Abstract
Triple negative breast cancer (TNBC) accounts for about a fifth of all breast cancers and includes a diverse group of cancers. The heterogeneity of TNBC and the lack of target receptors on the cell surface make it difficult to develop specific therapeutic treatments. These aspects cause the high negative prognosis of patients with this type of tumor. The analysis of the molecular profiles of TNBC samples has allowed a better characterization of this tumor, supporting the search for new reliable diagnostic markers. To this end, we have developed a bioinformatic approach to integrate networks of genes differentially expressed in basal breast cancer compared to healthy tissues, with miRNAs able to regulate their expression. We studied the role of these miRNAs in TNBC subtype cell lines. We therefore identified two miRNAs, namely miR-135b and miR-365, with a central role in regulating the altered functional pathways in basal breast cancer. These two miRNAs are differentially expressed in human TNBC immunohistochemistry-selected tissues, and their modulation has been shown to play a role in the proliferation of tumor control and its migratory and invasive capacity in TNBC subtype cell lines. From the perspective of personalized medicine, we managed to modulate the expression of the two miRNAs in organotypic cultures, suggesting their possible use as diagnostic and therapeutic molecules. miR-135b and miR-365 have a key role in TNBC, controlling proliferation and invasion. Their detection could be helpful in TNBC diagnosis, while their modulation could become a new therapeutic tool for TNBC.
Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC) is one of the four molecular immunohistochemical subtypes of breast cancer (BC) described in comparison to normal epithelium1. It represents a percentage of 10–20% of all BC tumors2. The overall 5-year survival rate for TNBC is 50–60%. The recurrence peak occurs within the first 3–5 years from diagnosis. On the contrary, late recurrences decline over the following 5 years3.
What is described by immunohistochemistry (IHC) as TNBC subtype, defined by negative immunohistochemical staining for estrogen receptor (ER) and progesterone receptor (PR) and lack of human epidermal growth factor receptor 2 (Her2/neu) overexpression4, includes a heterogeneous group of tumors from a genetic point of view, when compared to both normal tissue and other types of BC5. The classification given by the IHC analysis is limited to the description of the tumor, based on the presence on the cell membrane of hormone receptors (ER, Pr and HER2). The molecular classification, analyzing the expression level of the mRNAs, indicates the triple negative subtype with the name of basal (see6), which is characterized by the expression mainly of cytokeratin 5/6, 14 and 17, of laminin and of lipid protein binding 27.
However, a single set of markers that specifically describes basal BC does not yet exist. Clustering algorithms applied on molecular profiles are able to find further subclasses also in the basal subtype, but these subcategorizations require a greater number of samples to be better defined. Moreover, numerous variables could affect the subcategories of the basal subtype, drawing attention to the need of a standardized method and substantially large databases to define molecular classes of basal breast cancer. Nevertheless, there is a good degree of overlap between the molecular definition of basal and the description of TNBC defined by the IHC (about 80%)8. This also justifies the fact that both classifications are able to predict the worse prognosis of this tumor subtype, when they encounter it: the basal subtype, as well as TNBC, are characterized by high histological grade, high mitotic index and low differentiation and are often associated with the presence of metastases and lymph node involvement9.
The higher heterogeneity of this tumor type and the lack of IHC target receptor (ER, PR, HER2) resulted in the absence of FDA-approved target therapies for TNBC10. Because of the poor prognosis observed in TNBC patients and the lack of approved therapeutic treatment (see Sacituzumab/govitecan approval for TNBC from FDA, May 2020), this subtype accounts for a disproportionate number of metastatic cases and breast cancer deaths.
The possibility of using molecular profiling to better characterize and diagnose the TNBC subtype with higher accuracy would allow the comprehension of the altered molecular mechanisms in this specific subtype of BC.
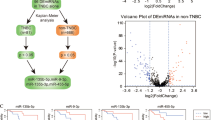
In order to identify new possible epigenetic molecules able to diagnose molecularly TNBC, we developed an in silico approach that identifies differentially expressed genes (DEGs) of basal BC compared to normal tissues; this information is integrated with functional pathways, and biological relationships among cellular components (network), also taking into account the microRNAs’ (miRNAs) regulatory role on the network (Fig. 1a). Indeed, miRNAs are small non-coding RNA with a post-transcriptional regulatory role on several mRNAs involved in coding proteins performing cancer hallmarks’ functions11,12. With this approach, we identified a small group of miRNAs able to control differentially expressed mRNA, specifically altered in basal BC, involved in specific functions of the tumor network. As miRNAs are cell- and tumor specific13, and as each miRNA belongs to a complex network of interactions, the alteration of miRNA expression levels could severely impact on the target mRNAs and, consequently, on the tumor behavior. In this study, we characterized the biological role of TNBC-associated miRNAs and found that Hsa-miR-135b (miR-135b) and Hsa-miR-365 (miR-365) have diagnostic properties, being able to classify IHC-selected TNBC samples versus normal tissues.
Overview of the data, methods (a) and results (b) for TBNC network identification (A) Workflow of the proposed computational approach. It consists of 6 steps. (1) identification of differentially expressed genes (DEGs) between basal and normal samples from TCGA data, (2) pathway enrichment analysis. In this step we identified the pathways enriched with DEGs, (3) construction of a classifier to distinguish basal versus normal samples using a metric able to reveal the interactions among pathways, (4) the classifier was evaluated using Area Under Curve (AUC) in the training and testing test, (5) we selected the pairs of pathways that achieved the best AUC values, (6) we applied an integration algorithm between miRNA and pathway in order to identify miRNAs regulating pathway network in basal breast cancer. (B) miRNAs regulating pathway network. The figure shows the top 10 pairs of pathways that achieved the best performance in the classification of basal versus normal samples and miRNAs regulating this network.
Furthermore, their altered expression, i.e. the increase of miR-135b and the decrease of miR-365, supported the aggressive, metastatic phenotype of TNBC cells, controlling their proliferation, migration and invasion. The efficacy of transfection of synthetic oligonucleotide in the cells could suggest their potential use as possible therapeutic agent.
Results
Computational approach
The computational analysis, performed to identify the pathways to which the basal-descriptive genes belong to and to integrate this information into a functional network, consists of 6 steps (Fig. 1a). In the first step, we identified and selected a set of DEGs between basal and NS; in the second step, we identified pathways enriched with DEGs; in the third step, we generated a classifier to distinguish basal versus NS using the pathway metric.
The first three analyses (identification of DEG, pathway enrichment analysis and the classification) were repeated 50 times on different generated training dataset.
In the fourth and fifth step, the classifier was evaluated with performance indices and the features that obtained the best results are selected and tested on the testing dataset. In the last step we applied an integration algorithm between miRNA and pathway network in order to identify a basal BC network consisting of miRNAs regulating a pathway network.
Step 1: Identification of differentially expressed genes
In order to identify differentially expressed genes between basal and NS, a statistical analysis using TCGA biolinks package was applied14, considering a gene differentially expressed when its |logFC|> 1 and p value < 0.01. The p values were adjusted by the Benjamin–Hochberg method for multiple testing corrections15.
Step 2: Pathway enrichment analysis
Once the DEGs were identified, the pathway enrichment algorithm was applied to verify in which of the 589 pathways from the Ingenuity Pathway Analysis (IPA) database the DEGs were inserted. The algorithm was performed using the Fisher's Exact Test; we considered a pathway to be enriched with DEGs if the associated adjusted p value was < 0.01.
Step 3: Build the classifier
The strength of the interactions among pathways were indicated by calculating a discriminating score, as reported in16. This score, which indicates the relationship between pairs of pathways, was calculated among all possible pairs of pathways enriched by the differentially expressed genes.
A machine-learning model (Random Forest classification) was developed using this score for each sample in order to build a network able to classify basal and NS.
Step 4 and 5: Evaluation and features selection
AUC was estimated as performance index. We considered for each of the 50 bootstraps the top 10 pairs of pathways that achieved the best performance in the training dataset. The testing dataset was used to validate the top 10 pairs of pathways obtained in the training step. At the end of all 50 bootstraps we selected the best top 10 pairs of pathways.
Step 6: Network identification
Once the network consisted of 10 pairs of pathways was obtained, by applying the mutual information and Fisher's exact test we identified those miRNAs that are able to regulate a significant number of genes in the pathway network. To each of these miRNAs we associated an index, namely the degree centrality (DC): miRNAs with higher DC (HDC miRNAs) were those that regulate the highest number of genes within functional coupled-pathways.
miR-365 and miR-135b have a high degree centrality in TNBC
The couple pathways that achieved the best performance in the classification of basal versus normal samples (NS) were listed in Fig. 1b. The interactions among pathways were identified by a discriminating score able to distinguish with the best performances basal versus NS, while miRNAs regulating the network were obtained in the last step of computational approach using mutual information and Fisher's exact test.
Ethanol degradation, DNA damage response, mismatch repair and cell cycle are the main functions emerging within the network. Indeed, in both training and testing dataset the network obtained a AUC with a median > 95%.
The in silico analysis revealed that 2 miRNAs are altered in TNBC, miR-365 and miR-135b with a key role in the network (Table 1). A schematic view of the regulatory interaction among miRNAs and coupled pathways is depicted in Fig. 1b. In particular, miR-365 has a degree centrality of 42, controlling 42 over 96 genes within the 2 couple pathways (Putrescin degradation, Role of BRCA1 in DNA damage response and Tryptophan degradation X), while miR-135b has a degree centrality of 12, controlling 12 over 35 genes within 1 couple pathways (Ethanol degradation X and Mismatch repair in eukaryotes).
Regarding the expression levels of these two miRNAs, in silico analysis suggested that miR-365 is downregulated in basal tumor cells compared to normal tissue17, while miR-135b is upregulated in basal tumor cells compared to normal tissue18.
miR-135b and miR-365 target analysis
Looking in more depth to the genes controlled by these two miRNAs in each pathway using mutual information, we identified ALDH family members (ALDH1A1/1A3, ALDH2), IL4I1, MAOA, SMOX belonging to ‘Tryptophane degradation X’ and ATR, CHEK2, E2F1, FANC family members (FANCA/B/C/E), MLH1, MSH6, NBN, PLK1, RB1, RBBP8, RBL1, RFC5 belonging to ‘Role of BRCA1 in DNA damage response’ as possible targets of miR-365, and ACSL1, ACSL3, ALDH1A1/1B1/3A2/4A1 belonging to ‘Ethanol degradation IV’ and FEN1, MSH6, RFC2, RFC4, RPA1, SLC19A1 belonging to ‘Mismatch repair in eukaryotes’ as possible targets of miR-135b (Table 2).
miR-135b and miR-365 are differentially expressed in basal cell line
The in silico analysis predicted miR-135b upregulation and miR-365 downregulation in basal breast cancer compared to normal tissue. To validate the level of expression of these two miRNAs, we performed a RT-qPCR analysis of both miRNAs in two TNBC cell lines, namely BT20 and MDA-MB-231, compared to MCF10A (Fig. 2). The results confirm miR-135b upregulation in BT20 and MDA-MB-231 compared to MCF10A normal-like cell line, with fold change values of 4.70 ± 0.89 in BT20, 2.93 ± 0.56 in MDA-MB-231 compared to MCF10A normal-like cell line (t test p value = 0.018 in BT20 and 0.019 in MDA-MB-231 compared to MCF10A, respectively). RT-qPCR analysis confirmed also miR-365 downregulation in BT20 and MDA-MB-231, compared to MCF10A normal-like cell line, with fold change values of 0.044 ± 0.010 in BT20, 0.011 ± 0.007 in MDA-MB-231, respectively (t test p value = 3.78 × 10–5 in BT20 and p value = 0.0032 in MDA-MB-231 compared to MCF10A). A further confirm was obtained by in silico analysis of different GEO datasets on several cell lines (supplementary Figures 1–3).
Validation of miRNA expression in MDA-MB-231 basal cell line model. miR-135b (a), and miR-365-2 (b) expression levels were evaluated by RT-qPCR analysis in BT20 and MDA-MB-231 cell lines compared with normal-like MCF10A cells. The results are the average of three independent experiments. T test p value < 0.05 (*), 0.01 (**), 0.001 (***).
miR-135b and miR-365 control cell proliferation
As several of the putative identified targets of miR-135b and miR-365 have a role in cell proliferation, such as Checkpoint Kinase 2 (CHEK2)19, E2F Transcription Factor 1 (E2F1)20, the tumor suppressor retinoblastoma protein (RB1)21, or the genes of the FANC family22, we first analysed the effect of miR-365 upregulation or miR-135b suppression on proliferation of the cells. In order to perform the in vitro experiment on cell lines, we used MDA-MB-231 cells as they grow faster than BT20 cells. After confirming that the treatment with 50 nM As miR-135b or S miR-365 affected miRNAs’ expression compared to scramble treated cells (Fig. 3a,b, respectively), we observed that both treatments were able to slow down proliferation by MTT assay in a significant way at 48 and 72 h, in respect to scramble-treated cells (Fig. 3c).
Proliferation of the cells after miRNA modulation. MTT assay (Absorbance at 540 nm) revealed that the MDA-MB-231 treated with 50 nM As miR-135b (a) or S miR-365 (b) reduced significantly the rate of proliferation (c). The graph represent the average of the absorbance at 540 nm of three counts performed in duplicate experiments (T test compared to untreated cells, p value < 0.05, *).
miR-135b and miR-365 control cell migration and invasion
Among the possible target identified with in silico analysis, we found some genes that are involved in DNA repair23 (i.e., Ataxia Telangiectasia And Rad3-Related Protein ATR), migration (E2F1)24 or invasion (i.e., Replication factor C5, RFC5)25.
We tested if the modulation of miR-135b or miR-365 was able to affect the MDA-MB-231 cell migration, by wound healing test. The treatment with As miR-135b or S miR-365 on MDA-MB-231 cells decreased their ability to fill up the wound, as shown by the increase of the ratio between wound area at time 24 h/wound area at time 0 h (Fig. 4a), with a percentage reduction of 35% for As miR-135b (t test p value = 0.035, *) and of 59% for S miR-365 (t test, p value = 0.001,**) in respect to scramble-treated cells (Fig. 4b).
miR-135b and miR-365 are able to control cell migration, invasion and MET. (a) Result of the wound healing assay revealed that both reduction of miR-135b or induction of miR-365 reduced the capability of MDA-MB-231 cells to close the wound. The area of wounds at 24 h normalized on the same area at 0 h was measured for 50 nM scramble-treated, 50 nM As miR-135b-treated and 50 nM S miR-365 treated cells and compared to that of untreated cells. The measures (average ± sd) were performed in 4 different point of the wound, and the experiment was performed in duplicate (T test compared to scramble-treated cells, p value < 0.05, *; < 0.01, **). (b) The percentage of wound closure efficacy has been calculate for each sample. (c) The counts of MDA-MB-231 cells, migrated through the Boyden chambers, were taken after 24 h of treatment with 100 nM scramble, As miR-135b or S miR-365 oligonucleotides. Data were normalized on scramble treated cells. The bars are the average ± sd of three counts performed in duplicate experiments (T test, p value < 0.05*; p value < 0.01 **). (d) The percentage of invasion efficacy is also calculated compared to scramble-treated cells. (e,f) The MDA-MB-231 treated with As miR-135b or S miR-365 oligonucleotides (50 nM) for 72 h showed significant increased levels of E-cadherin (E-Cad, epithelial marker) (e) and a reduced capability of MS formation (f). Reduction of average MS size after 50nMtreatment of As miR-135b or S miR-365 for 14d was quantified. Average of surface MS size ± sd is indicated in the table (t test versus scramble-treated cells, p value is indicated) (g).
To test the ability of miR-135b and miR-365 to control the invasion ability of the cells, we performed the Boyden’s chamber test. The treatment with 50 nM of As miRNA-135b or S miR-365 decrease significantly the capability of the cells to heal the wound (Fig. 4c). In particular, the modulation of both miRNAs reduced the invasion ability of the cells of about 13.2% for As miR-135b (t test, p value = 0.003, **) and of 22.7% for S miR-365 (t test p value = 0.003, **) in respect to untreated cells (Fig. 4d).
miR-135b reduction induced mesenchymal-to-epithelial transition (MET)
The reduction of miR-135b expression, obtained by As treatment, or the increase of miR-365, obtained by mimic (S) treatment, lead to increased in E-Cadherin expression (Fig. 4e). Moreover, As miR-135b and S miR-365 treatments reduced the ability of the cells to form mammospheres (MS), compared to both untreated or scramble treated cells (Fig. 4f). The reduced ability of MS formation considers the size of the MS (Fig. 4g).
miR-135b and miR-365 are differentially expressed in human sample
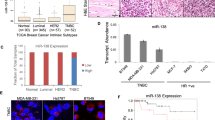
The RT-qPCR analysis revealed that also in human TNBC, miR-135b relative expression is upregulated (Fig. 5a; values of 3.14 ± 3.61 in TNBC tissues vs. 0.83 ± 1.70 in normal samples) and miR-365 relative expression is downregulated (Fig. 5b; values of 150.7 ± 220.3 in TNBC tissues vs 445.5 ± 852.8 in normal samples) compared to normal surrounding tissues. An in silico validation was performed on independent GEO datasets, GSE38167 and GSE58606. The results confirm the upregulation of miR135b and the downregulation of miR-165 in TNBC compared to normal tissues (see supplementary Figure 2).
miR-135b and miR-365 are differentially expressed in human basal BC samples. miR-135b (a), and miR-365 (b) expression levels were evaluated by RT-qPCR analysis in human basal BC (TNBC) or surrounding healthy tissue samples (normal samples, NS). The average of three independent experiments is depicted in red. T test p value < 0.05 (*), 0.01 (**).
In vitro treatment on organotypic TNBC cell culture is effective in modulating miRNAs expression
In order to evaluate the efficacy of oligonucleotide treatment in in vitro cultured organotypic slices of tumor, mimicking an organotypic-like culture, and to propose this treatment as a possible tool for TNBC therapy, we treated MDA-MB-231 tumor slices in vitro for 72 h with 50 nM As miR-135b or S miR-365 oligonucleotides. RT-qPCR analysis revealed that indeed the As oligonucleotide treatment was effective in reducing miR-135b (the fold change miR-135b expression was 0.67 ± 0.12 in As miR-135-treated tumor culture, compared to 0.76 ± 0.16 in scrambled-treated group) (Fig. 6a; t test p value = 0.05 compared to scramble and 0.037 compared to untreated organotypic-like culture) and the S oligonucleotide treatment induced miR-365 expression (Fig. 6b; the fold change miR-365 expression was 1.26 ± 0.04 in S miR-365-treated tumor culture, compared to 0.98 ± 0.13 in scrambled-treated group).
Therapeutic treatment with miR-135b and miR-365 oligonucleotides in MDA-MB-231 organoids modulates their expression. miR-135b (a), and miR-365 (b) fold change expression levels were evaluated by RT-qPCR analysis in human organotypic-like cultured slices obtained from MDA-MB-231 tumors, in vitro treated with 50 nM 72 h of As miR-135b or S miR-365 oligonucleotides. The average of two independent experiments is depicted. T test, p value < 0.05(*), 0.01 (**).
Discussion
In this study we identified and characterized two miRNAs with higher degree centrality in TNBC. In silico analysis of basal breast cancer profiles compared to normal breast epithelial tissue profile revealed that miR-135b is upregulated and miR-365 is downregulated in tumoral tissues compared to normal tissues. We validated these expressions in both cell lines and human BC tissue samples.
The integration of basal mRNA profile with miRNA profile revealed that miR-135b and miR-365 are involved in the control of several differentially expressed genes of TNBC, belonging to ‘Ethanol degradation IV’ and ‘Mismatch repair in eukaryotes’, and ‘Putrescine degradation III’, ‘Role of BRCA1 in DNA damage response’, and ‘Tryptophan degradation X’ pathways, respectively. A detailed analysis of the role of the two miRNAs within functional networks allowed us to suggest that both miRNAs are involved in the control of proliferation (predicted targets: CHEK219, E2F120, RB121, or the genes of the FANC family22), migration and invasion. Indeed, the in vitro modulation of miR-135b, by transfection of As oligonucleotide, or of mimic miR-365, by transfection of S oligoucleotide, confirmed that both miRNAs are able to regulated cell proliferation rate, without inducing apoptosis of the cells (not shown). The antiproliferative effect is due to the regulatory role of both miRNAs on their targets. For miR-365, this could involve the regulation on putrescine degradation pathway, as suggested by26. The antiproliferative effect of miR-135b could be due to its activity on mismatch repair genes, such as in colorectal cancer27.
The experiments with miRNAs’ modulation also revealed that they are both involved in the control of invasion and migration, as suggested by the results of the wound healing and Boyden’s chamber. Liu et al.28 confirmed that miR-365 overexpression inhibits cell proliferation, migration and invasion, possibly by direct targeting ADAM10, a membrane-bound cell surface glycoprotein involved in cell adhesion of TNBC29. Among the possible target of miR-365, we found Ataxia Telangiectasia And Rad3-Related Protein (ATR) serine/threonine kinase, a mediator of the expression of several cell surface proteins of the extracellular matrix (ECM) required for adhesion and invasion of the cells30. The silencing of miR-135b or the overexpression of mimic miR-365 are able to reduce cellular mobility, perhaps due to the increased mesenchymal-to-epithelial (MET) differentiation process.
Indeed, both miRNAs are involved in the regulation of ALDH genes (ALDH1A1, ALDH1A3 and ALDH2 genes) belonging to the Tryptophan (Trp) degradation pathway, but also related to stemness. Cancer stem cell (CSC) populations have elevated ALDH activity31, and the expression of these genes is associated to chemoresistance in breast cancer32 and negative prognosis33. The downregulation of miR-365 in TNBC, could lead to the increase in ALDH genes expression guiding the cells to a more undifferentiated and aggressive phenotype. In our experiments, we observed a tendency towards the increase in epithelial marker expression (E-Cad) with miR-365 overexpression. The shift of As miR-135b- or S miR-365-treated cells toward epithelial phenotype is also suggested by the test of MS formation: the silencing of miR-135b or the increase of miR-365 led to a reduction of number and size of MS, compared to scramble treated cells. This partially confirm a possible role of miR-135b and miR-365 in controlling genes, involved in the maintenance of stemness. The study of Liu and colleagues demonstrated that the expression of ALDH1A1 mRNA, one of predicted target mRNAs, in tumor tissues may be an independent predictor of TNBC outcome34.
Nowadays the treatment of TNBC is based on the combination of surgery removal of the cancer, radiation therapy and chemotherapy. Nevertheless, some patients don’t benefit from the chemotherapeutic treatment, leading to the development of relapse or metastasis in other tissues. Therefore, we also evaluated the possibility to use miRNA modulation in 3D structures, i.e. the organotypic culture of MDA-MB-231 cells. We succeeded in the reduction of miR-135b and in increase of miR-365 expression, suggesting a potential use of the two miRNA oligonucleotides in following experiments (that are beyond the objective of this work) aimed to deeper understand and validate these molecules and treatment strategy as new therapeutic drugs. Considering the anti-proliferative effects of the oligonucleotide treatment, their effect on blocking migration and invasion, and on favoring MET, their use could be helpful in counteracting the high metastatic potential of TNBC, alone or in combination with conventional therapy. No clinical trial is nowadays present on the use of miRNAs in TNBC treatment, although several publications propose their use as potential diagnostic or prognostic biomarkers35,36. The data need to be analysed in the light of a recent paper from Bao et al.37 who suggested that miR-135b-5p is downregulated in TNBC and this reduced expression would be important to increase proliferation and migration of the cell. These contradictory results could be partially justified by the fact that Bao et al. found miR-135b-5p downregulation in the comparison between TNBC and non-TNBC samples, correlated with the patient survival. On the contrary, our analysis identified miR-135b-5p as a possible regulator of a network of genes differentially expressed in TNBC versus normal tissues. We can’t exclude that miR-135b could be differentially expressed in non-TNBC compared to TNBC, as we didn’t perform this analysis on human samples.
Regarding miR-365, it has been already proposed as a tumor suppressor. Indeed, it has been found downregulated also in sera from early-stage breast cancer patients38. On the contrary, in HER2-positive breast cancer circulating exosomes, miR-365 was found to be significantly upregulated, suggesting that this miRNA could be expressed under the control of HER2 pathway39.
Although we propose miR135b and miR-365 as helpful biomarkers of TNBC diagnosis and for the development of new therapeutic tool, our study presents some limits: the first is linked to the fact that some sub-classifications of TNBC have been proposed. The paper of Perou highlighted that TNBC is also characterized by strong expression of basal markers such as cytokeratins 5, 6 and 1740. More recently, a study by Lehman et al.41 used histopathology and laser-capture micro-dissection to define 4 new subtypes of TNBC: basal subtypes 1 and 2 (BL1, BL2), characterized by elevated DNA damage response, higher expression of growth factor signaling pathways, glycolysis and gluconeogenesis; mesenchymal (M), enriched in components involved in cell motility, extracellular receptor interaction, and cell differentiation pathways; and luminal androgen receptor (LAR), enriched in hormonally regulated pathways such as steroid synthesis, porphyrin metabolism, and androgen/estrogen metabolism. In our analysis, we have considered what is defined as TNBC on the The Cancer Genome Atlas (TCGA) database, which included all the subtypes described before, without sub-classifying different molecular subtypes.
Our analyses possess the advantage to identify the network of genes that would be modulated by the miRNAs and not a single direct target since each of the two miRNAs analysed will be able to directly and indirectly modulate the entire network and no only the specific target. This approach will allow us to foresee a wider cellular outcome of their modulation. The identification of the specific direct targets will be done in the near future. Moreover, since the significant results of the analysis on human and organoid samples were obtained using a limited specimen number, further studies are needed for translating those miRNAs to clinical use. The use of miRNAs could be useful during the biopsy characterization phase. Besides, many miRNAs appear to be secreted in biological fluids, such as blood, plasma and serum. In this view, our study will pave the way for secreted miR-135b and miR-365 evaluation in different samples for their use in TNBC diagnosis or patient follow-up.
Conclusions
By integrative in silico approach we identified two diagnostic miRNAs, miR-135b and miR-365, with a high degree centrality in basal breast cancer, involved in the control of proliferation, invasion and migration of tumoral cells and human samples. The modulation of these two miRNAs, obtained by oligonucleotide treatment, has an impact in their expression, leading to MET, characterized by a lower aggressive, epithelial phenotype. These two miRNAs could be proposed as tissue diagnostic molecules and as tools for the development of new therapeutic approaches for TNBC.
Methods
In silico analysis
RNA-Seq data
In order to obtain the gene profile altered in basal BC, we applied the computational approach on a IlluminaHiSeq RNASeqV2 dataset derived from TCGA. We used basal BC matched samples of mRNA and miRNAselecting 74 basal BC samples classified by PAM50 methods16. We used the available data from TCGA for the normal samples (NS), namely 113 NS for gene expression analysis and 87 NS for miRNA analysis. The expression levels of 1046 miRNAs and 15243 genes (excluding genes with a small variance) were considered. The data were pre-processed and normalized using the protocols described in TCGAbiolinks14.
Computational approach
The computational analysis consists of 6 steps: (1) identification of differentially expressed genes between basal and NS; (2) pathway enrichment analysis; (3) construction of classifier; (4) evaluation of classifier; (5) features selection able to classify with the best performance basal versus normal samples; (6) integration of miRNA in order to identify miRNAs regulating a pathway network in basal BC.
This computational approach was based on a Monte Carlo Cross-Validation16, dividing the original dataset into training (60%) and testing (40%) dataset. To avoid problems of unbalanced classes, we generated randomly an equal number of samples for each class, for both basal and NS.
Cell culture
For in vitro studies, we used MDA-MB-231 cells and BT20 cell lines (ICLC, Genova, Italy), that are both referred as Basal, human, BC epithelial cell lines, and MCF10A cell line, as human, normal-like epithelial breast cells42. We maintained all the cell lines within a humidified atmosphere containing 5% CO2 at 37 °C. MDA-MB-231 cells were cultured in Advanced DMEM, while BT20 were culture in MEM cell culture medium (both Gibco, Life Technologies), added with 10% fetal bovine serum (FBS) (Lonza, Euroclone), 1% penicillin/streptomycin, 2 mM l-Glutamine following the manufacturer’s recommendation. MCF10A cells were cultured on Advanced DMEM with 10% FBS, 10 μg/ml insulin, 20 ng/ml hEGF, 0.5 μg/ml hydrocortisol, 2 mM l-Glutamine, 20 nM HEPES. Dulbecco Phosphate-Buffered Saline (D-PBS), and trypsin were obtained by Lonza (Euroclone).
miRNA modulation
miRNA increase or decrease was obtained by transfection of 50 or 100 nM mimic or antisense oligonucleotide (Sigma Aldrich) with metafectene reagents (Biontex, Germany), following manufacturer’s instruction. The sequences for miRNA modulation were the following:
-
miR-135b antisense (As): 5′- TCACATAGGAATGAAAAGCCATA-3′;
-
miR-365-2-5p mimic (S): 5′-GGGACTTTCAGGGGCAGCT-3′.
A scramble sequence, non-codifying for any known miRNA, was used as a control of the experiments (Scramble miRNA: 5′-ATGATGTCCTTCTAGTACGCATC-3′).
All miRNAs were designed on the sequences from miRbase database 43.
MTT assay
MDA-MB-231 cells were seeded in 96 wells at a confluence of 5000 cells/well. 24 h after seeding the cells were treated with mimic or antisense oligonucleotide for miRNA modulation. At 24, 48 and 72 h after treatment, the cells were stained with 50ug/ul of thiazolyl blue tetrazolium bromide (MTT) (Sigma Aldrich) for 4 h at 37 °C44. The cells were then lysed in dimethyl sulfoxide (DMSO; Euroclone, Italy) and the staining was quantified at 540 nm in FLUOstar Omega microplate reader (Euroclone). The presented results are the average ± sd of three independent experiments.
RNA extraction and real time- quantitative PCR analysis
Total RNA was isolated using TRIzol reagent (Life Technologies) and it was reverse transcribed for the miRNA quantification, using MystiCq microRNA cDNA synthesis kit (Sigma Aldrich), following manufacturer’s recommendations and as already described in44. miRNAs were amplified in Eco real time-quantitative PCR (RT-qPCR) (Illumina, Euroclone) using Power Up Sybr green mix (Applied Biosystem), in combination with homemade designed primers (all from IDT, Tema Ricerca, Italy): miR-365-2-5p Fw primer: 5′-GGGACTTTCAGGGGCAGCT-3′; miR-135b Fw primer 5′-TATGGCTTTTCATTCCTATGTGA-3’.
For each RT-qPCR analysis, the results are presented as values of relative expression 2^-ΔC(T) or as fold change expression (2^-ΔΔCt) using real-time qPCR (Eco, Illumina, Euroclone, Italy) normalizing the results on the expression of a housekeeping miRNA, miR-103-3p (5′-AGCAGCATTGTACAGGGCTATGA-3′) or the positive control present in the MystiCq microRNA cDNA synthesis kit.
Cell migration: wound healing
To study directional cell migration, we performed a wound-healing study. The MDA-MB-231cells were seeded at a confluence of 150–200,000/well in 24-well plate (Euroclone). After 48 h, necessary to reach 80% confluence, a scratch was performed in the middle of each well45. After washing the cells, new medium was added, and miRNAs were modulated, as described before. Pictures of each well were taken every 24 h. The effect on cell migration was quantified by ImageJ software46. The quantification of the wound was performed calculating the area of the scratch at 24 h normalized on the area in the same point of the well at time 0. This means that if a treatment reduced the capability of the cells to restore and fill up the wound, the ratio of the area 24 h/0 h should increase in respect to untreated or scramble treated cells, following the method described in46. Experiments were performed in triplicate (n = 9). A t test was calculated among untreated, scramble-treated, S miR-365-2-5p, or As miR-135b-treated MDA-MB-231 cells.
Cell invasion: Boyden’s chamber
A standard Boyden’s chamber test was used for the invasion study. The test was performed and quantified as described previously44.
Mammosphere formation test
For mammosphere (MS) generation, MDA-MB-231 cells were cultured on non-adherent plates (Corning, Sigma, Italy) at 100 single cells/ml. The culture medium was DMEM supplemented with B27 (Invitrogen), 20 ng/ml EGF (ThermoFisher Scientific, Life Technologies, Italy), 20 ng/ml bFGF and 4 μg/ml heparin (both Sigma, Italy) added, when required, with 50 nM scramble, As miR-135b or S miR-365 oligonucleotide (IDT, Tema Ricerca; Italy) every 2 days. Pictures of MS growth were taken every 2 days. We counted how many surface cells are present for every 10 MS (picture 20 ×) 47.
BC human tissue samples validation
Eight samples of TNBC human tissue and corresponding non-tumoral clear margins were selected from the collection of the “Bruno Boerci” Oncological Biobank for research applications (Istituti Clinici Scientifici Maugeri IRCCS, Pavia), a biobank certified to the ISO 9001:2015 international standard and member of BBMRI.it (the italian node of the BBMRI-ERIC, Biobanking and BioMolecular resources Research Infrastructure—European Research Infrastructure Consortium)44. Surgical resections were performed from 2011 to 2013 at the Breast Unit of Istituti Clinici Scientifici Maugeri IRCCS, Pavia, Italy. Fresh tumor and normal breast specimens from surgical resections have been collected immediately after breast-conserving surgery or total mastectomy performed at Breast Unit of Istituti Clinici Scientifici Maugeri IRCCS, Pavia, Italy. A written and oral informed consent to participate in the present protocol was informed and obtained from each patient. This study was approved by the Maugeri Ethical Committee. This study was performed in accordance with the relevant guidelines and regulations. Tissues are snap-frozen in liquid nitrogen within 30–60 min of surgery excision and stored at − 80 °C at the “Bruno Boerci” Oncological Biobank until further use, according to the guidelines of the European research infrastructure for biobanking BBMRI-ERIC (the Bruno Boerci Oncological Biobank Standard Operating Procedure is certified ISO 9001:2015 and ISO 20387:2018 standards).
At the time of collection, TNBC characterization was assessed through immunohistochemical analysis (IHC) by the Pathology Service (Istituti Clinici Scientifici Maugeri IRCCS, Pavia), according to the clinical guidelines on BC (ASCO—American Society of Clinical Oncology). IHC characterization of the tumor samples is shown in Table 2 and supplementary Table 1. The samples were used for the isolation of total RNA and miRNA analysis in real-time quantitative PCR (as described in the methods below).
Organotypic tissue culture and in vitro treatment
Primary tumors obtained from Athymic Nude-Foxn1 nu/nu mice implanted with MDA-MB-231 cells were isolated in sterile condition. Slices were cut at the vibratome (50 nu each) and put in culture on Millicent supports (Millipore) in the Epicult Medium, supplemented by 10% FBS, 1% Penicillin/Streptomycn, 100 μg/ml kanamycin. Fifty nanomolar mimic miR365 or As miR135b were added to the medium in the presence of GenomOne-Neo Ex (CosmoBio) transfection reagent, following manufacturer’s protocol. The organotypic-like culture tissues were maintained in culture for 48–72 h, as suggested by48; RNA was then isolated and RT-qPCR analyses for miR-135b and miR-365 were performed, as described before. Animal experiments were carried out in compliance with the institutional guidelines for the care and use of experimental animals (European Directive 2010/63/UE and the Italian law 26/2014) and with the ARRIVE guidelines (https://arriveguidelines.org), authorized by the Italian Ministry of Health and approved by the Animal Welfare Organisation of the University of Milan.
Ethics approval and consent to participate
As reported in the section “Methods”, TNBC human tissue and corresponding non-tumoral clear margins were selected from the collection of the “Bruno Boerci” Oncological Biobank for research applications (Istituti Clinici Scientifici Maugeri IRCCS, Pavia), a biobank certified to the ISO 9001:2015 international standard and member of BBMRI.it (the Italian node of the BBMRI-ERIC, Biobanking and BioMolecular resources Research Infrastructure—European Research Infrastructure Consortium). Surgical resections were performed from 2011 to 2013 at the Breast Unit of Istituti Clinici Scientifici Maugeri IRCCS, Pavia, Italy. Written informed consents from patients were obtained in advance and tissue samples were collected, processed and stored at − 80 °C as snap-frozen aliquots immediately after surgery, according to the best practices in biobanking (ISO 9001:2015 and ISO 20387:2018 standards). At the time of collection, TNBC characterization was assessed through immunohistochemical anlysis (IHC) by the Pathology Service (Istituti Clinici Scientifici Maugeri IRCCS, Pavia), according to the clinical guidelines on BC (ASCO—American Society of Clinical Oncology). IHC characterization of the tumor samples is shown in Table 1. The samples were used for the isolation of total RNA and miRNA analysis in real-time PCR (as described in the “Methods” below).
Data availability
In order to obtain the gene profile altered in TNBC, we applied the computational approach on a IlluminaHiSeq RNASeqV2 dataset derived from The Cancer Genome Atlas (TCGA), https://www.cancer.gov/tcga.
References
Cardoso, F. et al. 4th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 4)dagger. Ann. Oncol. 29, 1634–1657. https://doi.org/10.1093/annonc/mdy192 (2018).
Foulkes, W. D., Smith, I. E. & Reis-Filho, J. S. Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948. https://doi.org/10.1056/NEJMra1001389 (2010).
Pistelli, M. et al. Prognostic factors in early-stage triple-negative breast cancer: lessons and limits from clinical practice. Anticancer Res. 33, 2737–2742 (2013).
Goldhirsch, A. et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 24, 2206–2223. https://doi.org/10.1093/annonc/mdt303 (2013).
Brenton, J. D., Carey, L. A., Ahmed, A. A. & Caldas, C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application?. J. Clin. Oncol. 23, 7350–7360. https://doi.org/10.1200/JCO.2005.03.3845 (2005).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98, 10869–10874. https://doi.org/10.1073/pnas.191367098 (2001).
Hubalek, M., Czech, T. & Muller, H. Biological subtypes of triple-negative breast cancer. Breast Care (Basel) 12, 8–14. https://doi.org/10.1159/000455820 (2017).
Pusztai, L., Mazouni, C., Anderson, K., Wu, Y. & Symmans, W. F. Molecular classification of breast cancer: limitations and potential. Oncologist 11, 868–877. https://doi.org/10.1634/theoncologist.11-8-868 (2006).
Badowska-Kozakiewicz, A. M. & Budzik, M. P. Immunohistochemical characteristics of basal-like breast cancer. Contemp. Oncol. (Pozn) 20, 436–443. https://doi.org/10.5114/wo.2016.56938 (2016).
Thakur, V. & Kutty, R. V. Recent advances in nanotheranostics for triple negative breast cancer treatment. J. Exp. Clin. Cancer Res. 38, 430. https://doi.org/10.1186/s13046-019-1443-1 (2019).
Van Roosbroeck, K. & Calin, G. A. Cancer hallmarks and microRNAs: the therapeutic connection. Adv. Cancer Res. 135, 119–149. https://doi.org/10.1016/bs.acr.2017.06.002 (2017).
Bertoli, G., Cava, C. & Castiglioni, I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5, 1122–1143. https://doi.org/10.7150/thno.11543 (2015).
Yu, Y. et al. A tumor-specific microRNA recognition system facilitates the accurate targeting to tumor cells by magnetic nanoparticles. Mol. Ther. Nucleic Acids 5, e318. https://doi.org/10.1038/mtna.2016.28 (2016).
Colaprico, A. et al. TCGAbiolinks: an R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71. https://doi.org/10.1093/nar/gkv1507 (2016).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57, 289–300 (1995).
Colaprico, A., Cava, C., Bertoli, G., Bontempi, G. & Castiglioni, I. Integrative analysis with Monte Carlo cross-validation reveals miRNAs regulating pathways cross-talk in aggressive breast cancer. Biomed. Res. Int. 2015, 831314. https://doi.org/10.1155/2015/831314 (2015).
Cava, C. et al. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinf. 17, 348. https://doi.org/10.1186/s12859-016-1196-1 (2016).
Fan, C. & Liu, N. Identification of dysregulated microRNAs associated with diagnosis and prognosis in triplenegative breast cancer: an in silico study. Oncol. Rep. 41, 3313–3324. https://doi.org/10.3892/or.2019.7094 (2019).
Manic, G., Obrist, F., Sistigu, A. & Vitale, I. Trial Watch: targeting ATM-CHK2 and ATR-CHK1 pathways for anticancer therapy. Mol. Cell. Oncol. 2, e1012976. https://doi.org/10.1080/23723556.2015.1012976 (2015).
Xiong, Z. et al. ANP32E induces tumorigenesis of triple-negative breast cancer cells by upregulating E2F1. Mol. Oncol. 12, 896–912. https://doi.org/10.1002/1878-0261.12202 (2018).
Giacinti, C. & Giordano, A. RB and cell cycle progression. Oncogene 25, 5220–5227. https://doi.org/10.1038/sj.onc.1209615 (2006).
Ren, B. et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245–256. https://doi.org/10.1101/gad.949802 (2002).
Ali, R., Rakha, E. A., Madhusudan, S. & Bryant, H. E. DNA damage repair in breast cancer and its therapeutic implications. Pathology 49, 156–165. https://doi.org/10.1016/j.pathol.2016.11.002 (2017).
Hollern, D. P. et al. E2F1 drives breast cancer metastasis by regulating the target gene FGF13 and altering cell migration. Sci. Rep. 9, 10718. https://doi.org/10.1038/s41598-019-47218-0 (2019).
Li, Y. et al. Multifaceted regulation and functions of replication factor C family in human cancers. Am. J. Cancer Res. 8, 1343–1355 (2018).
Bae, D. H., Lane, D. J. R., Jansson, P. J. & Richardson, D. R. The old and new biochemistry of polyamines. Biochim. Biophys. Acta Gen. Subj. 2053–2068, 2018. https://doi.org/10.1016/j.bbagen.2018.06.004 (1862).
Xiang, S., Fang, J., Wang, S., Deng, B. & Zhu, L. MicroRNA135b regulates the stability of PTEN and promotes glycolysis by targeting USP13 in human colorectal cancers. Oncol. Rep. 33, 1342–1348. https://doi.org/10.3892/or.2014.3694 (2015).
Liu, F., Zhuang, L., Wu, R. & Li, D. miR-365 inhibits cell invasion and migration of triple negative breast cancer through ADAM10. J. Buon 24, 1905–1912 (2019).
Mullooly, M. et al. ADAM10: a new player in breast cancer progression?. Br. J. Cancer 113, 945–951. https://doi.org/10.1038/bjc.2015.288 (2015).
Lee, S. H., Cheng, H., Yuan, Y. & Wu, S. Regulation of ionizing radiation-induced adhesion of breast cancer cells to fibronectin by alpha5beta1 integrin. Radiat. Res. 181, 650–658. https://doi.org/10.1667/RR13543.1 (2014).
Tanei, T. et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 15, 4234–4241. https://doi.org/10.1158/1078-0432.CCR-08-1479 (2009).
Pal, D. et al. Targeting aberrant expression of Notch-1 in ALDH(+) cancer stem cells in breast cancer. Mol. Carcinog. 56, 1127–1136. https://doi.org/10.1002/mc.22579 (2017).
Brochez, L., Chevolet, I. & Kruse, V. The rationale of indoleamine 2,3-dioxygenase inhibition for cancer therapy. Eur. J. Cancer 76, 167–182. https://doi.org/10.1016/j.ejca.2017.01.011 (2017).
Liu, Y. et al. ALDH1A1 mRNA expression in association with prognosis of triple-negative breast cancer. Oncotarget 6, 41360–41369. https://doi.org/10.18632/oncotarget.6023 (2015).
Paszek, S. et al. Dysregulation of microRNAs in triple-negative breast cancer. Ginekol. Pol. 88, 530–536. https://doi.org/10.5603/GP.a2017.0097 (2017).
Arigoni, M. et al. miR-135b coordinates progression of ErbB2-driven mammary carcinomas through suppression of MID1 and MTCH2. Am. J. Pathol. 182, 2058–2070. https://doi.org/10.1016/j.ajpath.2013.02.046 (2013).
Bao, C. et al. Exploring specific prognostic biomarkers in triple-negative breast cancer. Cell. Death Dis. 10, 807. https://doi.org/10.1038/s41419-019-2043-x (2019).
Kodahl, A. R. et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol. Oncol. 8, 874–883. https://doi.org/10.1016/j.molonc.2014.03.002 (2014).
Stevic, I. et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC. Med. 16, 179. https://doi.org/10.1186/s12916-018-1163-y (2018).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752. https://doi.org/10.1038/35021093 (2000).
Lehmann, B. D. et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS ONE 11, e0157368. https://doi.org/10.1371/journal.pone.0157368 (2016).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767. https://doi.org/10.1172/JCI45014 (2011).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68-73. https://doi.org/10.1093/nar/gkt1181 (2014).
Cava, C. et al. Theranostic application of miR-429 in HER2+ breast cancer. Theranostics 10, 50–61. https://doi.org/10.7150/thno.36274 (2020).
Bertoli, G. et al. MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res. Treat. 161, 605–616. https://doi.org/10.1007/s10549-016-4079-2 (2017).
Grada, A., Otero-Vinas, M., Prieto-Castrillo, F., Obagi, Z. & Falanga, V. Research techniques made simple: analysis of collective cell migration using the wound healing assay. J. Invest. Dermatol. 137, e11–e16. https://doi.org/10.1016/j.jid.2016.11.020 (2017).
Cantini, L. et al. Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer. Nucleic Acids Res. 47, 2205–2215. https://doi.org/10.1093/nar/gkz016 (2019).
Carranza-Torres, I. E. et al. Organotypic culture of breast tumor explants as a multicellular system for the screening of natural compounds with antineoplastic potential. Biomed. Res. Int. 2015, 618021. https://doi.org/10.1155/2015/618021 (2015).
Acknowledgements
The authors would like to thank for the financial support: INTEROMICS flagship project (http://www.interomics.eu/it/home), National Research Council CUP Grant B91J12000190001, and the project Grant SysBioNet, Italian Roadmap Research Infrastructures 2012.
Author information
Authors and Affiliations
Contributions
G.B., C.C. and I.C. conceived of and designed the experiments. G.B., C.C., C.M., V.V. performed the experiments and data analysis. All the authors wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bertoli, G., Cava, C., Corsi, F. et al. Triple negative aggressive phenotype controlled by miR-135b and miR-365: new theranostics candidates. Sci Rep 11, 6553 (2021). https://doi.org/10.1038/s41598-021-85746-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85746-w
This article is cited by
-
CMA mediates resistance in breast cancer models
Cancer Cell International (2023)
-
Circular RNA hsa_circ_0044234 as distinct molecular signature of triple negative breast cancer: a potential regulator of GATA3
Cancer Cell International (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.