Abstract
Plasmodium knowlesi is the main cause of malaria in Sarawak, where studies on vectors of P. knowlesi have been conducted in only two districts. Anopheles balabacensis and An. donaldi were incriminated as vectors in Lawas and An. latens in Kapit. We studied a third location in Sarawak, Betong, where of 2169 mosquitoes collected over 36 days using human-landing catches, 169 (7.8%) were Anopheles spp. PCR and phylogenetic analyses identified P. knowlesi and/or P. cynomolgi, P. fieldi, P. inui, P. coatneyi and possibly novel Plasmodium spp. in salivary glands of An. latens and An. introlatus from the Leucosphyrus Group and in An. collessi and An. roperi from the Umbrosus Group. Phylogenetic analyses of cytochrome oxidase subunit I sequences indicated three P. knowlesi-positive An. introlatus had been misidentified morphologically as An. latens, while An. collessi and An. roperi could not be delineated using the region sequenced. Almost all vectors from the Leucosphyrus Group were biting after 1800 h but those belonging to the Umbrosus Group were also biting between 0700 and 1100 h. Our study incriminated new vectors of knowlesi malaria in Sarawak and underscores the importance of including entomological studies during the daytime to obtain a comprehensive understanding of the transmission dynamics of malaria.
Similar content being viewed by others
Introduction
Plasmodium knowlesi, primarily a simian malaria parasite, was reported to have caused a large number of human malaria infections in 2004 in the Kapit Division of Sarawak, Malaysian Borneo1,2. Subsequently knowlesi malaria cases have been reported throughout Southeast Asia and they continue to be a public health concern, particularly in Malaysia. From 2017 to 2019, a total of 10,968 knowlesi malaria cases were reported in Malaysia, with 87% occurring in the Malaysian Borneo states of Sabah and Sarawak (unpublished data, Ministry of Health Malaysia)3. Studies investigating the epidemiology of the parasite and the identity of its mosquito vectors have seen significant progress since the initial report in 2004. For example, long-tailed and pig-tailed macaques were identified as the reservoir hosts for P. knowlesi and other simian malaria parasites (P. coatneyi, P. cynomolgi, P. fieldi, and P. inui) in Sarawak4. Furthermore, phylogenetic analyses of the mitochondrial genomes have shown previously unreported species of Plasmodium in long-tailed macaques in Sarawak; P. inui-like parasites and P. simiovale5. In addition, microsatellite typing of P. knowlesi derived from humans and monkeys and whole genome sequencing of clinical samples have demonstrated the existence of at least three P. knowlesi subpopulations, two of which occur in Malaysian Borneo and the others in Peninsular Malaysia6,7. In terms of entomological studies, several species from the Leucosphyrus Group, two species from the Barbirostris Group, and a species from the Sundaicus Complex have been implicated as vectors of P. knowlesi in India (An. sundaicus), Vietnam (An. dirus), Peninsular Malaysia (An. hackeri, An. cracens and An. introlatus), Sabah (An. balabacensis and An. donaldi), and Sarawak (An. latens, An. balabacensis, and An. donaldi)8,9,10,11,12,13,14,15,16,17,18.
An effective vector control strategy is key to successful malaria prevention and control, but it will have to be devised based on accurate understanding of the identity and bionomics of the vectors at the location of transmission. This was previously demonstrated in Vietnam where a non-vector (An. varuna) was misidentified as the vector An. minimus and subsequently wrongly targeted for vector control19, causing the ineffective use of valuable and limited resources. Misidentification however, is relatively common in vector studies and especially where vectors comprise species complexes which are difficult to distinguish morphologically20,21,22,23. For this reason, molecular methods based on the cytochrome oxidase subunits I and II (COI and COII), second internal transcribed spacer (ITS2) within the ribosomal DNA, and NADH dehydrogenase sub-unit six (ND6) have been developed as a complementary tool to improve accuracy in identification of vectors20,22,24,25,26,27,28,29,30,31,32.
There remain gaps in knowledge regarding the vectors involved in the transmission of P. knowlesi and other simian malaria parasites in Malaysian Borneo. Most of the entomological studies were conducted following a similar pattern where mosquitoes were first collected in the field for a 6- or 12-h duration beginning at 1800 h or 1900 h. The mosquitoes were morphologically identified, in certain studies they were dissected, and DNA that was extracted was then analysed by PCR assays. These assays were designed to only detect the five simian malaria parasites initially described in macaques4. Consequently, it is unknown if other Plasmodium species could be transmitted by these mosquitoes. It is also not known if other species of vectors could be responsible for malaria transmission if a different collection period was adopted. Furthermore, previous studies have been carried out only in the Kapit and Lawas Districts of Sarawak, so entomological studies need to be conducted in many other localities in this large state which has a land area of 124,450 km2. Therefore, the aims of the current study were to identify vector(s) and the species of Plasmodium they transmit in the Betong District of Sarawak and to determine whether there are any vectors that feed earlier in the day.
Results
Species composition of mosquitoes
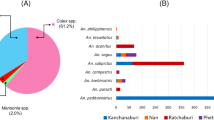
A total of 2169 mosquitoes were collected from all the collection sites from April 2015 to November 2016. Only 169 (7.8%) were identified morphologically as anophelines while the rest were culicines (92.2%) from other genera (Fig. 1a). Fourteen different species were identified and members of the Leucosphyrus and Umbrosus Groups were found to comprise more than 90% of the total Anopheles mosquitoes collected (Fig. 1b). Anopheles collessi and An. roperi were found to be the predominant species in site SM and both species were not collected during the ten collections in site B4 (Fig. 1c). An. latens was found to be the most common species found in site B4 and it was also present in site SM.
Bar chart representing the number of mosquitoes, according to morphological identification, collected in this study. Percentages of each species are shown above each bar. (a) All mosquitoes from all sites. (b) Anopheles mosquitoes from all sites. (c) Anopheles mosquitoes collected from sites B4 (blue) and SM (purple).
Identification of Plasmodium species
Only 6 out of 85 cases of malaria at Betong District in the 2 years preceding the current study were due to infections with human malaria parasites and 79 were caused by P. knowlesi. Since all 6 were infections acquired from overseas, we did not include PCR primers which could specifically detect human malaria parasites in our PCR screening. The PCR assays showed that An. latens is the vector of P. cynomolgi, P. fieldi, P. inui, and P. knowlesi while two members of the Umbrosus Goup had sporozoites of P. coatneyi, P. cynomolgi, and P. knowlesi (Table 1). Plasmodium inui and P. fieldi were detected only in An. latens and not in the other Plasmodium-positive mosquitoes. Two An. latens and two An. roperi were also each found to have two or more species of simian malaria parasites in their salivary glands. Two An. roperi and one An. collessi infected with simian malaria parasites were caught landing on humans during the morning collections. The 11 Anopheles mosquitoes which were found to have sporozoites of one or more simian malaria parasites comprised 28.2% of the 39 samples which were Plasmodium-positive by nested PCR assays.
Following PCR assays, the Plasmodium SSUrRNA genes were sequenced from these 11 samples for phylogenetic analysis. For six of these samples, Plasmodium species identified by both the PCR and ML tree were consistent but discrepancies between the two methods were observed for the remaining five samples. For example, only P. knowlesi was detected by PCR assays in samples B0870 and B1388, but P. inui- and P. fieldi-like SSUrDNA sequences, respectively, were derived from these mosquitoes. Furthermore, P. coatneyi and P. cynomolgi SSUrDNA sequences were not derived from samples BB1991 and B2000, respectively, even though these parasites were detected in these samples via nested PCR assays (Fig. 2). Multiple sequences derived from samples B0362, B1056, and B1057 which were identified by PCR assays as P. fieldi and/or P. inui were found to be closely related to those two species but the bootstrap values of the nodes were below 70%. In addition, a number of sequences were also derived from samples B1056 and B2000 which were genetically distinct from the referral sequences. Consistent with the PCR results, P. coatneyi sequences were only isolated from An. roperi.
Phylogenetic tree based on the Plasmodium SSUrRNA genes using the ML method. The analysis was based in the Tamura-3 + G + I substitution model. GenBank accession numbers are in brackets and only bootstrap values > 70% are shown at the nodes. Sequences derived from mosquitoes morphologically identified as An. collessi, An. latens and An. roperi are labelled in purple, blue and green respectively. The tree was generated with MEGA 7.0.21 (https://www.megasoftware.net/)57.
Phylogenetic analysis of partial COI gene sequences
In order to confirm morphological identification of the vectors, part of the COI gene was sequenced from the 11 Anopheles mosquitoes that were positive for simian malaria parasites by PCR assays (Table 1). In addition to these samples, 24 more An. collessi and An. roperi were also subjected to sequencing due to the lack of previous molecular characterisation of mosquitoes of the Umbrosus Group. These COI sequences were subsequently submitted to Barcode of Life Database (BOLD) for identification. When the COI gene of six morphologically identified An. latens were submitted to BOLD, three of them were identified as An. latens while the other three were identified as An. introlatus, a closely-related species within the same species complex as An. latens. The phylogenetic tree (Fig. 3) constructed with these sequences also supports the identification by BOLD, indicating that the An. introlatus (B0362, B1056, and B1283) were misidentified through taxonomic keys as An. latens. Anopheles latens were found to be infected with only P. knowlesi while An. introlatus had sporozoites of multiple simian malaria species (Table 1). None of the sequences of the members of the Umbrosus Group could be identified by BOLD. When they were aligned with the other An. leucosphyrus s.l. to infer phylogeny, these sequences formed a distinct monophyletic clade which is closely related to An. letifer (KF564694 and KF564695) collected from Singapore (Fig. 3). Mosquitoes which were morphologically identified as An. collessi and An. roperi did not form distinct clades in the ML phylogenetic tree.
Phylogenetic tree based on the Anopheles COI gene using the ML method. The analysis was based in the Tamura-3 + G substitution model. Accession numbers of An. introlatus, An. latens, and An. letifer sequences obtained from GenBank are labelled in black while those derived from mosquitoes morphologically identified as An. collesi, An. latens and An. roperi are labelled in purple, blue and green respectively. The tree was generated with MEGA 7.0.21 (https://www.megasoftware.net/)57.
Biting behaviour of vector species
As some of the morphologically identified An. latens may have been An. introlatus, their biting rates at specific time interval are not presented here but it is important to note that among the 46 collected from different sites, only one (B1444) from site SM started biting as early as 1600 h; the remaining 45 only started landing and feeding between 1800 and 2100 h. Anopheles collessi (n = 67) and An. roperi (n = 34) were found to bite throughout the collection period with peak biting times between 1700 and 1800 h and 1800–1900 h, respectively at site SM (Fig. 4). The only period where An. collessi was not landing on humans to feed was between 2000 and 2100 h while for An. roperi, it was from 0900 to 1000 h and 2000–2100 h. Despite being abundantly found in site SM, neither of these two genetically closely-related species were found in site B4, which was approximately 500 m from site SM.
Discussion
With regards to the detection of Plasmodium, observations including a higher sporozoite rate in vectors compared to other studies8,9,11,13,14,15; discrepancy between PCR and sequencing results, and the presence of Plasmodium species which could not be detected with the primers specific for the five simian malaria parasites that are reported in the current study, are consistent with the observations from our previous study in Lawas, Sarawak18. We have suggested that the high sporozoite rates observed could be attributed to a higher sensitivity of the sampling and detection methods since we used all the salivary glands for DNA extraction and we detected Plasmodium with molecular methods. Furthermore, since the Plasmodium-specific PCR assay uses primers which can amplify both the A and S types of SSU rDNA as opposed to the species-specific primers that are specific for either the A or S type, this indicates that either the number of sporozoites were low and/or the current primers could not detect other species of Plasmodium present in 28 of the Plasmodium-positive Anopheles mosquitoes. Plasmodium infection of diverse species with varying densities in these mosquitoes could cause the other two observations. It is also possible that the high Plasmodium detection rate observed is due to the close proximity of sites B4 and SM to wildlife such as long-tailed macaques, which were noted near site SM during one of the early collections.
Some of the sequences generated in our study did not fall into any particular clade following phylogenetic analysis and the reasons for this could be either that the SSUrRNA gene is not a suitable marker for identification of species of Plasmodium or that there are species of Plasmodium circulating among wildlife in Betong and Lawas that have not previously been characterised18. It would be necessary to sequence additional genes in order to confirm that there are novel species of Plasmodium carried by these mosquitoes as was done recently with non-nuclear genes5. Phylogenetic analyses of the Plasmodium mitochondrial genome and apicoplast caseinolytic protease M gene revealed the existence of two subpopulations of P. inui-like parasites in long-tailed macaques5 in Sarawak and that these macaques are new hosts for P. simiovale. In addition, microsatellite genotyping and whole genome sequencing has shown that there are three P. knowlesi subpopulations present in human patients and laboratory-maintained P. knowlesi strains from macaques6,33. Sequences labelled as P. cf. inui, P. cf. fieldi, and P. sp. in Fig. 2 were all unidentified sequences derived from long-tailed macaques in the Kapit Division of Sarawak34. These observations suggest that sequences which did not fall into a particular clade following phylogenetic analysis in the current study possibly represent novel Plasmodium species. However, it is not known at this stage, if these Plasmodium species would pose any threat to human health.
Phylogenetic analysis revealed three An. introlatus had probably been misidentified through taxonomic keys as An. latens and it was not possible to distinguish between the samples which were morphologically identified as An. collessi and An. roperi. This inability to differentiate An. collessi from An. roperi could be due to the short lengths of the COI sequences that were used for the phylogenetic analysis. Although the primers used in the present study generated fragments of approximately 1400 bp of the COI gene following PCR amplification, only 700 nucleotides were aligned during phylogenetic analyses because longer reference sequences were not available in GenBank. If a larger fragment of the gene was used in the alignment, the phylogenetic analysis may have had a greater capacity in determining the identity of the species of these mosquitoes. It is also possible that the COI gene is not a suitable marker for distinguishing different species in certain species complexes. This was demonstrated by Carter et al.35 in their recent attempt to use both the ITS2 and COI sequences to identify mosquito species in Ethiopia. They successfully distinguished two distantly related species, An. arabiensis and An. pretoriensis by using ITS2 but not with COI sequences. In a separate study conducted in South Sulawesi, Anopheles mosquitoes were identified morphologically to their genera and an approximately 700 nucleotide fragment of the COI gene was sequenced from each individual (n = 392)36. The sequences were aligned and were found to have separated into nine different contigs, suggesting the presence of nine species/species groups among the samples collected. By comparing the consensus sequences of the contigs to GenBank and BOLD, the identities of seven groups were determined while each of the remaining two groups had high sequence similarities (> 95%) to two distantly related species. This further demonstrates that parts of the COI gene of distantly related species could have high sequence similarity and that the gene could not be used by itself to reliably infer phylogeny. No other DNA barcodes (e.g. ITS2, COII, etc.) were sequenced for the An. collessi and An. roperi in the present study however, as sequences in the Umbrosus Group were not available in GenBank for use as reference sequences. Nevertheless, our data indicates that An. latens, An. introlatus, An. collessi, and An. roperi are vectors of P. knowlesi and/or other simian malaria parasites in the Betong District of Sarawak.
We have incriminated An. roperi and An. collessi, mosquitoes of the Umbrosus Group, as novel vectors of P. knowlesi and other simian malaria parasites in Malaysian Borneo. From the limited studies that have been undertaken, both these species have been found in several regions in Asia: An. collessi37,38 in Peninsular Malaysia, Malaysian Borneo, and Brunei, and An. roperi39 in Peninsular Malaysia, Malaysian Borneo, Thailand, Cambodia, and the Nicobar Island of India. There is a need for further studies on the geographic distribution of these vectors in order to provide specific guidelines for vector control strategies40,41,42. While P. knowlesi has been previously detected in the salivary glands and midguts of An. latens and An. introlatus12,15, respectively, this is the first time the parasite was identified in the salivary glands of An. introlatus, An. collessi and An. roperi. This is also the first time members of the Umbrosus Group are incriminated as vectors for P. knowlesi. Previous studies have indicated the role of the Umbrosus Group in the transmission of human and non-human malaria parasites in Malaysia. In Peninsular Malaysia in 1918, Barber43 had noticed "in some specimens of An. umbrosus s.l. the sporozoites appeared abnormally thick and short, and in other specimens they were apparently normal" while Hodgkin in 195644 suspected some of the An. umbrosus s.l. might have been infected with monkey malaria. Plasmodium traguli, the malaria parasite of the mouse deer (Tragulus kanchil), was able to develop into sporozoites in Anopheles umbrosus s.s., An. letifer, An. baezai, and An. roperi that had been bloodfed using infected mouse deer45. Two other investigations have also reported the presence of oocysts and/or sporozoites (parasite identity was not mentioned) from dissected An. baezai, An. collessi, An. letifer, An. separatus, An. roperi, and An. umbrosus caught in nature41,46. In Sarawak, the salivary glands of An. letifer have been found to be infected with what the authors described as malaria parasites of non-human and human origin in Baram in the 1950s and in Miri in 1997, respectively, but they did not provide descriptions of the morphological differences between these sporozoites47,48. Taken together, these studies show that some members of the Umbrosus Group are able to transmit simian and other malaria parasites, although the exact identity of the species of Plasmodium was not stated in certain studies.
Several members of the Leucosphyrus Group (An. cracens, An. latens, An. introlatus, and An. balabacensis in Malaysia; An. dirus in Vietnam) and An. donaldi (Malaysia) have previously been incriminated in the transmission of P. knowlesi and other simian malaria parasites in Southeast Asia8,9,11,12,13,14,15,16,18,49. In all the studies where peak biting times were described, An. balabacensis8,11,49, An. cracens13,14, An. donaldi11, and An. introlatus12 were reported to be mostly landing on humans between 1800 and 2100 h but were already biting within the first hour of the start of the collection period in the evenings. Anopheles latens was reported to have peak biting activity between 1900–2000 h and 0100–0200 h in the forest and farm settings, respectively, in the Kapit district of Sarawak, Malaysian Borneo but hourly peak biting rates were not provided50. The peak biting rates for An. balabacensis (approximately 0.18 bites/man/h at 1900–2000 h) and An. donaldi (approximately 0.35 bites/man/h at 1800–1900 h) determined from 6-h night collections in a study at Ranau and Keningau in Sabah, Malaysian Borneo11 appear to be lower than our peak biting rates for An. collessi (approximately 0.94 bites/man/h at 1700–1800 h and 0.60 at 0800–0900 h) and An. roperi (0.50 bites/man/h at 1800–1900 h and approximately 0.29 at 0700–0800 h). However, we only collected mosquitoes from 0600 to 1100 h and 1600–2100 h, so we cannot rule out the possibility that at other time periods, An. collessi and An. roperi may have peak biting rates greater or lower than those we observed. The other investigations for vectors of knowlesi malaria involved 12 h-long mosquito collections at night, where the peak biting times were described as bites/man/night for each month and ranged from approximately 0–13 for An. latens in Kapit, Sarawak50, approximately 1–8 for An. cracens in Kuala Lipis, Peninsular Malaysia13, and approximately 2–28 for An. balabacensis in Banggi Island and Kudat, Sabah8. It is necessary to conduct further studies involving longer durations of collection times and monthly mosquito collections lasting 3 days each for over a year before valid comparisons of peak biting rates of mosquitoes at Betong can be made with those described by other investigators. Such studies will lead to a greater understanding of the bionomics of An. collessi and An. roperi.
Previous entomological surveys for P. knowlesi vectors have all been conducted between dusk and dawn (for a 6- or 12-h period starting from 1800 h or 1900 h) to coincide with what was widely accepted as the period when most malaria vectors were seeking a bloodmeal8,9,11,12,13,14,15,16,49,50. However, that widely held belief was derived mostly from indoor collections which reported the absence of endophagic Anopheles mosquitoes decades ago when human-to-human malaria transmission was the prevalent mode of transmission41,51,52. This assumption is therefore not applicable to vectors of P. knowlesi where transmission predominantly occurs outdoors2,8,9,11,13,14,15,50. Among the malaria vectors in Malaysia, An. letifer, An. umbrosus s.s., An. roperi, and An. donaldi have been previously shown to feed on humans in forested shade when disturbed at their resting sites between 0600 and 1200 h41,53,54. While the dusk-to-dawn sampling period has enabled the incrimination of An. leucosphyrus s.l., and more recently An. donaldi11,18, as vectors of knowlesi malaria, it is possible that other vectors could have been discovered if collections were also conducted during the 12-h period between dawn to dusk. With the incrimination of An. collessi and An. roperi, which have peak biting times at 0700–0900 h and 1700–1900 h in Betong, dawn-to-dusk mosquito collections need to be utilised in future studies for a better understanding of the vectors and the transmission dynamics of knowlesi malaria.
In conclusion, this study has incriminated two members of the Umbrosus Group (An. collessi and An. roperi) and two members of the Leucosphyrus Complex (An. latens and An. introlatus) as vectors of P. knowlesi and other simian malaria parasites in Betong, Sarawak, Malaysian Borneo. Phylogenetic analyses of the Plasmodium SSUrRNA sequences derived from the salivary glands of these mosquitoes have indicated these vectors may also transmit novel species of Plasmodium. This is the first time members of the Umbrosus Group, which feed on humans in the evening as well as in the morning, were shown to harbour simian malaria parasites, underscoring the importance of also conducting entomological surveys during the daytime in order to obtain a comprehensive understanding of vectors and the transmission dynamics of malaria.
Methods
Study sites and collection periods
The study was carried out in the Betong District of Sarawak where 93% (79/85) of human malaria infections were caused by P. knowlesi in the two years preceding the start of the study in 2015. The other six were caused by human malaria parasites and all these infections had been acquired overseas (Betong Health Department, unpublished data). Mosquito collection sites were selected during the initial stage of the study based on the following criteria: (a) occurrence of P. knowlesi infection in the longhouse community one month prior to collection; (b) within close proximity to areas where long-tailed and/or pig-tailed macaques had been sighted and; (c) farm/hunting ground where the person infected with P. knowlesi had potentially acquired the infection. Mosquitoes were collected for 36 days during 8 field trips in the months of April, August, and October 2015, and in March, April, May, August, and November 2016. Although multiple collection sites near to 5 longhouses (see Supplementary Table S1 and Fig. 5) were selected in the beginning of the study, most of the local guides (previously infected with P. knowlesi) from the longhouses were not always available to guide the team to the collection sites. Sites B4 and SM close to the Bungkang longhouse were therefore selected as the main collection sites due to the availability of a local guide and the presence of a higher number of anophelines compared to the other sites from the initial surveys. These two sites were also the only sites where both the early (0600–1100 h) and late (1600–2100 h) collections were carried out, while in the other sampling locations only the late collections were conducted. Site B4 was a slope on a hill situated approximately 600 m southeast of the Bungkang longhouse. The area was surrounded by Eugeissona insignis, a flowering plant in the palm family which produces fruits that macaques feed on, according to the local communities. Located between site B4 and the longhouse was an approximately 5-m long stream which most villagers had to cross to get to their farms. Site SM, approximately 1 km east of Bungkang, was situated near Sungai Malaban, a slow-moving stream. This site was a favourite hunting area for the villagers and long-tailed macaques were sighted during one of the early collections. Both sites B4 and SM were secondary forests with little vegetation on the forest ground and were approximately 500 m apart. A total of 22 mosquito collections were undertaken at these two sites (B4: eight late and two early collections; SM: six late and six early collections).
Map showing the five villages (black circles) of mosquito collection sites relative to Betong town and inset map showing where previous studies (yellow triangles) on vectors of knowlesi malaria were conducted in Sarawak. The images were obtained from Google Earth Pro 7.3.3.7786 (https://www.google.com/intl/en_uk/earth/versions/#earth-pro)58.
Mosquito collection, identification and dissection
Prior to each collection, tissue moistened with distilled water was placed on the base of each 50 mm × 19 mm cylindrical specimen tubes (SAMCO, UK) followed by plugging the tube opening with a ball of cotton wool. All mosquitoes were collected using the bare-leg catch method where any mosquitoes found landing on the legs of the collectors were trapped using the specimen tubes. The mosquitoes were brought to the field laboratory for morphological identification where the non-anophelines were identified to their genera while the anophelines were identified to their species/group using taxonomic keys41,42,55. Salivary glands of each of the Anopheles were dissected and preserved in individual 1.5-mL microcentrifuge tubes (AXYGEN, USA) containing 0.5 mL absolute alcohol. The preserved salivary glands were transported to the main laboratory at Universiti Malaysia Sarawak under room temperature. This study was approved by the Medical Ethics Committee of Universiti Malaysia Sarawak and by the Medical Research and Ethics Committee, Ministry of Health Malaysia (NMRR-10-1194-7854). All field staff and volunteers who carried out mosquito collections were provided with antimalarial prophylaxis.
DNA extraction and PCR detection of Plasmodium species
Absolute alcohol preserving the salivary glands was first dried prior to DNA extraction. Genomic DNA of the dried salivary glands was extracted using DNeasy Blood and Tissue Kit (QIAGEN, Germany) according to the manufacturer's protocol and stored at 4 °C until required. Nested PCR assays to detect Plasmodium DNA were initially undertaken and Plasmodium-positive samples were then subjected to PCR assays with primers specific for P. coatneyi, P. cynomolgi, P. fieldi, P. inui and P. knowlesi as described previously18.
Generating Plasmodium SSUrRNA and Anopheles COI gene amplicons for cloning and sequencing
The SSUrRNA genes were amplified by PCR assays using primers rPLU1 and rPLU556 while amplicons for Anopheles COI were generated with primers SCOIF (5′-GGA TTT GGA AAT TGA TTA GTT CCT T-3′) and AnCOX1R (5′-CCT AAA TTT GCT CAT GTT GCC-3′) using the Phusion High-Fidelity DNA Polymerase (THERMO FISHER SCIENTIFIC, USA) as described previously18 to produce ~ 1405 bp-long blunt-ended amplicons for blunt-end cloning. Amplicons generated were then cloned and sequenced according to the protocol described by Ang et al.18. For the COI inserts, internal primer AnCOX1F (5′-CTA GTG TGC TTC CCA TGG AGA TAG-3′) was used for sequencing.
Sequence alignment and phylogenetic analysis
Multiple sequences generated from this study and those obtained from GenBank were aligned using the default parameters of ClustalW within the LaserGene 7.1 programme (DNASTAR, USA). Reference sequences obtained from GenBank are listed in the additional file (see Supplementary Tables S2 and S3). The best nucleotide substitution models were calculated using MEGA 7.0.21 and the models with the lowest Bayesian Information Criterion (BIC) score were selected. Subsequently, phylogenetic trees were constructed by the Maximum Likelihood (ML) method using MEGA 7.0.21 with bootstrap values calculated from 1000 replicates57.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Coatney, G. R., Collins, W. E., Warren, M. & Contacos, P. G. The Primate Malarias (CDC, 2003).
Singh, B. et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363, 1017–1024 (2004).
Hussin, N. et al. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar. J. 19, 55 (2020).
Lee, K. S. et al. Plasmodium knowlesi: Reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 7, e1002015 (2011).
Nada Raja, T. et al. Malaria parasites of long-tailed macaques in Sarawak, Malaysian Borneo: A novel species and demographic and evolutionary histories. BMC Evol. Biol. 18, 1–12 (2018).
Divis, P. C. S. et al. Admixture in humans of two divergent Plasmodium knowlesi populations associated with different macaque host species. PLoS Pathog. 11, 1–17 (2015).
Divis, P. C. S. et al. Three divergent subpopulations of the malaria parasite Plasmodium knowlesi. Emerg. Infect. Dis. 23, 616–624 (2017).
Wong, M. L. et al. Seasonal and spatial dynamics of the primary vector of Plasmodium knowlesi within a major transmission focus in Sabah, Malaysia. PLoS Negl. Trop. Dis. 9, 1–15 (2015).
Chua, T. H., Manin, B. O., Daim, S., Vythilingam, I. & Drakeley, C. Phylogenetic analysis of simian Plasmodium spp. infecting Anopheles balabacensis Baisas in Sabah, Malaysia. PLoS Negl. Trop. Dis. 11, 1–13 (2017).
Wharton, R. H. & Eyles, D. E. Anopheles hackeri, a vector of Plasmodium knowlesi in Malaya. Science 134, 279–280 (1961).
Hawkes, F. M. et al. Vector compositions change across forested to deforested ecotones in emerging areas of zoonotic malaria transmission in Malaysia. Sci. Rep. 9, 1–12 (2019).
Vythilingam, I. et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): Epidemiologic and entomologic analysis. Parasites Vectors 7, 436 (2014).
Jiram, A. I. et al. Entomologic investigation of Plasmodium knowlesi vectors in Kuala Lipis, Pahang, Malaysia. Malar. J. 11, 213 (2012).
Vythilingam, I. et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasites Vectors 1, 1–10 (2008).
Vythilingam, I. et al. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans. R. Soc. Trop. Med. Hyg. 100, 1087–1088 (2006).
Marchand, R. P., Culleton, R., Maeno, Y., Quang, N. T. & Nakazawa, S. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes, Southern Vietnam. Emerg. Infect. Dis. 17, 1232–1239 (2011).
Vidhya, P. T., Pulikkottil, I., Maile, A. & Zahid, A. K. Anopheles sundaicus mosquitoes as vector for Plasmodium knowlesi, Andaman and Nicobar Islands, India. Emerg. Infect. Dis. 25, 817–820 (2019).
Ang, J. X. D. et al. New vectors in northern Sarawak, Malaysian Borneo, for the zoonotic malaria parasite, Plasmodium knowlesi. Parasites Vectors 13, 472 (2020).
Van Bortel, W. et al. Confirmation of Anopheles varuna in Vietnam, previously misidentified and mistargeted as the malaria vector Anopheles minimus. Am. J. Trop. Med. Hyg. 65, 729–732 (2001).
Paredes-Esquivel, C., Donnelly, M. J., Harbach, R. E. & Townson, H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris Subgroup reveals cryptic species: Implications for identification of disease vectors. Mol. Phylogenet. Evol. 50, 141–151 (2009).
Sungvornyothin, S., Garros, C., Chareonviriyaphap, T. & Manguin, S. How reliable is the humeral pale spot for identification of cryptic species of the Minimus Complex?. J. Am. Mosq. Control Assoc. 22, 185–191 (2006).
Dusfour, I. et al. Polymerase chain reaction identification of three members of the Anopheles sundaicus (Diptera: Culicidae) Complex, malaria vectors in Southeast Asia. J. Med. Entomol. 44, 723–731 (2007).
Manguin, S., Garros, C., Dusfour, I., Harbach, R. E. & Coosemans, M. Bionomics, taxonomy, and distribution of the major malaria vector taxa of Anopheles subgenus Cellia in Southeast Asia: An updated review. Infect. Genet. Evol. 8, 489–503 (2008).
Chan, A. et al. DNA barcoding: Complementing morphological identification of mosquito species in Singapore. Parasites Vectors 7, 12 (2014).
Takano, K. T. et al. Partial mitochondrial DNA sequences suggest the existence of a cryptic species within the Leucosphyrus Group of the genus Anopheles (Diptera: Culicidae), forest malaria vectors, in northern Vietnam. Parasites Vectors 3, 1–16 (2010).
Walton, C. et al. Identification of five species of the Anopheles dirus Complex from Thailand, using allele-specific polymerase chain reaction. Med. Vet. Entomol. 13, 24–32 (1999).
Sallum, M. A. M., Foster, P. G., Li, C., Sithiprasasna, R. & Wilkerson, R. C. Phylogeny of the Leucosphyrus Group of Anopheles (Cellia) (Diptera: Culcidae) based on mitochondrial gene sequences. Ann. Entomol. Soc. Am. 100, 27–35 (2007).
Phuc, H. K. et al. Multiplex PCR assay for malaria vector Anopheles minimus and four related species in the Myzomyia Series from Southeast Asia. Med. Vet. Entomol. 17, 423–428 (2003).
Surendran, S. N. et al. Molecular characterization and identification of members of the Anopheles subpictus Complex in Sri Lanka. Malar. J. 12, 1 (2013).
Goswami, G. et al. Identification of all members of the Anopheles culicifacies Complex using allele-specific polymerase chain reaction assays. Am. J. Trop. Med. Hyg. 75, 454–460 (2006).
Ma, Y., Li, S. & Xu, J. Molecular identification and phylogeny of the Maculatus Group of Anopheles mosquitoes (Diptera: Culicidae) based on nuclear and mitochondrial DNA sequences. Acta Trop. 99, 272–280 (2006).
Brosseau, L. et al. A multiplex PCR assay for the identification of five species of the Anopheles barbirostris Complex in Thailand. Parasit. Vectors 12, 223 (2019).
Assefa, S. et al. Population genomic structure and adaptation in the zoonotic malaria parasite Plasmodium knowlesi. Proc. Natl. Acad. Sci. U. S. A. 112, 13027–13032 (2015).
Divis, P. C. S. Identification and Molecular Characterisation of Malaria Parasites of Macaques in Kapit, Sarawak. (Universiti Malaysia Sarawak, 2007).
Carter, T. E., Yared, S., Hansel, S., Lopez, K. & Janies, D. Sequence-based identification of Anopheles species in eastern Ethiopia. Malar. J. 18, 135 (2019).
Davidson, J. R. et al. Characterization of vector communities and biting behavior in South Sulawesi with host decoy traps and human landing catches. Parasit. Vectors 13, 329 (2020).
Cheong, W. H., Loong, K. P., Mahadevan, S., Mak, J. W. & Kan, S. K. Mosquito fauna of the Bengkoka Peninsula, Sabah, Malaysia. Southeast Asian. J. Trop. Med. Public Health 15, 19–26 (1984).
Walter Reed Biosystematics Unit. Anopheles collessi species page. http://wrbu.si.edu/vectorspecies/mosquitoes/collessi (2021).
Walter Reed Biosystematics Unit. Anopheles roperi species page. http://wrbu.si.edu/vectorspecies/mosquitoes/roperi (2021).
Das, M. K., Nagpal, B. N. & Ansari, M. A. Mosquito fauna and breeding habitats of anophelines in Little Andaman Island, Andaman and Nicobar Islands, India. Indian J. Malariol. 39, 83–95 (2002).
Reid, J. A. Anophelines Mosquitoes of Malaya and Borneo (Staples Printers Limited, 1968).
Rattanarithikul, R., Harrison, B. A., Harbach, R. E., Panthusiri, P. & Coleman, R. E. Illustrated keys to the mosquitoes of Thailand IV. Anopheles. Southeast Asian J. Trop. Med. Public Health 37, 1–128 (2006).
Barber, M. A. Some observations and experiments on Malayan Anopheles with special reference to the transmission of malaria. Phillipine J. Sci. 13, 1–46 (1918).
Hodgkins, E. P. The Transmission of Malaria in Malaya (Caxton Press, 1956).
Wharton, R. H., Eyles, D. E., Warren, M., Moorhouse, D. E. & Sandosham, A. A. Investigations leading to the identification of members of the Anopheles umbrosus Group as the probable vectors of mouse deer malaria. Bull. World Health Organ. 29, 357–374 (1963).
Hodgkins, E. P. The Anopheles Umbrosus Group (Diptera: Culicidae). Part II: Biology and transmission of malaria. Trans. R. Entomol. Soc. Lond. 101, 319–334 (1950).
Zulueta, J. D. Malaria in Sarawak and Brunei. Bull. World Health Organ. 15, 651–671 (1956).
Chang, M. S., Hii, J., Buttner, P. & Mansoor, F. Changes in abundance and behaviour of vector mosquitoes induced by land use during the development of an oil palm plantation in Sarawak. Trans. R. Soc. Trop. Med. Hyg. 91, 382–386 (1997).
Manin, B. O. et al. Investigating the contribution of peri-domestic transmission to risk of zoonotic malaria infection in humans. PLoS Negl. Trop. Dis. 10, e0005064 (2016).
Tan, C. H., Vythilingam, I., Matusop, A., Chan, S. T. & Singh, B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar. J. 7, 52 (2008).
Gater, B. A. R. Notes on Malayan mosquitoes. II: Seasonal distribution. Med. J. Malaya 8, 43–45 (1930).
Wallace, R. B. Insecticides and A. maculatus. Med. J. Malaya 3, 5–33 (1948).
Moorhouse, D. E. & Wharton, R. H. Studies on Malayan vectors of malaria; methods of trapping, and observations on biting cycles. J. Med. Entomol. 1, 359–370 (1965).
Thevasagayam, E. S. & Fah, L. C. Studies on the biology of Anopheles letifer Sandosham (Diptera, Culicidae) and its response to residual spraying, carried out in Sarawak, Malaysia. Med. J. Malaysia 34, 254–264 (1980).
Rattanarithikul, R., Harrison, B. A., Panthusiri, P. & Coleman, R. E. Illustrated keys to the mosquitoes of Thailand I. Background; geographic distribution; lists of genera, subgenera, and species; and a key to the genera. Southeast Asian. J. Trop. Med. Public Health 36, 59–69 (2005).
Singh, B. et al. A genus-and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am. J. Trop. Med. Hyg. 60, 687 (1999).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Google Earth Pro. Map data of Betong and Sarawak. https://www.google.com/intl/en_uk/earth/versions/#earth-pro (2020).
Acknowledgements
We would like to thank staff (especially Walter ak Gary and Rahynold Rehengia ak Gagat) from the Sarawak Health Department for their assistance during the field trips and Jingong ak Jackson from Betong who guided us into the forest for most of the field trips. We also thank the Director General of Health in Malaysia for permission to publish this article. This study was supported by research grants from the Ministry of Higher Education in Malaysia (FRGS/1/2014/SKK01/UNIMAS/03/1), Universiti Malaysia Sarawak (Grant no.: F05/SpFRC/1433/16/1, F05/DPP/1508/2016), and by a postgraduate scholarship to JXDA from the Ministry of Higher Education in Malaysia.
Funding
Open Access funding provided by Universiti Malaysia Sarawak.
Author information
Authors and Affiliations
Contributions
B.S. and K.Y. designed the research. J.X.D.A. and K.Y. carried out the fieldwork. J.X.D.A. and B.S. analysed the results and wrote the manuscript. J.X.D.A. and K.A.K., under the supervision of B.S., carried out the molecular studies. K.Y. organised the fieldwork logistics while A.M. organised and supervised staff from Sarawak Department of Health involved in the field trips. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
De Ang, J.X., Yaman, K., Kadir, K.A. et al. New vectors that are early feeders for Plasmodium knowlesi and other simian malaria parasites in Sarawak, Malaysian Borneo. Sci Rep 11, 7739 (2021). https://doi.org/10.1038/s41598-021-86107-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-86107-3
This article is cited by
-
Spatial analyses of Plasmodium knowlesi vectors with reference to control interventions in Malaysia
Parasites & Vectors (2023)
-
The potential for zoonotic malaria transmission in five areas of Indonesia inhabited by non-human primates
Parasites & Vectors (2023)
-
Natural vectors of Plasmodium knowlesi and other primate, avian and ungulate malaria parasites in Narathiwat Province, Southern Thailand
Scientific Reports (2023)
-
Population genetic analysis of Plasmodium knowlesi reveals differential selection and exchange events between Borneo and Peninsular sub-populations
Scientific Reports (2023)
-
Reimagining zoonotic malaria control in communities exposed to Plasmodium knowlesi infection
Journal of Physiological Anthropology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.