Abstract
Elizabethkingia anophelis is an emerging multidrug resistant pathogen that has caused several global outbreaks. E. anophelis belongs to the large family of Flavobacteriaceae, which contains many bacteria that are plant, bird, fish, and human pathogens. Several antibiotic resistance genes are found within the E. anophelis genome, including a chloramphenicol acetyltransferase (CAT). CATs play important roles in antibiotic resistance and can be transferred in genetic mobile elements. They catalyse the acetylation of the antibiotic chloramphenicol, thereby reducing its effectiveness as a viable drug for therapy. Here, we determined the high-resolution crystal structure of a CAT protein from the E. anophelis NUHP1 strain that caused a Singaporean outbreak. Its structure does not resemble that of the classical Type A CATs but rather exhibits significant similarity to other previously characterized Type B (CatB) proteins from Pseudomonas aeruginosa, Vibrio cholerae and Vibrio vulnificus, which adopt a hexapeptide repeat fold. Moreover, the CAT protein from E. anophelis displayed high sequence similarity to other clinically validated chloramphenicol resistance genes, indicating it may also play a role in resistance to this antibiotic. Our work expands the very limited structural and functional coverage of proteins from Flavobacteriaceae pathogens which are becoming increasingly more problematic.
Similar content being viewed by others
Introduction
Flavobacteriaceae is a large family of Gram-negative, mostly aerobic bacteria found in a wide variety of environments1. Within the family, some genera contain species that are known to be pathogenic toward plants, fish, birds and humans2. For example, the genus Flavobacterium has three species, F. psychrophilum, F. columnare and F. branchiophilum, that cause disease within fish, and one species, F. johnsoniae, that infects plants2,3. Tenacibaculum maritimum also infects fish by causing tenacibaculosis4. In ducks, geese and turkeys, Riemerella anatipestifer causes serositis and septicaemia5. Additionally, Ornithobacterium rhinotracheale, Coenonia anatina, and Elizabethkingia meningoseptica cause respiratory diseases within birds6,7,8. Human pathogens from Flavobacteriaceae include Capnocytophaga canimorsus and Elizabethkingia spp. The former is found within the saliva of dogs and cats and causes sepsis, gangrene, meningitis, endocarditis and eye infections; it is transmitted to humans primarily through bites9. Currently, the transmission of Elizabethkingia pathogens remains unclear, but the bacterium resides in water, in soil, on hospital surfaces, hospital water service lines, and in human patients.
Elizabethkingia is an opportunistic, emerging pathogen that has caused recent outbreaks among the general population and immunocompromised patients in Asia and North America, specifically in Singapore, Hong Kong, Taiwan, and the United States (Wisconsin, Illinois and Michigan)10,11. It was first isolated in 1949 from infants with septicemia or meningitis and was initially classified under the genus Flavobacterium12. Later it was reclassified into the genus Chryseobacterium, and in 2005 it was given its own genus Elizabethkingia13. Currently, the genus comprises three aerobic, non-motile rod-shaped Gram-negative species, including E. miricola, E. meningoseptica, and E. anophelis. Recent studies have identified potential new species that could be added to this genus, but further investigation is required14. Multiple Elizabethkingia strains have been isolated from a variety of environments, including human patients and mosquitoes11. Interestingly, E. miricola was isolated in 2003 from condensed water samples obtained from the space station Mir15. It has since been reported to cause pneumonia and lower respiratory tract infections, but the mechanism of transmission of the bacterium is unknown16. E. meningoseptica, previously identified as Chryseobacterium meningoseptica, is a hospital acquired pathogen that causes neonatal meningitis, pneumonia, and endocarditis17,18. E. anophelis is a relatively newly identified bacterium, and some investigations have suggested that it originated from the midgut of the Anopheles mosquito, Anopheles gambiae19,20; however, it has also been isolated from Anopheles stephensi21. It causes similar infections as E. meningoseptica, which has made it challenging to clinically differentiate between these two organisms. As a result, E. anophelis infections have been underestimated due to their misclassification22.
The E. anophelis bacterium responsible for the Singaporean outbreak was isolated from a human patient and designated as the NUHP1 strain. NUHP1 is differentiated from other E. anophelis strains because it acquired an ICEEa1 integrative conjugative element (ICE) within its mutY gene. ICE is a mobile genetic element that integrates into the host chromosome, replicates, excises, and forms a plasmid in order to be transferred to other bacterial cells via horizontal conjugation. These mobile genetic elements are used by bacteria to enhance survival in diverse environments and often contain antibiotic resistance genes. Within the Flavobacteriaceae family, only E. anophelis and R. anatipestifer have been shown to contain ICE23,24. Notably, only strains of E. anophelis with ICE have caused outbreaks. While ICE contain antibiotic resistance genes, additional antibiotic resistance genes are found outside these regions on the chromosome and contribute to bacterial survival. Within the E. anophelis NUHP1 genome, 14 antibiotic resistance genes have been identified, including ones required for resistance to aminoglycosides, beta-lactams, macrolides, tetracycline, trimethoprim, and chloramphenicol (Cm)25. One example of an antibiotic resistance gene that is conserved in all Elizabethkingia strains is the chloramphenicol acetyltransferase (CAT) gene26. To our knowledge this gene has not been described as being located in an ICE.
Since the E. anophelis NUHP1 strain has caused significant outbreaks in recent years, the Seattle Center for Structural Genomics of Infectious Diseases selected antibiotic resistance proteins from this pathogen for structural determination. One of these proteins is CatB, which is predicted to be a chloramphenicol acetyltransferase. Homologs of this protein are important for bacterial Cm antibiotic resistance because they acetylate the 3′-hydroxyl group of Cm (Fig. 1A). This covalent chemical modification prevents Cm from inhibiting bacterial protein synthesis in ribosomes. CAT enzymes are classified based on their origin and their sequence and structural homology. They have been categorized into three predominant types: Type A, Type B and Type C27. These are also sometimes referred to as CatA, CatB, and CatC. A defining characteristic between these CATs is that Type B and C display a lower apparent affinity for Cm than Type A27. Additionally, Type A CATs form a distinct structural group compared to Type B and C. All three types of proteins are trimers, but Type B and C adopt a hexapeptide repeat structural fold, which is not found in Type A (Fig. 1B–D). Since the E. anophelis catB gene is conserved across all Elizabethkingia strains and CatB proteins are critical for Cm resistance in important human bacterial pathogens, we explored the structural characteristics of this protein in more detail. This study provides further insight into antibiotic resistance proteins from this important emerging pathogen and adds to the fundamental structural knowledge of CatB proteins.
Chloramphenicol acetyltransferase (CAT) reaction and representative types of CAT structures. (A) CATs catalyse the O-acetylation transfer reaction of an acetyl group from acetyl coenzyme A (AcCoA) to the 3′-position of chloramphenicol. Representative single monomer structures of different types of CAT proteins are also shown. (B) Type A CAT from Escherichia coli (PDB ID: 3U9F28) in green. C) Type B CAT from Pseudomonas aeruginosa (PDB ID: 2XAT29) in red. D) Type C CAT from Allivibrio fischeri (PDB ID: 5UX927) in blue.
Materials and methods
The E. anophelis NUHP1 catB gene (UniProt ID: A0A077EJ45) was cloned, expressed, and purified as described30. In preparation for crystallography, E. anophelis CAT was concentrated to 21 mg/ml in 25 mM HEPES/NaOH, pH = 7.0, 500 mM NaCl, 5% glycerol, 2 mM DTT, 0.025% NaN3 (SSGCID batch ID ElanA.01572.a.B1.PW38419). The protein was further diluted to 19.5 mg/ml upon the addition of 5 mM MgCl2 and 2.5 mM acetyl-CoA and 2.5 mM chloramphenicol and incubated for 10 min at 287 K. Crystals were then grown at 287 K by sitting drop vapor diffusion in XJR trays. A volume of 0.4 μl of protein/ligand complex was mixed with 0.4 μl of JCSG + , well A11 reservoir solution (Rigaku Reagents, Bainbridge Island, WA): 50% (v/v) MPD, 0.1 M Tris base/HCl, pH = 8.5, 0.2 M ammonium phosphate monobasic. The reservoir volume was 80 μl. Crystals were harvested and flash‐frozen in liquid nitrogen. Data were collected at 100 K on a Rayonix MX‐300 mm CCD detector at a wavelength of 0.97872 Å on beamline 21‐ID‐F at Life Sciences Collaborative Access Team (LS‐CAT) at the Advanced Photon Source (APS, Argonne, IL). Data were reduced with the XDS/XSCALE package31. The structure was solved using molecular replacement with MorDA32 and PDB ID: 1XAT29 as a starting model. Iterative rounds of manual model building and automated refinement were carried out using Coot33 and Phenix34. The quality of the structure was checked by Molprobity35, and the final structure was deposited into the Protein Data Bank (PDB) using the code 6MFK. However, the protein was produced with a non-cleavable N-terminal polyhistidine tag (MAHHHHHH) and two C-terminal histidine residues of the tag were partially ordered. The backbone atoms of these His residues and residues 1 and 2 of the protein were ordered. Both the backbone and the side chains of the rest of the residues (3-208) were ordered and included in the model.
Results
Structural analysis of E. anophelis NUHP1 CAT protein
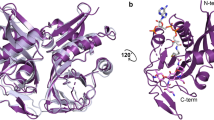
To characterize the type of CAT E. anophelis NUHP1 harbors, we determined the structure of the protein encoded by the catB gene using X-ray crystallography (Table 1). The electron density map allowed all residues of the protein (1-208) to be modeled. A single protein monomer was present in the asymmetric unit of the crystal with the topology shown in Fig. 2A. The monomer contained two main domains: (1) a left-handed β-helix core (15 β-strands spanning residues 12-163) forms a prism at the center of the monomer and contains two extended loop regions (44-56 and 72-110), and (2) a C-terminal α-helical domain comprised of three α-helices (spanning residues 164-208) (Fig. 2B). To determine whether the E. anophelis CAT was likely to form a multimer, we examined the crystal structure by expanding the asymmetric unit and investigated whether large interfaces may be present using Proteins, Interfaces, Structures and Assemblies (PISA)24. The only biological assembly predicted from PISA for the E. anophelis CAT enzyme was a trimer, which is identical to all other structurally characterized CAT enzymes. (Fig. 2C).
Elizabethkingia anophelis chloramphenicol acetyltransferase (CatB) protein structure. (A) Topology map. Cyan arrows indicate beta strands, red cylinders indicate 3/10 alpha helices, yellow cylinders represent alpha helices, and lines between shapes represent loops. The N-terminus is shown in pink and the C-terminus is in yellow. (B) Tertiary structure of a single monomer. Each domain is coloured as in (A). (C) Trimeric quaternary structure.
To characterize the interfacial residues that are important for this biological assembly, and to reveal the conservation of these residues in other CAT enzymes, we compared these interfaces with other structures of CATs . In our E. anophelis CAT structure, we found one interface mediating the trimer formation (Supplementary Figure 1). This interface is mediated through a large number of polar and non-polar interactions. In total, the interface forms 21 hydrogen bonds, two salt bridges (Supplementary Tables 1 and 2), and buries 1699.8 Å2 of solvent accessible surface area (ASA). Key binding sites include (1) binding residues within the N-terminal tail A:Met1/B:Met1 (A represents one monomer, B represents another monomer within the biological assembly); (2) seven interactions within the beta-sheet prism; A:Tyr35/B:Ala44,Tyr46, A:Ser32/B:Tyr46, A:Lys162/B:Tyr81, A:Arg140/B:Glu122,Asn156, A:Asn156/B:Asn156, A:Glu122/B:Arg45, A:Asp116/B:Ser85, Ser86,Phe87; and (3) three interactions within the α-helix C-terminal domain (yellow) A:Arg164/B:Tyr81,Tpr83,Ile84, A:Leu193/B:Ile84,Ser85, A:Ser195/B:Ile84. We also identified a large number of hydrophobic and non-bonded contacts at the interface (listed in Supplementary Table 3). Next, we compared this biological interface with related CAT enzymes containing acetyl-CoA (PDB ID: 6U9C27), and Cm (PDB ID: 2XAT29). We found very similar interfaces in these ligand and cofactor bound structures, with CAT bound to acetyl-CoA exhibiting 23 hydrogen bonds, 2 salt bridges, and 1612 Å2 ASA, and CAT bound to Cm exhibiting 16 hydrogen bonds, 2 salt bridges, 1391 Å2 ASA (Supplementary Tables 1 and 3).
The E. anophelis protein exhibits greatest structural homology with Type B CATs
To determine which type of CAT the E. anophelis protein most closely resembles, we performed a pairwise sequence alignment and a structural comparison with representative sequences and structures of different categories of CATs. To begin, we selected sequences of three different types of CAT proteins that had been structurally characterized: Type A from E. coli (catI; UniProt ID: P62577), Type B from P. aeruginosa (catB7; UniProt ID: P26841), and Type C from A. fischeri (UniProt ID: Q5DZD6). We performed a multiple sequence alignment with the E. anophelis CAT and these three proteins and found the E. anophelis CAT shared 17%, 62%, and 54% identity with Type A, Type B, and Type C CATs, respectively (Supplementary Figure 2). The insertion typically found in Type C CATs was not present in the E. anophelis protein. Therefore, the E. anophelis protein most closely resembled Type B CAT protein sequences. To further explore whether the E. anophelis protein structure also resembled Type B CATs, we performed a structural comparison of this protein with other types of CATs that had been structurally characterized. We specifically compared the E. anophelis 6MFK structure with the E. coli 3U9F28, P. aeruginosa 2XAT29, and A. fischeri 5UX927 structures. The Type A protein from E. coli adopts a completely different fold than the Type B and C proteins from P. aeruginosa and A. fischeri, respectively; Type B and C CATs adopt a hexapeptide repeat fold (Fig. 1). The E. anophelis 6MFK structure superimposed well with the Type B 2XAT structure29 (rmsd 0.6 Å) and Type C 5UX927 structure (rmsd 1.1 Å). Therefore, the results of the sequence and structural comparisons indicate the E. anophelis protein is most likely a Type B CAT.
Putative active site residues of the E. anophelis CAT protein and other Type B CATs are conserved
Next, we examined the putative active site residues of the E. anophelis CAT protein and compared them to previously determined Type B CAT structures in complex with substrates or substrate analogs. CAT proteins have two binding sites for each substrate (Cm and AcCoA), which are located at the interface between monomers of the trimer. To determine which residues are located in both sites of the E. anophelis CAT, we superimposed its structure with the P. aeruginosa catB7 (PDB ID: 2XAT29; rmsd 0.6 Å) structure in complex with Cm and desulfo-coenzyme A and the V. cholerae catB9 (PDB ID: 6U9C27; rmsd 0.6 Å) structure in complex with AcCoA. The trimeric E. anophelis CAT structure was built based on the 6U9C27 structure and ligands from the 2XAT29 and 6U9C27 structures were modeled into the trimer to compare putative active site residues. The residues that could potentially hydrogen bond with Cm in the binding site of the E. anophelis structure include Pro8, Gly11, Tyr30, and Ser32 from one monomer and His79 from a second monomer (Fig. 3). These residues are all identical in the CAT enzyme from P. aeruginosa (PDB ID: 2XAT29) (Fig. 3).
Comparison of the Cm acceptor sites from E. anophelis CatB and P. aeruginosa catB7. (A) Superimposed structure of E. anophelis CatB (PDB ID: 6MFK) and P. aeruginosa catB7 (PDB ID: 2XAT29) trimer. Zoomed view to show H-bonding interactions between putative active site residues and Cm. (B) Cm binding site residues of E. anophelis and P. aeruginosa CAT proteins. Cm is shown in gray sticks. (C) Diagram of hydrogen bonds between residues of the E. anophelis and P. aeruginosa CAT proteins and Cm molecule. (D) E. anopheles CatB putative Cm acceptor site cavity. Cm is in gray sticks and residues important for H-bonding are coloured in green. Cm was modeled from the P. aeruginosa 2XAT29 structure.
Residues critical for mediating H-bonding interactions with AcCoA in the 6U9C27 structure included Ser137 and Lys160 from the first monomer and Thr142 in the second monomer. The corresponding residues in the 6MFK structure are Ser139, Lys162, Thr144 (Fig. 4).
Comparison of the AcCoA donor sites of E. anophelis CatB and V. cholerae catB7 proteins. (A) Superimposed structure of E. anophelis CatB (6MFK) and V. cholerae catB9 (6U9C27) trimer. Zoomed view to show residues surrounding AcCoA, shown in yellow sticks. (B) Putative active site residues of E. anophelis and V. cholerae CAT proteins found within the AcCoA binding site. (C) Diagram of hydrogen bonds between putative active site residues of the E. anophelis and V. cholerae CAT proteins and AcCoA molecule. (D) CatB putative active site cavity where AcCoA binds; residues important for H-bonding are coloured in red. AcCoA is in yellow sticks and is modeled from the V. cholerae 6U9C27 structure.
Once we identified the key putative active site residues in the E. anophelis CAT structure, we examined the conservation of these residues across sequences of other Flavobacteriaceae pathogens (Fig. 5). To begin, we aligned the E. anophelis CAT protein sequence to others identified by BLASTp against the genomes of the following pathogens: E. miricola, E. meningoseptica, Chryseobacterium sp., and R. anatipestifer. We found the E. anophelis CAT protein shares 95% sequence identity with E. miricola, 86% identity with E. meningoseptica, 83% identity with Chryseobacterium sp., and 81% identity with R. anatipestifer Type B CAT proteins. Additionally, the putative active site residues for both Cm and AcCoA sites identified in the E. anophelis CAT protein were highly conserved across all of the CAT sequences from these pathogens.
Multiple sequence alignment of E. anophelis NUHP1 CAT with various Type B CATs from Flavobacteriaceae and validated Cm resistance proteins. The secondary structural elements of the E. anophelis NUHP1 protein (PDB ID: 6MFK) are shown above the multiple sequence alignment. Conserved residues are highlighted in red. Residues of the Cm binding site are denoted with cyan (H-bonds) and blue (hydrophobic interactions) symbols, where residues from one monomer are indicated with triangles and residues of the second monomer are shown as inverted triangles. Residues of the AcCoA binding site are denoted with magenta (H-bonds) and purple (hydrophobic) symbols, where residues from one monomer are indicated with triangles and residues of the second monomer are shown as inverted triangles. Green and orange symbols indicate interfacial residues that form H-bonds or salt bridges between protomers. Protein sequences within the alignment include: Elizabethkingia anophelis catB (UniProt ID A0A077EJ45), Elizabethkingia miricola catB (NCBI Accession KGO08276), Elizabethkingia meningoseptica catB (NCBI Accession QDZ61149), Chryseobacterium sp. catB (UniProt ID A0A3DMIM7), Riemerella anatipestifer catB (UniProt ID E5D2K4), Klebsiella oxytoca KONIH1 catB11 (NCBI Accession AID93387), Pasterurella multocida catB2 (UniProt ID Q83ZX9), Salmonella enterica subsp. enterica serovar Typhi catB8 (UniProt ID Q79PD0), Enterobacter cloacae catB3 (UniProt ID C1IUN4), Pseudomonas aeruginosa catB10 (UniProt ID A2Q6I9), Pseudomonas aeruginosa catB6 (UniProt ID Q9R818), Agrobacterium fabrum str. C58 catB (UniProt ID P23364), Vibrio cholerae catB9 (UniProt ID H9L3X9), and Pseudomonas aeruginosa PAO1 catB7 (UniProt ID P26841). The multiple sequence alignment was generated with ESpript (http://espript.ibcp.fr/ESPript/ESPript/).
E. anophelis CatB is similar to clinically validated Cm resistance Proteins
To determine whether the E. anophelis CAT protein was similar to known Cm resistance proteins in different pathogens, we searched the Comprehensive Antibiotic Resistance Database (CARD)36 using the E. anophelis CAT protein sequence. The search yielded nine proteins from a variety of bacterial pathogens with validated Cm resistance profiles (Table 2; Fig. 5). All of these proteins were Type B CATs and they shared between 62–73% sequence identity with the E. anophelis CAT. When we compared their active site residues with the E. anophelis CAT protein, we saw all Cm binding site residues were conserved. The only variation in residue conservation was in the AcCoA binding site. The majority of the residue substitutions were of similar chemical properties (e.g. substituting Val for Ile). However, one exception to this pattern was observed: the CatB7 protein from P. aeruginosa had a Gly residue in the corresponding location of Lys145 in the E. anophelis protein. Based on these results, it is highly likely that the E. anophelis CAT and its homologs in other Flavobacteriaceae pathogens could also catalyze the acetylation of Cm and confer resistance to this antibiotic.
Discussion
E. anophelis has been discovered in diverse environmental and host associations, including its presence in soil and water and its isolation from the African and Asian malaria vector mosquitoes Anopheles gambiae and Anopheles stephensi. Currently it is unclear whether the mosquito itself can be a source of E. anophelis transmission to humans, or if other mechanisms occur, including potential reservoirs within hospitals such as sink basins and water faucets37. In a study examining different E. anophelis strains, those isolated from the midgut of a mosquito contained genes that encode a xylose isomerase and xylulose kinase, which human isolates lacked. This suggests that different environments may have specific requirements for sugar metabolism, and clinical and environmental strains of E. anophelis differ from one another19. Some other Flavobacteriaceae pathogens have been shown to be transmitted in the air, on surfaces, in water, and by ingesting contaminated food38. They can also be part of the normal flora of the throat of some duck species39.
E. anophelis harbors a variety of antibiotic resistance genes along with several multidrug efflux pumps, which are speculated to contribute to the shape and stabilization of the microbial community within the gut of the mosquito40. In addition to antibiotic resistance genes, multiple virulence factors contribute to Elizabethkingia species survival and persistence. For example, E. meningoseptica contains various virulence factors, including those that contribute to proteolysis, iron uptake and transport, biofilms, and capsule formation. Bacterial persistence on surfaces, including cellular adhesion and medical devices, is facilitated by these virulence factors via biofilm, capsule and sometimes curli formation. Biofilms have been shown to be critical for E. meningoseptica infections and also allow these bacteria to resist surface disinfection41.
Cm has anti-biofilm properties and is effective at killing a variety of bacteria and preventing colony growth42; however, this has not been investigated for Elizabethkingia species. Cm is not typically used to treat internal bacterial infections due to serious side-effects and toxicity, but it is more widely used as a topical treatment or for some ocular infections43. Because Cm is not widely prescribed, some current bacterial pathogens are still susceptible to this antibiotic. Therefore, renewed interest in salvaging or repurposing this drug to treat bacterial infections is also being considered as a viable path forward44. While it is unlikely this drug would be used to treat an E. anophelis infection in the current pharmaceutical climate, it is possible Cm would be considered in the future as our arsenal of effective antibiotics becomes reduced. Moreover, Cm is an effective drug that can efficiently pass the blood–brain barrier45, which is important for treating meningitis infections like those associated with E. anophelis or E. meningoseptica. Thus, knowing the E. anophelis NUHP1 bacterium contains a Cm resistance gene may be important for future therapeutic considerations.
Our analysis of the E. anophelis CAT protein has shown it adopts a similar structure and retains conserved putative active site residues as clinically validated Type B CAT proteins in other bacterial pathogens. While we recognize in vitro enzyme kinetics assays are needed to ensure the E. anophelis enzyme can actually acetylate Cm, the near 100% conservation of putative active site residues compared to other Type B CATs that have been kinetically characterized suggests they are functionally similar. Even though Type B CATs acetylate Cm and are important for Cm resistance, their native functions remain elusive. Several other hexapeptide repeat acetyltransferases have been explored and have been shown to acylate different sugars or cell wall polysaccharides46,47. However, these characterized enzymes are still divergent in sequence and structure from the Type B CATs. Therefore, further investigation into the native function of the Type B CAT enzymes is warranted, especially since these proteins are widely found in opportunistic and colonizing pathogens. It would be particularly interesting to explore the native function of the CatB protein from E. anophelis since it has an extraordinarily flexible biological capacity to live in a wide array of environments and at times can colonize humans and cause disease.
Despite our lack of knowledge of the native function of the E. anophelis CAT protein, the gene is conserved in all identified environmental and nosocomial E. anophelis strains48. Our analysis of the Type B CAT protein sequence across Elizabethkinigia species showed most of the protein sequence is conserved; however, there were some key amino acid changes between the proteins from different species. Due to the difficulty in accurately differentiating Elizabethkingia species during genome sequencing22, this gene may be used as a tool to improve species identification of isolates and propagation of different strains around the world.
References
McBride, M. J. The family flavobacteriaceae. In The Prokaryotes (eds Rosenberg, E. et al.) 643–676 (Springer, Berlin, 2014).
Bernardet, J.-F. & Bowman, J. P. The genus flavobacterium. In The prokaryotes Vol. 7 (eds Dworkin, M. et al.) 481–531 (Springer, Berlin, 2006).
Loch, T. P. & Faisal, M. Emerging flavobacterial infections in fish: a review. J. Adv. Res. 6(3), 283–300 (2015).
Avendaño-Herrera, R., Toranzo, A. E. & Magariños, B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis. Aquat. Org. 71(3), 255–266 (2006).
Huang, L. et al. Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China. Front. Microbiol. 8, 297 (2017).
Van Empel, P. & Hafez, H. Ornithobacterium rhinotracheale: A review. Avian Pathol. 28(3), 217–227 (1999).
Vancanneyt, M. et al. Flavobacterium meningosepticum, a pathogen in birds. J. Clin. Microbiol. 32(10), 2398–2403 (1994).
Vandamme, P. et al. Coenonia anatina gen. nov., sp. nov., a novel bacterium associated with respiratory disease in ducks and geese. Int. J. Syst. Evolut. Microbiol. 49(2), 867–874 (1999).
Butler, T. Capnocytophaga canimorsus: an emerging cause of sepsis, meningitis, and post-splenectomy infection after dog bites. Eur. J. Clin. Microbiol. Infect. Dis. 34(7), 1271–1280 (2015).
Janda, J. M. & Lopez, D. L. Mini review: New pathogen profiles: Elizabethkingia anophelis. Diagn. Microbiol. Infect. Dis. 88(2), 201–205 (2017).
Perrin, A. et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 8, 15483 (2017).
King, E. O. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am. J. Clin. Pathol. 31(3), 241–247 (1959).
Ceyhan, M. & Celik, M. Elizabethkingia meningosepticum (Chryseobacterium meningosepticum) infections in children. Int. J. Pediatr. 2011, 215237 (2011).
Nicholson, A. C. et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov.. Antonie Van Leeuwenhoek 111(1), 55–72 (2018).
Li, Y. et al. Chryseobacterium miricola sp. nov., a novel species isolated from condensation water of space station Mir. Syst. Appl. Microbiol. 26(4), 523–528 (2003).
Zdziarski, P., Paściak, M., Rogala, K., Korzeniowska-Kowal, A. & Gamian, A. Elizabethkingia miricola as an opportunistic oral pathogen associated with superinfectious complications in humoral immunodeficiency: A case report. BMC Infect. Dis. 17(1), 1–6 (2017).
Hsu, M.-S. et al. Clinical features, antimicrobial susceptibilities, and outcomes of Elizabethkingia meningoseptica (Chryseobacterium meningosepticum) bacteremia at a medical center in Taiwan, 1999–2006. Eur. J. Clin. Microbiol. Infect. Dis. 30(10), 1271–1278 (2011).
Bloch, K. C., Nadarajah, R. & Jacobs, R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine 76(1), 30–41 (1997).
Chen, S., Bagdasarian, M. & Walker, E. D. Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl. Environ. Microbiol. 81(6), 2233–2243 (2015).
Lau, S. K. et al. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg. Infect. Dis. 21(2), 232 (2015).
Garay, J. A. R., Hughes, G. L., Koundal, V., Rasgon, J. L. & Mwangi, M. M. Genome sequence of Elizabethkingia anophelis strain EaAs1, isolated from the Asian malaria mosquito Anopheles stephensi. Genome Announc. 4(2), e00084-e116 (2016).
Doijad, S., Ghosh, H., Glaeser, S., Kämpfer, P. & Chakraborty, T. Taxonomic reassessment of the genus Elizabethkingia using whole-genome sequencing: Elizabethkingia endophytica Kämpfer et al. 2015 is a later subjective synonym of Elizabethkingia anophelis Kämpfer et al. 2011. Int. J. Syst. Evol. Microbiol. 66(11), 4555–4559 (2016).
Xu, J., Pei, D., Nicholson, A., Lan, Y. & Xia, Q. In silico identification of three types of integrative and conjugative elements in Elizabethkingia anophelis strains isolated from around the world. Msphere. 4(2), e00040-19 (2019).
Zhu, D. et al. First report of integrative conjugative elements in Riemerella anatipestifer isolates from ducks in china. Front. Vet. Sci. 6, 128 (2019).
Li, Y. et al. Complete genome sequence and transcriptomic analysis of the novel pathogen Elizabethkingia anophelis in response to oxidative stress. Genome Biol. Evol. 7(6), 1676–1685 (2015).
Breurec, S. et al. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci. Rep. 6, 30379 (2016).
Alcala, A. et al. Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species. Protein Sci. 29(3), 695–710 (2020).
Biswas, T., Houghton, J. L., Garneau-Tsodikova, S. & Tsodikov, O. V. The structural basis for substrate versatility of chloramphenicol acetyltransferase CATI. Protein Sci. 21(4), 520–530 (2012).
Beaman, T. W., Sugantino, M. & Roderick, S. L. Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa. Biochemistry 37(19), 6689–6696 (1998).
Bryan, C. M. et al. High-throughput protein production and purification at the Seattle Structural Genomics Center for Infectious Disease. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 67(9), 1010–1014 (2011).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66(2), 133–144 (2010).
Vagin, A., Lebedev, A. (ed.) MoRDa, an automatic molecular replacement pipeline. Acta Crystallographica A-Foundation and Advances (Int Union Crystallography 2 Abbey Sq, Chester, Ch1 2hu, England, 2015).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60(12), 2126–2132 (2004).
Adams, P. D. et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66(2), 213–221 (2010).
Chen, V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66(1), 12–21 (2010).
McArthur, A. G. et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57(7), 3348–3357 (2013).
Seong, H. et al. Risk Factors for mortality in patients with Elizabethkingia infection and the clinical impact of the antimicrobial susceptibility patterns of Elizabethkingia species. J. Clin. Med. 9(5), 1431 (2020).
Ren, X. et al. Riemerella anatipestifer AS87_RS09170 gene is responsible for biotin synthesis, bacterial morphology and virulence. Sci. Rep. 8(1), 1–13 (2018).
Ryll, M. et al. Studies on the prevalence of Riemerella anatipestifer in the upper respiratory tract of clinically healthy ducklings and characterization of untypable strains. J. Vet. Med. Ser. B 48(7), 537–546 (2001).
Kukutla, P. et al. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS ONE 9(5), e97715 (2014).
Chen, S. et al. Comparative genomic analyses reveal diverse virulence factors and antimicrobial resistance mechanisms in clinical Elizabethkingia meningoseptica strains. PLoS ONE 14(10), e0222648 (2019).
Singh, R., Sahore, S., Kaur, P., Rani, A. & Ray, P. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain-and antibiotic-specific differences. Pathog. Dis. 74(6), ftw056 (2016).
Drago, L. Chloramphenicol resurrected: A journey from antibiotic resistance in eye infections to biofilm and ocular microbiota. Microorganisms. 7(9), 278 (2019).
Dinos, G. P. et al. Chloramphenicol derivatives as antibacterial and anticancer agents: historic problems and current solutions. Antibiotics. 5(2), 20 (2016).
Nagabhushan, T., Miller, G. H., Varma, K. J. Chloramphenicol and analogues. Kirk‐Othmer Encyclopedia of Chemical Technology (2000).
Lo Leggio, L., Dal Degan, F., Poulsen, P., Andersen, S. M. & Larsen, S. The structure and specificity of Escherichia coli maltose acetyltransferase give new insight into the LacA family of acyltransferases. Biochemistry 42(18), 5225–5235 (2003).
Pauly, M. & Ramírez, V. New insights into wall polysaccharide O-acetylation. Front. Plant Sci. 9, 1210 (2018).
Wang, M. et al. The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate. MicrobiologyOpen. 8(11), e804 (2019).
Acknowledgements
Student support for AMR was provided through the NIH MARC T34-GM008574 grant and a CSU Sally Casanova Pre-doctoral scholarship to AMA. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases (contract no. HHSN272201700059C to Peter J. Myler). We thank the SSGCID cloning and protein production groups at the Center for Infectious Disease Research and at the University of Washington.
Author information
Authors and Affiliations
Contributions
S.M.G., P.S., and J.K.F. performed data analysis, created figures, wrote and revised significant portions of the manuscript, and constructed the final draft. A.M.R., A.M.A., and M.L.K. performed literature searches, data analyses, created figures and tables, wrote and revised significant portions of the manuscript, and approved the final draft. M.D. and S.J.M. acquired the crystallographic data, solved the structure and contributed to the analysis and manuscript. D.D.L., P.J.M., and T.E.E. are the site manager and primary investigators for SSGCID and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ghafoori, S.M., Robles, A.M., Arada, A.M. et al. Structural characterization of a Type B chloramphenicol acetyltransferase from the emerging pathogen Elizabethkingia anophelis NUHP1. Sci Rep 11, 9453 (2021). https://doi.org/10.1038/s41598-021-88672-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-88672-z
This article is cited by
-
Whole-genome sequence and resistance determinants of four Elizabethkingia anophelis clinical isolates collected in Hanoi, Vietnam
Scientific Reports (2024)
-
Methicillin-resistant Staphylococcus aureus: novel treatment approach breakthroughs
Bulletin of the National Research Centre (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.