Abstract
Pigeon pea, a grain legume of the semiarid tropics, is a rich source of high-quality protein. The productivity of this pulse is seriously affected by lepidopteron insect pests. To generate a sustainable insect-resistant plant, synthetically prepared bioactive key constituents of a crystal protein (Syn Cry1Ab) of Bacillus thuringiensis were expressed in pigeon pea under the guidance of a tissue-specific promoter of the RuBP carboxylase/oxygenase small subunit (rbcS) gene. Regenerated transgenic plants with the cry1Ab expression cassette (cry1Ab-lox-bar-lox) showed the optimum insect motility rate (90%) in an in vitro insect bioassay with second instar larvae, signifying the insecticidal potency of Syn Cry1Ab. In parallel, another plant line was also generated with a chimaeric vector harbouring a cre recombinase gene under the control of the CaMV 2 × 35S promoter. Crossing between T1 plants with a single insertion of cry1Ab-lox-bar-lox T-DNA and T1 plants with moderate expression of a cre gene with a linked hygromycin resistance (hptII) gene was performed to exclude the bialaphos resistance (bar) marker gene. Excision of the bar gene was achieved in T1F1 hybrids, with up to 35.71% recombination frequency. Insect-resistant pigeon pea plants devoid of selectable marker genes (syn Cry1Ab- bar and cre-hptII) were established in a consecutive generation (T1F2) through genetic segregation.
Similar content being viewed by others
Introduction
Pigeon pea is one of the most popular protein-rich grain legumes consumed as a pulse in the semiarid tropics (SAT) region1. Compared to other pigeon pea-producing countries, India contributes 90% to global production. This crop has social, economic and medicinal importance in developing countries2. It can grow in a variety of environmental conditions throughout the year due to its adaptive nature. This adaptive quality minimizes the cultivation cost, which ultimately increases the profit of marginal farmers3. The seed of this plant contains approximately 20–22% protein, mostly sulfur-containing amino acids, viz., cysteine and methionine, which is almost three times more than that of cereals4. Moreover, seeds also contain carbohydrates (57.3–58.7%), crude fibres (1.2–8.1%), and lipids (0.6–3.8%)5. In the Caribbean region, pigeon pea is consumed as a green vegetable due to its balanced nutritional quality6,7. Despite its high demand, the productivity of pigeon pea has increased only 1% during the past several years. This caused a serious scarcity in the per capita availability of this pulse, mainly in India8. The major factors for this low production are biotic and abiotic stresses and a lack of cultivation management practices. One of the most serious biotic stresses associated with pigeon pea cultivation is the infestation of Helicoverpa armigera, a lepidopteran pest9,10. Its larvae attack green parts such as the leaves, flowers, and pods of growing plants, resulting in considerable damage of approximately 40–50% and annual yield losses equivalent to 400 million US$ worldwide11. Farmers frequently use different kinds of chemical pesticides to fight this pest. Although this method has immense potential to control pests even in adverse conditions, it contaminates food and food products with insecticide residues12 and allows for the emergence of secondary pest problems. In addition, the unaccountable use of these chemicals is also responsible for the development of insecticide-resistant pests, which makes it more complex to address this problem only through chemical control. Elite pigeon pea cultivars with insect resistance could be produced through conventional breeding methods to overcome this pest problem. Unfortunately, this method has not been successful due to the incompatibility with wild species and the presence of limited genetic variation in the cultivated germplasm6. Recently, the plant genetic engineering approach has shown immense potential to overcome this type of difficulty. Genes conferring resistance to insect pests have been introduced successfully in a wide array of crop plants13. Hence, the development of insect-resistant cultivars through transgenic approaches has become an effective alternative to the integrated pest management program14,15. Efficient regeneration and transformation systems, a prerequisite to introducing any novel trait for successful development of transgenic plants, have been achieved in pigeon pea4,16,17,18,19. Some success in the development of pod borer-resistant pigeon pea/chickpea has already been demonstrated by using synthetic cry1E-C, cry1Ab, cry1Ac, and chimaeric cry1AcF as insecticidal genes11,20,21; however, there is inadequate evidence validating transgenic pigeon pea events in terms of the stability of protein expression and the rate of insect mortality22. Despite these preliminary successes, more positive events with adequate expression of the toxin gene resulting in a more significant impact on pod borer-resistant plants under field conditions are needed. The marker gene, which is required for screening transformed plants under in vitro conditions, raises public and regulatory concerns23; therefore, removal of the marker gene should also be taken into account to prevent gene flow and introgression. One of the straightforward ways to eliminate the possibilities of marker gene transfer is the excision of the marker gene after the development and selection of transgenic plants using a site-specific recombinase system (e.g., cre-lox system). This could be possible in a hybrid plant resulting from a cross between two lines. Establishment of an efficient crossing procedure is essential to create successful hybrid plants though crossing; however, it is tedious and requires specialised skills along with the knowledge of various stages of flower development 24,25,26.
Considering all the previous reports and requirements, an effort has been made to develop a marker gene-free transgenic pigeon pea plant expressing the cry1Ab gene for resistance against the legume pod borer H. armigera.
Results
Development of a green tissue-specific, insect resistance syn cry1Ab and cre line in pigeon pea
Generation of the cry1Ab line
To achieve sustainable resistance against the lepidopteran insect pest H. armigera, the synthetically prepared bioactive core element of the cry1Ab gene of B. thuringiensis was expressed in pigeon pea. The toxin protein was expressed in a tissue-specific manner and showed insecticidal potency against H. armigera under laboratory conditions. The results are elaborated in the following section.
Generation of pLBRCAb transgenic pigeon pea lines harbouring the syn cry1Ab transgene
The cotyledonary node of the Asha variety was cocultivated with A. tumefaciens EHA105 harbouring the pLBRCAb vector, with three experimental attempts. Agrobacterium-mediated transformation and regeneration were performed following the established protocol reported by Sarkar et al17. Transformed regenerated plants were selected using medium containing bialaphos (4 mg/L) as a selection agent. Under selection pressure, ~ 16% of the regenerated plants showed resistance to bialaphos and further developed into healthy plants after 50 days of recovery. Surviving plants were rooted under selection pressure.
Transformation efficiency of T0 transformants
Drug-resistant T0 plants were further analysed by dot blot to determine the transformation efficiency of the transformed genes. For this, three sets of experiments were performed, wherein a total of 27 plants (T0) showed resistance against the selection medium out of 300 infected explants. Among these 27 plants, 10 showed positive results for the dot blot analysis, indicating 3.33% transformation efficiency (Table 1).
Progeny analysis of transgenic pigeon pea lines harbouring syn cry1Ab
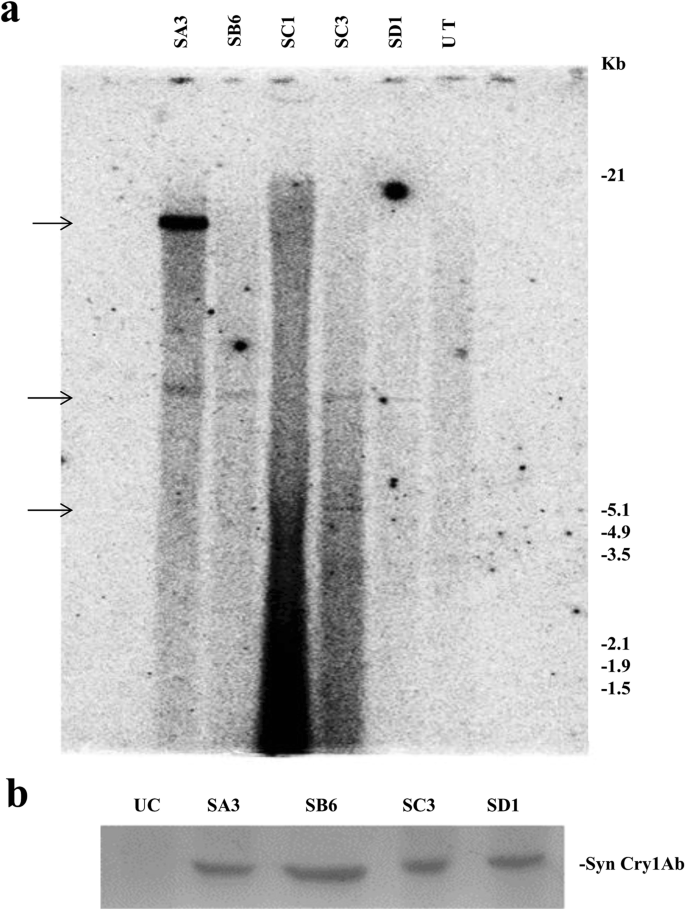
Out of several putative transformants generated using pLBRCAb, only 7 healthy plants were selected for further analysis after stringent screening. Six individual progenies of each of the 7 T0 plant lines (total 42 plants) were randomly selected to carry out dot blot hybridization. Among these plants, 5 (SA3, SB6, SC1, SC3, and SD1) showed positive results (Fig. 1a). Densitometry analysis of dot blots was also carried out by ImageJ software (Fig. 1b) for comparison with the positive and negative controls. SD1 and SC1 plants showed the highest and lowest intensity, respectively. Furthermore, the presence of the cry1Ab gene in the T1 generation was confirmed by Southern blot analysis (Fig. 2a). Of these 5 individual transformants selected through dot blot analysis, 4 plants contained cry1Ab based on Southern blot analysis, among which 2 showed a single integration and 2 showed a double integration of the cry1Ab gene, while the untransformed control plant did not show any band. This experiment provided evidence of stable integration of the transgene and its transfer to consecutive generations.
(a) Dot blot hybridization of the cry1Ab gene in the T1 generation: Six randomly selected T1 plants from 7 individual events (T0 plants) were analysed. A replica blot of the genomic DNA of 42 individual plants was prepared and hybridized with the 800 bp PCR amplified cry1Ab gene as the probe. In this replica blot, SA3, SB6, SC1, SC3, and SD1 showed a positive result, where –C is the untransformed control and + C is the positive control. (b) Graphical representation of the densitometry analysis of dot blots with two replicas by ImageJ Software (Version: 1.50, https://imagej.nih.gov/ij/download.html).
(a) Southern blot hybridization of the cry1Ab gene in the T1 generation: Genomic DNA of dot blot-positive T1 plants was digested with HindIII. The blot was hybridized with the 800 bp PCR-amplified cry1Ab gene (α-32[P]dCTP labelled)) as the probe. Lanes labelled SB6 and SD1 showed a single integration, SA3 and SC3 show a double integration, and UT shows the untransformed control. Approximate molecular weight markers are indicated. (b) Western blot analysis of the Syn Cry1Ab toxin protein in T1 progeny plants with the anti-Cry1Ab antibody: Lane UC: Untransformed control. Lanes SA3, SB6, SC3, and SD1: Transgenic lines.
Expression analysis of the syn cry1Ab gene in T1 transgenic lines
All four Southern-positive transgenic pigeon pea lines of syn cry1Ab (SA3, SB6, SC3, and SD1) were subjected to expression analysis. Expression of the syn cry1Ab gene in four transgenic lines was analysed using an anti-Cry1Ab antibody by western blot (Fig. 2b); distinct bands were observed in all plants. No Cry1Ab protein band was observed in the untransformed control line.
Entomocidal activity of Syn Cry1Ab toxin present in the T1 transgenic pigeon pea
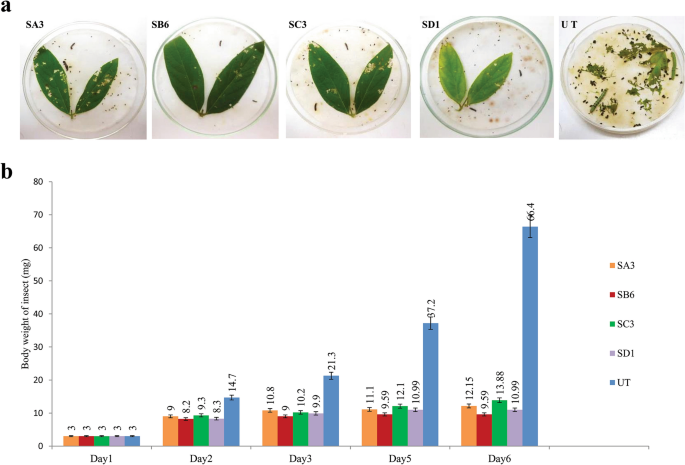
Transgenic plants were tested for insecticidal activity. Fully opened young, fresh leaves of four T1 transgenic pigeon pea lines were tested through an in vitro insect feeding bioassay to confirm the insecticidal activity of Syn Cry1Ab toxin against second instar larvae of H. armigera. The young leaves of the untransformed plants were used as a negative control. The transgenic plants showed very little damage after feeding by insects, while massive damage was observed with the untransformed control line (Fig. 3a). Extensive feeding of leaf tissue (> 90%) by the larvae was observed for untransformed control plants, and larvae were healthy, active, and showed regular developmental cycles. Larvae fed on transgenic plants SA3, SB6, SC3, and SD1 showed a 70–90% mortality rate with a 79.1–85.5% bodyweight reduction (Table 2). The gradual decreases in larval body weight after 6 days of incubation in the insect feeding bioassay are graphically presented in Fig. 3b.
In vitro insect (H. armigera) feeding assay: The T1 transgenic plants with 5 replica experiments were carried out with two insects for each individual plant. (a) SA3, SB6, SC3, and SD1 are transgenic plants that showed a positive result, and UT is an untransformed negative control plant. (b) Graphical presentation of insect weight loss during the feeding assay in comparison with the untransformed control.
Segregation analyses of the bar gene in T1 progeny
The segregation pattern of the bar transgene through seeds in the T1 progeny was monitored by germination on the selection medium. Randomly selected seeds of each of the four T0 transformants were screened for germination in 4 mg/l bialaphos-containing medium alongside seeds of untransformed plants. It was observed that some of the T1 seeds did not grow after germination and gradually turned brown in the presence of bialaphos. Most of the T1 seedlings could grow in the presence of bialaphos. The ratio of bialaphos resistant:sensitive seedlings was scored after 4 weeks, and it was observed that the ratio was close to the expected ratio (3:1) in each case, indicating a Mendelian mode of inheritance for the monohybrid cross (Table 3). The plants that survived on the screening medium confirmed the stable inheritance of the bar transgene, whereas the untransformed control plants ceased to germinate and gradually turned brown even after 10 days on the same medium. The germinated plantlets were transferred to pots in the greenhouse for growth and further analysis. Since the bar gene is linked to the cry1Ab gene, the inheritance pattern of the cry1Ab gene is expected to be the same as that of the bar gene.
Generation of cre lines
To eliminate the selectable marker (bar) gene from syn cry1Ab transgenic pigeon pea lines through Cre/lox-mediated recombination, cre pigeon pea lines were generated. To catalyse recombination, crossings were performed between the cre plant line and syn cry1Ab plant line, which carries lox-bar-lox. Finally, superfluous cre and hptII genes were excluded through consecutive genetic segregation. The results are elaborated in the following sections.
Generation of transgenic pigeon pea lines harbouring the cre recombinase gene
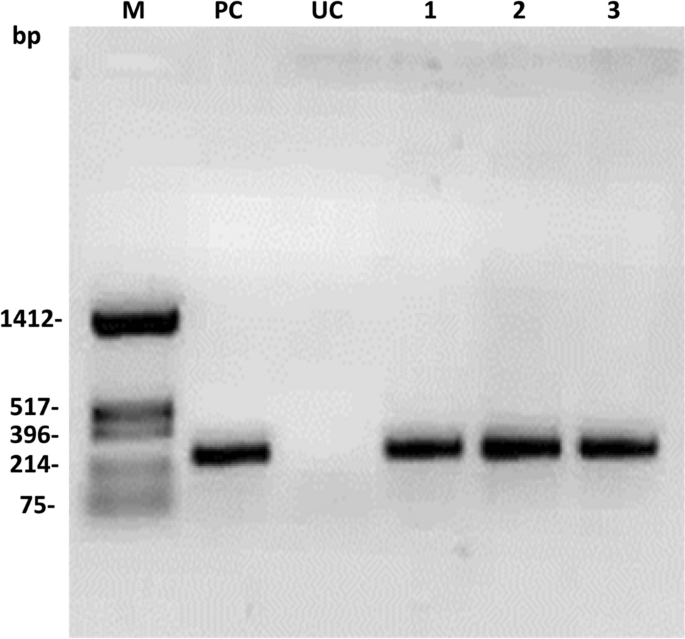
The pigeon pea transformants with pHC (H: hptII and C: cre) were generated following the Agrobacterium-mediated pigeon pea transformation protocol in Asha varieties. A total of 270 explants were cocultivated with Agrobacterium EHA105 containing pHC. Only 27 regenerated plants were retrieved after stringent hygromycin selection (50 mg/L) (Table 4). Among them, 13 plantlets showed a cre gene-specific PCR amplification product (275 bp) with cre gene-specific primers (CRFP2/CRRP2) (Supplementary Table S1). Representative PCR amplification of the cre gene is shown in Fig. 4.
Seed transmission of the cre gene through T1 generation and progeny analysis
A containment condition was created to prevent cross-pollination and allow T0 transgenic pigeon pea plants to self-pollinate, and T1 seeds were harvested. T1 seeds from representative transgenic plants were initially screened by growing on antibiotic (50 mg/L hygromycin)-supplemented media. The sensitive seedlings ceased to grow and gradually died, whereas resistant seedlings showed healthy growth. The data regarding transgene (hptII) segregation in T1 progenies were evaluated by the χ2 test. The results showed that the hptII gene segregated closely in a Mendelian fashion with a 3:1 segregation ratio (Table 5).
Southern blot analysis for the confirmation of the presence of the cre gene in the T1 line
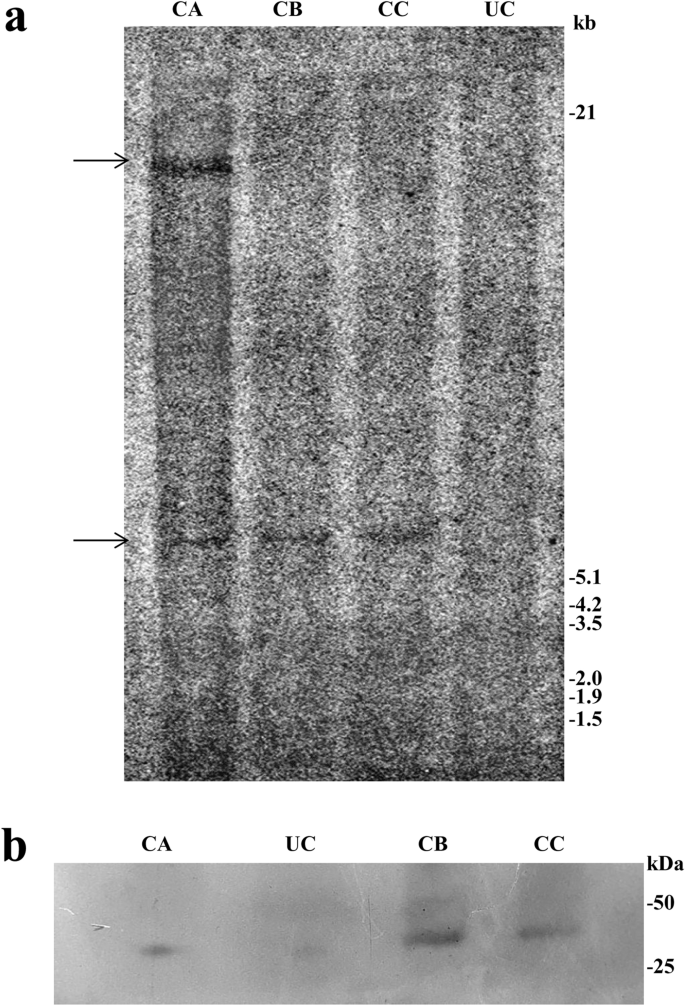
The integration of the cre recombinase gene into the pigeon pea transformants was further confirmed by Southern blot analysis. Randomly chosen PCR-positive seeds of transformants grown in the presence of hygromycin selection were selected for Southern blot analysis. The genomic DNA of T1 plants was digested with HindIII restriction endonuclease, and hybridization was performed with an α-[32P] dCTP radiolabelled probe of the cre gene (1202 bp). The developed blot of three transformed lines (CA, CB, and CC) showed transgene integration (Fig. 5a). Among these three plants, the presence of a single insertion of the cre gene was observed in two plants, viz., CB and CC, while double integrations were observed in one plant, viz., CA. No band was detected for the untransformed control (UC) plant.
(a) Southern blot analysis of PCR-positive plants in T1 using the cre gene as the probe. Genomic DNA was digested with HindIII. Lane UC: Genomic DNA from the untransformed control line, and lanes CA, CB and CC: genomic DNA of transformed plants. Approximate molecular weight markers are indicated. (b) Western blot analysis of T1 transgenic lines with the polyclonal Cre recombinase antibody raised in rabbit for the detection of cre recombinase gene expression. Lane UC: Untransformed control. Lanes CA, CB, and CC: Transgenic plant lines.
Western blot analysis of the T1 line to determine the presence of Cre recombinase
The expression of the cre gene was analysed by western blot analysis. Total leaf protein was isolated from these three Southern blot-positive plants (30 days old). The total soluble protein of transgenic plants and untransformed controlled plants was loaded and separated on a 12% SDS-PAGE gel. Upon western blot analysis using the anti-Cre antibody (Product no. ab41104, Abcam, UK), distinct bands were observed in all transgenic pigeon pea plants (Fig. 5b). The three individual plants showed specific expression of the 38 kDa Cre protein. No band was detected in the case of the untransformed control (UC) plant.
Elimination of selectable marker genes from syn cry1Ab pigeon pea lines through Cre/lox-mediated recombination and consecutive genetic segregation
Crosses between T1 syn cry1Ab and cre lines
To initiate the crossing between T1 syn cry1Ab and cre lines for the production of marker-free insect-resistant pigeon pea, we first identified the appropriate time of crossing. The outcomes of the findings are discussed in the following sections.
Identification of the appropriate timing for crossing in pigeon pea
Flowers at different stages were collected to observe the morphological development of the stigma and anther in the process of flower development. A flower fully covered with calyx was designated stage zero, whereas blooming of the flower was divided into four stages up to fully bloomed, depending upon the morphological appearance (Supplementary Fig. S3). The anther and stigma of the respective stages of flower development were separated and observed under a microscope. The stagewise development of anthers and stigmas and their functions are represented in Supplementary Fig. S4. In the present observation, it was found that stigma at stage 3 (Supplementary Fig. S4, 3a) had the highest pollen receptivity. In stage 2 (Supplementary Fig. S4, 2b), it was observed that the anthers started to release pollen, and no pollen was attached to the stigma (Supplementary S4, 2a) until that time. Stage 2 was found to be an appropriate stage for the emasculation of the anther, as it begins to open for discharging pollen. Stage 3 (1–3 PM) is also suitable for pollination in the emasculated flower; in this stage, stigmas show the highest pollen receptivity and become sexually active for fertilization. After that, the receptivity of the stigma decreases continuously over time.
The crossing of parental pigeon pea lines and screening of hybrid plants
Two independent T1 progeny plants (SB6 and SD1) harbouring pLBRCAb [considered the female parent (♀)] were crossed with a T1 progeny plant (CC) harbouring pHC [considered the male parent (♂)] with moderate cre gene expression. The hybrid progenies obtained from the cross between T1 pHC and the pLBRCAb line were named T1F1. The consecutive generation of the hybrid line was designated T1F2. All the seeds of two independent crossing experiments were harvested, air-dried, and stored. The hybrid seeds were surface sterilized and germinated on MS medium in the presence of 50 mg/L hygromycin. Drug-resistant seedlings were transferred to the greenhouse and allowed to grow to maturity for further detection of selectable marker gene (bar) elimination.
PCR-based analysis of marker gene elimination following Cre/lox-mediated recombination
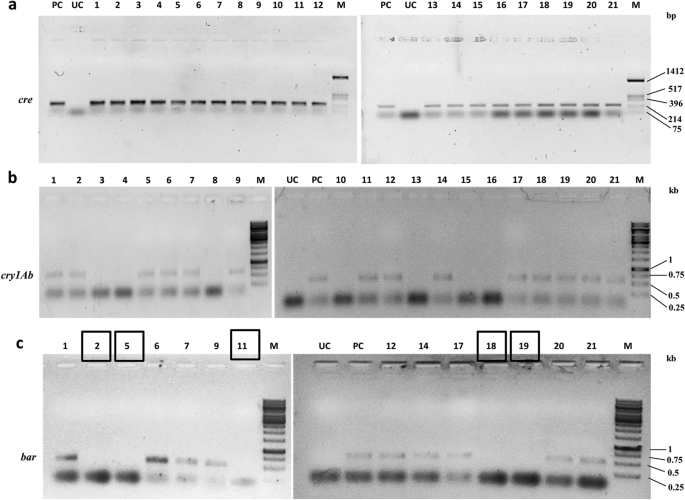
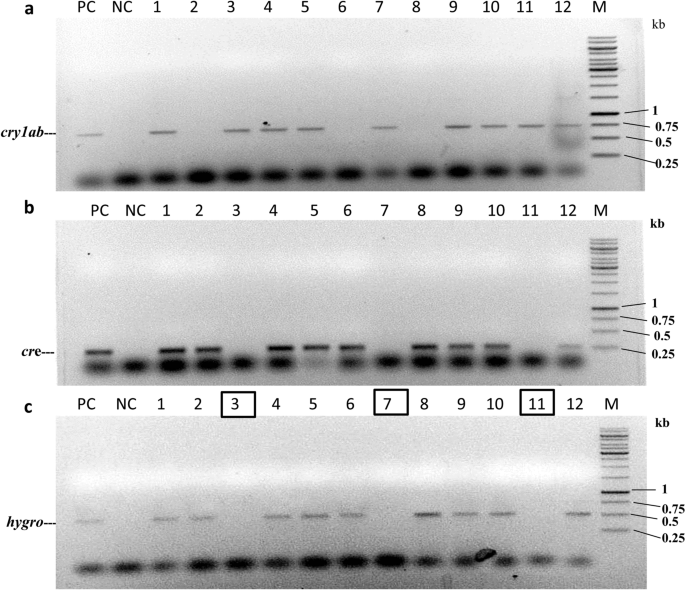
The T1F1 seeds (SB6CC) were collected separately from two independently crossed events (Table 6) and germinated on selection medium containing hygromycin (50 mg/L) to eliminate possible azygous plants for hptII and the linked cre transgene. Forty-two hybrid seeds from the SB6CC line were randomly selected for initial hygromycin screening. Among the hybrid seeds, 21 seeds grew into plantlets under selection pressure (Table 6). These initially screened hybrid plants were further screened (verified) by separate PCRs with cre, syn cry1Ab, and bar gene-specific primer pairs. The representative data from hybrid line SB6CC are shown in Fig. 6a–c. Among these 21 progeny plants, 14 plants were found to be positive for both cre and syn cry1Ab genes; 7 plants were positive for only the cre gene (Fig. 6a,b). As expected, no plant harbouring only the syn cry1Ab cassette was obtained, as these plants did not contain the hptII gene and thus were excluded from the screening process. Marker gene elimination among the 14 syn cry1Ab- and cre-positive hybrid plants of SB6CC was further verified through PCR using bar gene-specific primers (Fig. 6c). This PCR result showed five bar-negative plants, as the bar gene was expected to be eliminated through recombination due to the presence of the cre recombinase gene. Based on this finding, the frequency of Cre/loxP-mediated recombination was calculated to be 35.71% in the case of hybrid line SB6CC. Similarly, the other hybrid line SD1CC showed a 33.33% recombination frequency, with an average recombination frequency for these two lines of 34.52%.
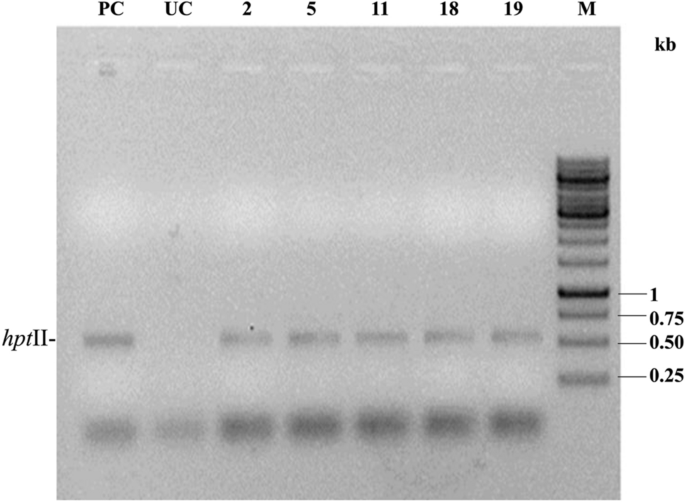
PCR analysis of the 21 T1F1 hybrid line for Cre-mediated bar gene elimination. (a) PCR amplification of hygromycin-selected progenies of line SB6CC with cre-specific primers. Lane PC: PCR from the pHC plasmid as a positive control; lanes 1–21: 21 T1F1 hybrid progenies. The 275 bp amplicon represented the cre recombinase. (b) PCR with syn cry1Ab-specific primers of 21 cre-positive plants of the T1F1 hybrid line SB6CC. Fourteen plants showed amplification of a representative 720 bp region of the syn cry1Ab gene. Lane PC: pLBRCAb as a template. (c) PCR with bar-specific primers of 14 syn cry1Ab- and cre-positive plants of the SB6CC line. Lanes 1–14: T1F1 hybrid plants, where plants no. 2, 5, 11, 18 and 19 showed the absence of bar. Lane PC: PCR from pLBRCAb; the 552 bp amplicon represents the bar gene. Plant lines boxed in black show successful elimination of the bar gene. Lane M in (a), HinfI-digested pUC18 DNA as a molecular weight marker. Lane M in (b) and (c), GeneRuler 1 kb DNA Ladder, Thermo Scientific. Lane UC represents PCR results from the genomic DNA of an untransformed plant. All figures of each independent gel are separated by a white border.
Establishment of completely selectable marker-free T1F2 transgenic pigeon pea plants
As the bar-negative T1F1 plants are cre positive, there is a high possibility that they still bear hptII as a linked gene. PCR-based screening with hptII gene-specific primers supported this fact (Fig. 7). Consequently, some of the T1F1 plants underwent self-fertilization to obtain T1F2 progeny plants and were analysed for complete excision of selectable marker genes through genetic segregation. Twelve T1F2 progenies of the bar-negative line SB6CC (19) were analysed for the presence of syn cry1Ab, cre, and hptII genes. Six plants [T1F2 plant SB6CC (19)- 1, 4, 5, 9, 10,12] contained syn cry1Ab, cre and hptII genes, three plants [SB6CC (19)- 2, 6, 8] showed the presence of cre and hptII, and three plants [SB6CC- 3, 7, 11] were found to contain only syn cry1Ab without the presence of the cre or hptII gene (Fig. 8a,b,c). Thus, the elimination of the selectable marker gene (hptII) and linked transgene (cre) in T1F1 plants was performed by genetic segregation in the consecutive generation (T1F2). Therefore, these three identified pigeon pea plants are now considered completely marker-free transgenic syn cry1Ab lines.
PCR analysis of 12 randomly selected bar-negative SB6CC T1F2 plants to establish complete elimination of the cre-hptII gene. (a) PCR with syn cry1Ab-specific primers, (b) PCR with cre-specific primers, and (c) PCR with hptII-specific primers. PC: positive control (amplification from syn cry1Ab or cre gene containing expression constructs); UC: no amplification from the untransformed control line. MF (Marker Free): represents T1F2 plants with complete elimination of all the selection markers (bar) and redundant transgenes (cre-hptII linked gene) with retention of the toxin gene. M HinfI-digested pUC18 as a molecular weight marker.
Finally, the insecticidal activity of Syn Cry1Ab toxin in T1F2 marker-free transgenic plants of the SB6CC line was analysed by an in vitro insect feeding assay. In this experiment, young transgenic leaves were incubated with second instar larvae and compared with insects fed untransformed control leaves. The transgenic plants showed very little damage, while massive damage was observed for the untransformed control line (Fig. 9). Extensive feeding of leaf tissue (> 90%) by the larvae was observed for the untransformed control plants, and all the larvae were healthy, active, and showed regular developmental cycles. Larvae fed on transgenic plants SB6CC(3), SB6CC(7), and SB6CC(11) showed > 90% mortality rates with drastic body weight reduction. The data were collected after 6 days of incubation in an insect feeding bioassay.
In vitro insect (H. armigera) feeding assay of T1F2 marker-free transgenic plants of the SB6CC line: Transgenic plants with 3 replica experiments were carried out by second instar larvae for each individual plant. Insect feeding assay of SB6CC(3), SB6CC(7) and SB6CC(11) transgenic plants with two insects showed a positive result; UC is an untransformed control plant (leaf), and the white arrow shows live and healthy larvae.
Discussion
Agricultural production loss due to insect pest damage is a severe problem worldwide. Extensive time and effort has already been spent to address this issue. The massive use of chemical pesticides to counter these losses not only poses a significant threat to the environment but also requires significant costs. This expense often cannot be afforded by marginal farmers in developing countries. Accordingly, that focus has shifted towards advances that have already been made in the field of genetic engineering regarding the generation of insect-resistant crops. Even though GM or Bt crops are controversial, biotechnologically produced plants have gained importance over those produced via conventional crop improvement through breeding, mainly for the development of insect resistant crops27. Thus, the comparatively less time-consuming transgenic technology would be the best choice for plant improvement over conventional breeding to address the growing problem of food scarcity. The development of transgenic plants expressing the insecticidal protein gene of B. thuringiensis has been one of the powerful techniques to develop insect-resistant crop plants offering protection against insect damage. To date, more than 30 insect-resistant crop plants have been developed using the Bt toxin gene.
However, in reality, an efficient regeneration and transformation method for the development of stable transgenic plants plays an important role. Pigeon pea is a leguminous plant that is not readily amenable to tissue culture for in vitro plant regeneration. However, by adopting a protocol for multiple shoot development from embryonic meristematic tissue of pigeon pea followed by lateral branching reported by Sarkar et al17, this constraint was suitably resolved.
The present experiment was performed according to our previously reported regeneration and transformation protocol17. The Agrobacterium-mediated transformation of the pLBRCAb cassette resulted in the generation of 27 prospective bialaphos-resistant plants (T0). Ten transformants were shown to be positive in terms of transgene integration with 3.33% transformation efficiency, as revealed by dot blot hybridization (Table 1). Furthermore, progeny analysis was performed on 7 well-grown/surviving transgenic plants (T0). Six drug-selected plantlets (T1) of seven T0 plants were randomly selected and subjected to dot blot analysis (Fig. 1a), which confirmed the integration of candidate genes. Single representative positive plants of each event (T1) were subjected to further Southern blot analysis, which revealed the integration pattern of the cry1Ab gene.
Integration of the cry1Ab transgene into the genome of pigeon pea was found to vary in number and loci (Fig. 2), indicating that all were separate events except SB6 and SD1, which might be the same event or siblings. In our present study, Southern-positive plants in the T1 generation were considered stably integrated transgenic events; western blot experiments revealed that these transgenic plants produced transgenic protein. The survival of T1 seeds in bialaphos-containing media indicated that the transgenes segregated according to the Mendelian 3:1 ratio and were transmitted to the next generation (Table 3). This observation also revealed that a single copy of the transgene could be integrated at a single site or two copies of the transgene could be integrated very close together on a chromosome. The presently described protocol took 122–127 days for the regeneration of putatively transformed pigeon pea.
Finally, in vitro insect feeding bioassays were carried out with larvae from the major lepidopteran insect pest H. armigera, which showed that the Syn Cry1Ab toxin present in the green leaves of transgenic plants offered significant protection against insect-inflicted damage when compared with the control untransformed plants. In this experiment, four transformants revealed that all the transformants could resist damage caused by insects to some degree. This small variation within the four transgenic plants in the protection against insect damage can be attributed to the differences in the expression levels of the syn cry1Ab gene. The transgenic lines SA3, SB6, SC3, and SD1 showed a drastic size reduction of approximately 79.1–85.5%, leading to the death of 70–90% of the insects within 6 days of incubation. Similarly, Ghosh et al9 observed up to 100% mortality of second instar larvae at the end of insect feeding bioassays (6 days after feeding) of T1 transgenic plants carrying the cry1Ac and cry2Aa insecticidal genes separately9. The deviation in the percentage of larval mortality from our findings might be due to the amount of leaves used for the experiment, the Bt gene used, and the time of data collection, per the agreement with previous observations28,29. The present insect bioassay experiment showed a significant reduction in larval weight followed by mortality compared to larvae fed on untransformed control plants, revealing the expression of an adequate amount of toxin protein.
Antibiotic or herbicide-resistant marker genes play a crucial role as plant selection markers in the process of transgenic plant development. However, retaining the selection marker within the genome of transgenic plants poses a significant obstacle to the development of consumer-friendly GM crops. The reason for this obstacle is that the transgene could horizontally transfer from plants to bacteria or from plant products consumed as foods to intestinal microbes, with the potential for emergence of antibiotic resistance in those organisms30. Therefore, the routine use of plant transgenic technology may be efficient in broad-spectrum pest mortality, but the biosafety of such transgenic plants remains a critical environmental concern. Therefore, there is a need to strategically assemble these genes to develop marker-free GM pigeon pea with broader acceptance by the consumer. Extensive research has been done to develop insect-resistant pigeon pea, but until now, no marker-free plant has been generated. Thus, in the current experiment, an attempt was undertaken to develop a marker-free pigeon pea plant using Cre/lox technology with insecticidal activity31,32,33,34. To perform this, we also developed a cre line with a pHC construct and crossed this line with the cry1Ab line. As an outcome of this experiment, we obtained marker-free pigeon pea plants with an average recombination frequency of 34.52%; the highest recombination frequency was achieved in the SB6CC hybrid (35.71%). The variation in the recombination frequency between the two lines could be due to the positional effect of the transgene (cre) influencing the expression level, resulting in different recombinase activity35,36.
According to Kalve and Tadege24, the low success rate in the production of hybrid plants (chickpea) is due to the lack of detailed information on the flowering stages chosen for crossing. Before starting the crossing events between transgenic plants carrying cry1Ab and cre genes, we performed microscopic analysis of flowers in different stages to understand the exact timing/stage of pollination. In our present investigation, we found that stage 2 was best for emasculation (became female) and stage 3 was appropriate for collecting pollen for crossing. Furthermore, we have also observed that stage 3 is the scientifically accurate stage for crossing24. The reason behind this selection is that the stigma in stage 3 showed the highest pollen acceptability, as at this stage, stigma became the most sexually active for pollination, resulting in hybridization37. Stage 2 is the right time for emasculation because the anther starts to burst, so there is a chance of gating the stigma without pollination. This type of microscopic analysis of the developing reproductive organ combined with precise crossing was performed for the first time in pigeon pea, which helped to increase the crossing efficiency24.
To develop the marker-free plant, two independent T1 plants (SB6 and SD1) harbouring pLBRCAb (as the female parent) were crossed with a T1 progeny plant (CC) harbouring pHC (as the male parent). The generated hybrid plants were designated TIF1 and did not show any phenotypic aberration due to the introgression of the cre transgene, indicating the feasibility of efficient recombination. Excision of the selectable marker gene from the syn cry1Ab expression cassette could not generate complete marker-free plants, as the cre and hptII genes remained in T1F1 hybrid plants. Further elimination of redundant transgenes was achieved via genetic segregation in the following generation (T1F2) through self-pollination. The successful complete marker gene elimination event (syn bar and cre-hptII linked genes) was confirmed by PCR analysis. Furthermore, an insect feeding assay on T1F2 transgenic plants showed the toxicity to insects, which was responsible for inhibiting growth and led to the death of larvae. Thus, the present study was successful in generating insect-resistant genetically modified pigeon pea plants without any antibiotic resistance marker or unnecessarily present transgenes, such as cre recombinase.
In conclusion, the present study could be considered a breakthrough in legume/pulse research concerning the establishment of marker-free insect-resistant pigeon pea plants free of other redundant genes, which makes it one step closer to commercialization.
Materials and methods
Experiments performed in the present study were conducted at the Advanced Laboratory for Plant Genetic Engineering, IIT-Kharagpur, West Bengal, India. The materials and methods of this work are elaborately described in the following section.
Plant material
For the present experiment, seeds of the C. cajan cultivar Asha (ICPL-87119) were obtained from Indian Institute of Pulses Research, Kanpur, India. The study complies with the National guidelines for using biological materials. Permission to use the Pigeon pea seeds in the study was obtained from ICAR, Govt. of India.
Bacterial strain
Agrobacterium-mediated plant transformations were carried out by the supervirulent EHA 105 strain carrying pCAMBIA1300.
Insect source
Eggs of H. armigera were purchased from the NBAIR (National Bureau of Agricultural Insect Resources), Bangalore, India, to perform insect bioassay experiments with second instar larvae under laboratory conditions.
Construction of the plant transformation vectors pLBRCAb and pHC
For the preparation of the transformation cassette, the synthetically prepared herbicide-resistant syn bar gene was PCR-amplified from pBAR17, and primers (Supplementary Table S1) were designed to have a loxP sequence in direct orientation. Then, the amplified syn bar gene flanked by the loxP sequence under the control of the CaMV35S prompter and terminator was cloned (between XhoI sites) into pCAMBIA1300 by replacement of the hptII gene to obtain pLB. A lepidopteran insect pest-specific toxin gene (synthetic cry1Ab prepared in our laboratory38) was cloned under the rubisco small subunit promoter (rbcS) and nos terminator in the pCAMBIA 1300 binary vector (between KpnI and XbaI) and named pLBRCAb (Supplementary Fig. S1a). The chimaeric gene construct was digested with the respective enzyme and analysed by agarose gel electrophoresis to confirm the presence and proper arrangement of the molecular components (Supplementary Fig. S1b). The construct of the cre gene cassette was prepared by cloning the cre gene in pCAMBIA1300 at the SacI and KpnI sites, and the CDS of the cre gene was PCR-amplified from the pX6-GFP plant DNA expression vector (Gene Bank AF 330636.1) by CRFP1/CRRP1 primers (FP: 5′ggccggtaccatgtccaatttactg3′/RP: 5′gaccgagctcctaatcgccatcttc3′) (Supplementary Table S1). The 2xCaMv35s promoter was cloned at the 5´ end of HindIII, the KpnI site and nos gene were incorporated at the 3′ end of the cre gene at the SacI and EcoRI sites, and the construct was named pHC (Supplementary Fig. S2a). The prepared chimaeric gene construct was digested with the respective enzyme and analysed by agarose gel electrophoresis to confirm the presence and proper arrangement of the molecular components (Supplementary Fig. S2b).
Generation of pLBRCAb and pHC plant lines (T0 and T1)
For the generation of two separate pigeon pea lines, pLBRCAb and pHC, a well-established Agrobacterium-mediated transformation and regeneration protocol was used as described by Sarkar et al17. Well-grown regenerated transformed plants were selected in selection medium with appropriate doses of drugs (bialaphos at 4 mg/L for pLBRCAb and hygromycin at 50 mg/L for the pHC plant line). After that, surviving plants with elongated roots were exposed to hardening, transferred to the greenhouse, and allowed to grow to maturity. The individual transgenic line (T0) harbouring the expression cassettes pLBRCAb and pHC was allowed to self-fertilize under controlled conditions (to prevent cross-pollination) to obtain T1 seeds. T1 seeds obtained from self-fertilized individual T0 plants of two separate lines were screened for antibiotic sensitivity on the respective drug-supplemented media. Two-week-old seedlings were scored for resistance or sensitivity. The segregation pattern of the transgene in T1 progeny plants was calculated and validated by comparing the data with the expected ratio by χ2-test.
Dot blot analysis
For the initial sorting of T1 progeny plants, dot blot analysis was performed. Total genomic DNA from young fresh leaves of transgenic plants was extracted following the CTAB method39 and transferred to the Hybond-N+ nylon membrane (GE Healthcare, US) using a dot blot apparatus (Bio-Rad Laboratories, USA). Hybridization, followed by the development of blots, was performed using a cry1Ab gene-specific probe according to the steps described by Sarkar et al17.
Southern blot analysis
Thirty-day-old dot blot-positive transgenic plants along with one untransformed control were selected to isolate total genomic DNA from young and fresh leaves and subjected to Southern blot hybridization, according to Sambrook et al40. Approximately 10 μg of genomic DNA from all plants was digested with a restriction enzyme (HindIII) in the respective buffer. Completely digested DNA samples were electrophoresed on a 0.8% (w/v) agarose gel and subsequently transferred to Hybond-N+ nylon membranes using a vacuum blot transfer apparatus (Bio-Rad Laboratories, model 785, USA) with 20X SSC for 2 h at 4 mmHg pressure. After complete transfer of DNA, the blot was washed with 6X SSC, air-dried, and UV cross-linked in a Crosslinker, according to the manufacturer's instructions (Amersham, UK). Moreover, the PCR-amplified product of the target gene (800 bp for syn cry1Ab and cre) was radiolabelled to prepare the probe. The purified DNA was radiolabelled with α-[32P] dCTP (3500 Ci/mmol) by random priming using a DNA labelling system, according to the manufacturer's protocol (Thermo Scientific, US). After labelling, the probe was purified by passing through a Sephadex G-50 column to remove free radionucleotides. Prehybridization and hybridization of Southern blots were performed under stringent conditions of 65 °C for 2 h and 18 h, respectively, in a roller tube inside the hybridization incubator/shaker (Amersham Biosciences, UK) with Church buffer. The blots were washed with under stringent conditions as follows: 20 min in SSC (2×) and SDS (0.1%) at 50 °C; 20 min in SSC (1×) and SDS (0.1%) at 55 °C; 15 min in SSC (0.5×) and SDS (0.1%) at 60 °C; and another 15 min in SSC (0.1×) and SDS (0.1%) at 65 °C. Autoradiographic exposure was performed at room temperature by exposing the membrane to Super-Resolution Screen (Perkin Elmer) inside a Hypercassette (Amersham Life Science, UK) for 5–30 min depending on the radioactive count. The screen was scanned in the Storage Phosphor System (Cyclone Plus, Perkin Elmer) at a 300 dpi resolution using a medium carousel type to generate the autoradiographic image.
Western blot analysis
Approximately 50 mg leaf tissues of the transgenic lines (30-day-old plants) and untransformed control plants were collected for protein extraction using extraction buffer [50 mM Tris–Cl (pH 8.0), 10 mM PMSF, 2 mM EDTA, 10 μM leupeptin, 0.02% Triton-X, and 1 μM pepstatin], and the protein concentration was estimated according to the Bradford method (Bradford 1976). Approximately 30 μg of protein was loaded for electrophoresis onto 12% SDS-PAGE gels and blotted to Hybond-N+ nylon membranes by a wet transfer procedure. The polyclonal antibody was raised in rabbits against bacterially expressed Cry1Ab38, and rabbit polyclonal anti-Cre recombinase antibody (Product no. ab41104, Abcam, UK) was used as a 1° antibody at a 1:1000 dilution. Anti-isotype IgG-horseradish peroxidase (HRP) conjugate (Sigma Aldrich, USA) was used as a 2° antibody at a 1:2000 dilution. This whole procedure was carried out using the Western Chromogenic Kit (Roche Applied Science, Germany) according to the manufacturer's protocol.
Insect feeding bioassay under laboratory conditions
The insecticidal activity of Southern-positive T1 transgenic plants expressing Cy1Ab protein was analysed through a leaf-feeding bioassay using second instar larvae of H. armigera. Fully open leaves of thirty-day-old pigeon pea plants were incubated with larvae of H. armigera. Each experiment was repeated two times with five replicas, and each replicate was carried out with two larvae. The body weight reduction of larvae was recorded at 24 h intervals, and the percentage of mortality was scored after 6 days of incubation.
The crossing of T1 parental transgenic pigeon pea lines for Cre/lox-mediated marker elimination
The T1 progeny plants harbouring the cry1Ab gene cassette were considered the female parent. Emasculation was performed during the early morning between 5 and 7 AM. The emasculation of the anther in the appropriate stage is very important; thus, all the flowering stages were critically examined, and an unopened flower at the early stage of anthesis (stage 2) was selected. The anthers present inside the flowers were removed by a needle without damaging the gynoecium. Then, emasculated flowers were covered with fine-mesh nylon cloth bags to prevent contamination from foreign pollen. Thus, the female parent was generated. On the other hand, the T1 progeny plant, harbouring the cre gene cassette with a moderate level of Cre recombinase expression, was considered the male parent. At approximately 1–3 PM, the flowers (stage 3) of the cre plant line were taken for crossing. The fine-mesh nylon cloth bags, which covered the emasculated female parent, were removed to expose the sigma, and then the staminal column of the pollen bud was used to brush pollen on the stigma of the female to disperse the pollen. Then, fine-mesh nylon cloth bags were again closed with proper tagging. The T1F1 hybrid progenies were derived from crossing between T1 pHC and pLBRCAb lines; consecutive generations were designated T1F2. The harvested seeds from each independent crossing experiment were collected and stored with proper labelling. Thereafter, per the requirements of our experiment, surface-sterilized seeds were allowed to germinate on MS medium in vitro followed by transfer to the greenhouse for further growth.
Polymerase chain reaction (PCR) analysis
Genomic DNA was extracted using the CTAB method from immature leaves of transgenic pigeon pea plants to conduct PCR. The PCR amplifications for various lines were performed in separate reaction tubes using gene-specific primers for each gene. The PCR conditions and thermal profiles were performed according to the steps reported by Sarkar et al17. Gene-specific forward and reverse primers (Supplementary Table S1) were designed to amplify specific transgenes. The amplification products of the respective genes were verified in an EtBr-stained agarose gel (1%) and observed on a transilluminator.
References
Saxena, K. B. Genetic improvement of pigeon pea-a review. Trop. Plant Biol. 1, 159–178 (2008).
Ramdas, V. R., Gaurav, A. D., Jaysing, C. R. & Arun, G. I. Rapid and highly competent shoot regeneration of pigeon pea (Cajanus cajan) using variable explants by in-vitro culture system. J. Pharmacogn. Phytochem. 4, 1–5 (2015).
FAOSTAT (Food and Agriculture Organization Corporate Statistical Database) Rome, Italy. http://www.fao.org/faostat/en/#home (2014).
Srivastava, J. In-vitro regeneration protocol for pigeon pea—A review. Int. J. Food Agric. Vet. Sci. 3, 63–76 (2013).
Sinha, K. S. Food legumes: Distribution, adaptability, and biology of yield, In FAO Plant Production and Protection Paper 3, 1–102. https://www.cabdirect.org/cabdirect/abstract/19786724643 (1977).
Nene, Y. L. & Sheila, V. K. Pigeon pea: Geography and importance. In: Nene, Y. L., Hall, S. D. & Sheila, V. K. The pigeon pea CAB International, Wallingford, UK, 1–14 (1990).
Saxena, K. B., Kumar, R. V. & Sultana, R. Quality nutrition through pigeon pea—A review. Health 2, 1335–1344 (2010).
Jhoshi, P. K., Parthasarathy, Rao V., Gowda, C. L. L., Jones, R. B., Silim, S. N., Saxena, K. B. & Kumar. J. The world chickpea and pigeon pea economies, facts trends, and outlook. ICRISAT, Patancheru, Andhra Pradesh, India, http://www.icrisat.org/PDF/Outlook%20rep-The%20World%20Chickpea.pdf (2001).
Ghosh, G. et al. Transgenic pigeon pea events expressing Cry1Ac and Cry2Aa exhibit resistance to Helicoverpa armigera. Plant Cell Rep. 36, 1037–1051 (2017).
Choudhary, A. K., Raje, S. R., Datta, S., Sultana, R. & Ontagodi, T. Conventional and molecular approaches towards genetic improvement in pigeon pea for insects resistance. Am. J. Plant Sci. 4, 372–385. https://doi.org/10.4236/ajps.2013.42A049 (2013).
Kaur, A. et al. Pod borer resistant transgenic pigeon pea (Cajanus cajan L.) expressing cry1Ac transgene generated through simplified Agrobacterium-transformation of pricked embryo axes. Plant Cell Tissue Organ Cult. 127, 717–727 (2016).
Durgad, G. A., Guledagudda, S. S., Kiresur, R. V. & Kariyanna, B. Pesticide usage practice by IPM and NON-IPM farmers in pigeon pea from Vijayapura district of Karnataka, India. J. Entomol. Zool. Stud. 6, 95–98 (2018).
Dunwell, J. M. Transgenic approaches to crop improvement. J. Exp. Bot. 51, 487–496 (2000).
Rao. R. G. V. & Shanower, T. G. Identification and management of pigeon pea and chickpea insect pests in Asia, Information Bulletin 57 ICRISAT. http://oar.icrisat.org/2432/ (1999).
Sharma, K. K., Lavanya, M. & Anjaiah, V. Agrobacterium-mediated production of transgenic pigeon pea (Cajanus cajan) expressing the synthetic cry1Ab gene. In-vitro Cell Dev. Biol. Plant 42, 165–173 (2006).
Rao, S. K., Sreevathsa, R., Sharma, P. D., Keshamma, E. & Kumar, M. U. In-planta transformation of pigeon pea: A method to overcome recalcitrance of the crop to regeneration in-vitro. Physiol. Mol. Biol. Plants 14, 321–328 (2008).
Sarkar, S., Roy, S., Ghosh, S. K. & Basu, A. Application of lateral branching to overcome the recalcitrance of in-vitro regeneration of Agrobacterium-infected pigeon pea (Cajanus cajan (L.) Millsp.). Plant Cell Tissue Organ Cult. 137, 23–32 (2019).
Singh, N. D., Sahoo, L., Saini, R., Sarin, N. B. & Jaiwal, P. K. In-vitro shoot organogenesis and plant regeneration from the cotyledonary node and leaf explants of pigeon pea (Cajanus cajan). Physiol. Mol. Biol. Plants 8, 133–140 (2002).
Yadav, A., Kumar, A., Yadav, R., Misra, J. P. & Kumar, R. In-vitro regeneration through organogenesis in pigeon pea (Cajanus cajan). J. Cell Tissue Res. 16, 5485–5490 (2016).
Ramu, S. V. et al. Expression of a synthetic cry1AcF gene in transgenic pigeon pea confers resistance to Helicoverpa armigera. J. Appl. Entomol. 136, 675–687 (2012).
Singh, S. et al. Expression of Cry2Aa, a Bacillus thuringiensis insecticidal protein in transgenic pigeon pea confers resistance to gram pod borer, Helicoverpa armigera. Sci. Rep. 8, 8820. https://doi.org/10.1038/s41598-018-26358-9 (2018).
Ghosh, G., Purohit, A., Ganguly, S., Chaudhuri, R. K. & Chakraborti, D. In-vitro shoot grafting on rootstock: An effective tool for Agrobacterium-mediated transformation of pigeon pea (Cajanus cajan). Plant Biotechnol. 31, 301–308 (2014).
Bawa, A. S. & Anilakumar, K. R. Genetically modified foods: Safety, risks, and public concerns-a review. J. Food Sci. Technol. 50, 1035–1046 (2013).
Kalve, S. & Tadege, M. A comprehensive technique for artificial hybridization in Chickpea (Cicer arietinum). Plant Methods 13, 52. https://doi.org/10.1186/s13007-017-0202-6 (2017).
Luo, H. R., Dalvi, A. V., Li, R. Y. & Saxena, B. K. A study on stigma receptivity of cytoplasmic-nuclear male-sterile lines of pigeon pea, Cajanus cajan (L.) Millsp. J. Plant Breed. Crop Sci. 1, 254–257 (2009).
Dalvi, A. V. & Saxena, B. K. Stigma receptivity in pigeon pea [Cajanus cajan (L.) Millspaugh]. Indian J. Genet. 69, 247–249 (2009).
Tamiru, A., Khan, R. Z. & Bruce, A. J. T. New directions for improving crop resistance to insects by breeding for egg induced defense. Curr. Opin. Insect Sci. 9, 51–55 (2015).
Kranthi, S. & Kranthi, R. K. Annual progress report: Baseline susceptibility of Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae) to Cry1Ac. Central Institute for Cotton Research, Nagpur, India. http://www.moef.gov.in/sites/default/files/geac/cicr0304.htm (2004).
Lacey, A. L. & Kaya, K. H. Field manual of techniques in invertebrate pathology: Application and evaluation of pathogens for control of insect and other invertebrate pests, vol. 1, 571–575. https://doi.org/10.1007/978-94-017-1547-8 (2000).
Darbani, B., Eimanifar, A., Stewart, N. C. & Camargo, N. W. Methods to produce marker-free transgenic plants. Biotechnol. J. 2, 83–90 (2007).
Bala, A., Roy, A., Das, A., Chakraborti, D. & Das, S. Development of selectable marker-free, insect resistant, transgenic mustard (Brassica juncea) plants using Cre/lox mediated recombination. BMC Biotechnol. 13, 88. https://doi.org/10.1186/1472-6750-13-88 (2013).
Chakraborti, D. et al. Cre/lox system to develop selectable marker-free transgenic tobacco plants conferring resistance against sap-sucking homopteran insect. Plant Cell Rep. 27, 1623–1633 (2008).
Hoa, T. T. C., Bong, B. B., Huq, E. & Hodge, T. K. Cre/lox site-specific recombination controls the excision of a transgene from the rice genome. Theor. Appl. Genet. 104, 518–525 (2002).
Meszaros, K. et al. Generating marker-free transgenic wheat using minimal gene cassette and cold-inducible Cre/lox system. Plant Mol. Biol. Rep. 33, 1221–1231 (2014).
Matzke, A. J. M. & Matzke, M. A. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1, 142-l48 (1998).
Russell, S. H., Hoopes, J. L. & Odell, J. T. Directed excision of a transgene from the plant genome. Mol. Genet. Genomics 234, 49–59 (1992).
Prasad, S., Prakash, R. & Haque, M. F. Floral biology of pigeon pea. Trop. Grain Legume Bull. 7, 12 (1977).
Dandapat, A. et al. Variant cry1Ab entomocidal Bacillus thuringiensis toxin gene facilitates the recovery of an increased number of lepidopteran insect-resistant independent rice transformants against yellow stem borer (Scirpophaga incertulus) inflicted damage. J. Plant Biochem. Biotechnol. 23, 81–92 (2014).
Doyle, J. J. & Doyle, J. L. A rapid total DNA preparation procedure for fresh plant tissue. Focus 12, 13–15 (1990).
Sambrook, J., Fritschi, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 1989).
Acknowledgements
Financial assistance in the form of the grant support to this laboratory from the Indian Council of Agricultural Research (NFBSFARA-PB-2010-2010-11-IITKGP), Government of India, is acknowledged.
Author information
Authors and Affiliations
Contributions
S.S. and S.R. conducted experiments. S.S., S.R., and S.K.G. conceived and designed research, analyzed data, and wrote the manuscript. S.K.G. made critical suggestions in conducting experiments and managed funds. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sarkar, S., Roy, S. & Ghosh, S.K. Development of marker-free transgenic pigeon pea (Cajanus cajan) expressing a pod borer insecticidal protein. Sci Rep 11, 10543 (2021). https://doi.org/10.1038/s41598-021-90050-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-90050-8
This article is cited by
-
Over-expression of Trigonella foenum-graecum defensin (Tfgd2) and Raphanus sativus antifungal protein (RsAFP2) in transgenic pigeonpea confers resistance to the Helicoverpa armigera
Plant Cell, Tissue and Organ Culture (PCTOC) (2023)
-
A DEAD box helicase Psp68 positively regulates salt stress responses in marker-free transgenic rice plants
Transgenic Research (2023)
-
Differential in vitro morphology exhibited by pigeonpea seedlings in response to plant growth regulators
Vegetos (2022)
-
Clean gene technology to develop selectable marker-free pod borer-resistant transgenic pigeon pea events involving the constitutive expression of Cry1Ac
Applied Microbiology and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.