. Orthop. Surg. Traumatol. 30, 955–967. https://doi.org/10.1007/s00590-020-02623-4 (2020).
Murray, I. R., Geeslin, A. G., Goudie, E. B., Petrigliano, F. A. & LaPrade, R. F. Minimum information for studies evaluating biologics in orthopaedics (MIBO): platelet-rich plasma and mesenchymal stem cells. J. Bone Joint Surg. Am. 99, 809–819. https://doi.org/10.2106/JBJS.16.00793 (2017).
Cavallo, C. et al. Platelet-rich plasma: the choice of activation method affects the release of bioactive molecules. Biomed. Res. Int. 2016, 6591717. https://doi.org/10.1155/2016/6591717 (2016).
Lee, A. et al. Tumor necrosis factor alpha induces sustained signaling and a prolonged and unremitting inflammatory response in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 65, 928–938. https://doi.org/10.1002/art.37853 (2013).
Yarilina, A., Park-Min, K. H., Antoniv, T., Hu, X. & Ivashkiv, L. B. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 9, 378–387. https://doi.org/10.1038/ni1576 (2008).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. https://doi.org/10.1093/bioinformatics/bty560 (2018).
Yaari, G., Bolen, C. R., Thakar, J. & Kleinstein, S. H. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucl. Acids Res. 41, e170. https://doi.org/10.1093/nar/gkt660 (2013).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. https://doi.org/10.1016/j.cels.2015.12.004 (2015).
Potter, H. G., Linklater, J. M., Allen, A. A., Hannafin, J. A. & Haas, S. B. Magnetic resonance imaging of articular cartilage in the knee. An evaluation with use of fast-spin-echo imaging. J. Bone Joint Surg. Am. 80, 1276–1284. https://doi.org/10.2106/00004623-199809000-00005 (1998).
Peterfy, C. G. et al. Whole-organ magnetic resonance imaging score (WORMS) of the knee in osteoarthritis. Osteoarthritis Cartilage 12, 177–190. https://doi.org/10.1016/j.joca.2003.11.003 (2004).
Singh, P., Lessard, S. G., Mukherjee, P., Rourke, B. & Otero, M. Changes in DNA methylation accompany changes in gene expression during chondrocyte hypertrophic differentiation in vitro. Ann. N. Y. Acad. Sci. https://doi.org/10.1111/nyas.14494 (2020).
Bendinelli, P. et al. Molecular basis of anti-inflammatory action of platelet-rich plasma on human chondrocytes: mechanisms of NF-kappaB inhibition via HGF. J. Cell Physiol. 225, 757–766. https://doi.org/10.1002/jcp.22274 (2010).
Hudgens, J. L. et al. Platelet-rich plasma activates proinflammatory signaling pathways and induces oxidative stress in tendon fibroblasts. Am. J. Sports Med. 44, 1931–1940. https://doi.org/10.1177/0363546516637176 (2016).
Yin, W. et al. Comparative evaluation of the effects of plateletrich plasma formulations on extracellular matrix formation and the NFkappaB signaling pathway in human articular chondrocytes. Mol. Med. Rep. 15, 2940–2948. https://doi.org/10.3892/mmr.2017.6365 (2017).
Moriuchi, H., Moriuchi, M. & Fauci, A. S. Nuclear factor-kappa B potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J. Immunol. 158, 3483–3491 (1997).
Pahl, H. L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18, 6853–6866. https://doi.org/10.1038/sj.onc.1203239 (1999).
Boswell, S. G., Cole, B. J., Sundman, E. A., Karas, V. & Fortier, L. A. Platelet-rich plasma: a milieu of bioactive factors. Arthroscopy 28, 429–439. https://doi.org/10.1016/j.arthro.2011.10.018 (2012).
Riboh, J. C., Saltzman, B. M., Yanke, A. B., Fortier, L. & Cole, B. J. Effect of leukocyte concentration on the efficacy of platelet-rich plasma in the treatment of knee osteoarthritis. Am. J. Sports Med. 44, 792–800. https://doi.org/10.1177/0363546515580787 (2016).
Smith, P. A. Intra-articular autologous conditioned plasma injections provide safe and efficacious treatment for knee osteoarthritis: an FDA-sanctioned, randomized, double-blind, placebo-controlled clinical trial. Am. J. Sports Med. 44, 884–891. https://doi.org/10.1177/0363546515624678 (2016).
Patel, S., Dhillon, M. S., Aggarwal, S., Marwaha, N. & Jain, A. Treatment with platelet-rich plasma is more effective than placebo for knee osteoarthritis: a prospective, double-blind, randomized trial. Am. J. Sports Med. 41, 356–364. https://doi.org/10.1177/0363546512471299 (2013).
Simental-Mendia, M. et al. Leukocyte-poor platelet-rich plasma is more effective than the conventional therapy with acetaminophen for the treatment of early knee osteoarthritis. Arch. Orthop. Trauma Surg. 136, 1723–1732. https://doi.org/10.1007/s00402-016-2545-2 (2016).
Burchard, R. et al. Efficiency of platelet-rich plasma therapy in knee osteoarthritis does not depend on level of cartilage damage. J. Orthop. Surg. Res. 14, 153. https://doi.org/10.1186/s13018-019-1203-0 (2019).
Mazzocca, A. D. et al. Platelet-rich plasma differs according to preparation method and human variability. J. Bone Joint Surg. Am. 94, 308–316. https://doi.org/10.2106/JBJS.K.00430 (2012).
Evanson, J. R. et al. Gender and age differences in growth factor concentrations from platelet-rich plasma in adults. Mil. Med. 179, 799–805. https://doi.org/10.7205/MILMED-D-13-00336 (2014).
Xiong, G. et al. Men and women differ in the biochemical composition of platelet-rich plasma. Am. J. Sports Med. 46, 409–419. https://doi.org/10.1177/0363546517740845 (2018).
Barata, J. T., Durum, S. K. & Seddon, B. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 20, 1584–1593. https://doi.org/10.1038/s41590-019-0479-x (2019).
Kim, S. J. et al. Macrophages are the primary effector cells in IL-7-induced arthritis. Cell Mol. Immunol. https://doi.org/10.1038/s41423-019-0235-z (2019).
Pickens, S. R. et al. Characterization of interleukin-7 and interleukin-7 receptor in the pathogenesis of rheumatoid arthritis. Arthritis Rheum. 63, 2884–2893. https://doi.org/10.1002/art.30493 (2011).
Heitzer, E. et al. IL-7, IL-18, MCP-1, MIP1-beta, and OPG as biomarkers for pain treatment response in patients with cancer. Pain Phys. 15, 499–510 (2012).
Singh, J. A., Noorbaloochi, S. & Knutson, K. L. Cytokine and neuropeptide levels are associated with pain relief in patients with chronically painful total knee arthroplasty: a pilot study. BMC Musculoskelet. Disord. 18, 17. https://doi.org/10.1186/s12891-016-1375-2 (2017).
Abrams, G. D., Frank, R. M., Fortier, L. A. & Cole, B. J. Platelet-rich plasma for articular cartilage repair. Sports Med. Arthrosc. Rev. 21, 213–219. https://doi.org/10.1097/JSA.0b013e3182999740 (2013).
Ruan, S., Deng, J., Yan, L. & Huang, W. Evaluation of the effects of the combination of BMP-2-modified BMSCs and PRP on cartilage defects. Exp. Ther. Med. 16, 4569–4577. https://doi.org/10.3892/etm.2018.6776 (2018).
Linke, B. et al. Activated platelets induce an anti-inflammatory response of monocytes/macrophages through cross-regulation of PGE2 and cytokines. Mediators Inflamm. 2017, 1463216. https://doi.org/10.1155/2017/1463216 (2017).
van Buul, G. M. et al. Platelet-rich plasma releasate inhibits inflammatory processes in osteoarthritic chondrocytes. Am. J. Sports Med. 39, 2362–2370. https://doi.org/10.1177/0363546511419278 (2011).
Barrat, F. J., Crow, M. K. & Ivashkiv, L. B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20, 1574–1583. https://doi.org/10.1038/s41590-019-0466-2 (2019).
Ahmed, A. S. et al. Activation of NF-kappaB in synovium versus cartilage from patients with advanced knee osteoarthritis: a potential contributor to inflammatory aspects of disease progression. J. Immunol. 201, 1918–1927. https://doi.org/10.4049/jimmunol.1800486 (2018).
Marcu, K. B., Otero, M., Olivotto, E., Borzi, R. M. & Goldring, M. B. NF-kappaB signaling: multiple angles to target OA. Curr. Drug Targets 11, 599–613. https://doi.org/10.2174/138945010791011938 (2010).
Hartung, J. E. et al. Nuclear factor-kappa B regulates pain and COMT expression in a rodent model of inflammation. Brain Behav. Immun. 50, 196–202. https://doi.org/10.1016/j.bbi.2015.07.014 (2015).
Acknowledgements
The authors are grateful to Dr. Lisa Fortier for her advice during the preparation of this study. The authors are also grateful to the Tow Foundation, which provided support for the David Z. Rosensweig Genomics Research Center, Giammaria Giuliani, the Derfner Foundation, the Ira W. DeCamp Foundation, and the Ambrose Monell Foundation. Technical support for the RNA-seq experiments was provided by the Epigenomics Core of Weill Cornell Medicine. Technical support to determine the complete blood count with differential was provided by the Department of Pathology and Laboratory Medicine at HSS.
Funding
This work was supported by the Feldstein Medical Foundation (M.O.) and the National Center for Advancing Translational Sciences of the National Institutes of Health under Award No. UL1TR002384 (M.O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. B.D. received support from the AOA Theta Chapter at Albany Medical College.
Author information
Authors and Affiliations
Contributions
L.D., L.I., S.R. and M.O. contributed to the conception and design of the study. H.Z., B.D., X.Y., Y.C., C.K., A.B., R.B., D.N., P.F., P.M., H.P., J.N., B.H., S.R. and M.O. performed the acquisition, analysis, and interpretation of data. H.Z., B.D., X.Y., Y.C., S.R. and M.O. drafted the article. H.Z., B.D., X.Y., Y.C., C.K., A.B., R.B., D.N., P.F., P.M., H.P., J.N., B.H., L.D., L.I., S.R., M.O. reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zahir, H., Dehghani, B., Yuan, X. et al. In vitro responses to platelet-rich-plasma are associated with variable clinical outcomes in patients with knee osteoarthritis. Sci Rep 11, 11493 (2021). https://doi.org/10.1038/s41598-021-90174-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-90174-x
This article is cited by
-
Is cell therapy no better than steroid injection?
Nature Reviews Rheumatology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.