Abstract
Immune system plays a dual role in cancer by either targeting or supporting neoplastic cells at various stages of disease, including metastasis. Yet, the exact immune-related transcriptome profiles of primary tumours (PT) and lymph node metastases (LNM) and their evolution during luminal breast cancer (BCa) dissemination remain undiscovered. In order to identify the immune-related transcriptome changes that accompany lymphatic spread, we analysed PT-LNM pairs of luminal BCa using NanoString technology. Decrease in complement C3—one of the top-downregulated genes, in LNM was validated at the protein level using immunohistochemistry. Thirty-three of 360 analysed genes were downregulated (9%), whereas only 3 (0.8%) upregulated in LNM when compared to the corresponding PT. In LNM, reduced expression was observed in genes related to innate immunity, particularly to the complement system (C1QB, C1S, C1R, C4B, CFB, C3, SERPING1 and C3AR1). In validation cohort, complement C3 protein was less frequently expressed in LNM than in PT and it was associated with worse prognosis. To conclude, local expression of the complement system components declines during lymphatic spread of non-metastatic luminal BCa, whilst further reduction of tumoral complement C3 in LNM is indicative for poor survival. This points to context-dependent role of complement C3 in BCa dissemination.
Similar content being viewed by others
Introduction
The metastatic disease remains the leading cause of cancer-related deaths. Metastasis is a multistep process that involves the action of both tumour microenvironment (TME), comprising immune cells and stromal components, and cancer cells. The current opinion states that cancer cells tend to spread either via lymph or blood, reaching their specific final destination—lymph nodes or distant organs1. In breast cancer (BCa), lymph nodes are the first site to be colonized through the lymphatic route, usually much earlier than the distant sites reached via haematogenic route. Recent research has demonstrated that distant metastases of BCa may be seeded from the metastatic foci in lymph nodes2,3,4. As lymph nodes play a key role in immune response, they may also contribute to the selection of the immune-evading phenotype of cancer cells, thus driving further metastatic spread. Still, the transcriptional changes that accompany the dissemination process remain unknown.
The immune system, although originally developed for defence against pathogens, is a key player in cancer development and progression. The interaction between the tumour and surrounding immune cells is constant and complex, leading either to inhibition or stimulation of tumour growth, as included in the hallmarks of cancer by Hanahan and Weinberg5. In contrary to common knowledge, luminal breast tumours have recently been demonstrated to exhibit heterogeneous immunogenicity reflected by distinct patterns of immune gene expression6,7.
Thus, in this study we aimed to explore the changes of immune-related transcriptome indicative for metastatic colonization in luminal BCa. We compared data on 360 immune-related genes expression in matched pairs of primary breast tumours (PT) and lymph node metastases (LNM). To compensate for the physiological differences between breast and lymph node tissue, we incorporated a healthy background normalization step based on normal tissue expression data from GeneCards database. The selected transcriptional changes were subsequently validated at the protein level using immunohistochemical (IHC) staining.
Results
Immune-related transcriptomic changes during metastatic colonization in luminal BCa
LNM vs. LYMPH NODE comparison
Owing to the physiological transcriptome differences between lymph node and breast tissue, we decided to include a healthy-background-normalization step in our analysis (Fig. 1). Using expression data for lymph nodes (LYMPH NODE) and breast tissue (BREAST) from GeneCards database (GTEx project data8), we calculated median normalized LNM/PT ratios for 360 genes linked with the immune system.
General flow of data analysis. 360 genes were analysed in 11 matched pairs of PT and LNM tissues (CANCER; NanoString data) with a healthy-background normalization based on expression data for healthy breast and lymph node tissues (HEALTHY; Illumina Body Map data); for each gene, normalized LNM/PT ratio was calculated for each matched LNM-PT pair separately, and the median of all normalized LNM/PT ratios was employed in further analyses.
Here we observed a substantial disproportion in the number of genes that were enriched or depleted in LNM in comparison to healthy lymph node. Eleven of 360 genes demonstrated increased expression in LNM, while a decrease was observed in 70 genes (Supplementary Table S1). LNM-enriched genes were primarily breast, breast cancer or pro-metastatic markers, while the LNM-depleted genes mainly contributed to innate immune response. These observations showed that in the LNM, the healthy lymphatic tissue—physiologically abundant in immune-related transcripts—was replaced by cancer, which putatively induced changes in stromal cells (TME) and was connected with decreased innate immune response.
LNM versus PT comparison
In the next step, we looked for transcriptional changes from PT to LNM. Based on the opposite ratios of gene expression levels in LYMPH NODE/BREAST and LNM/PT, we selected LNM-upregulated genes from LNM-enriched genes (3/11) and LNM-downregulated genes from LNM-depleted genes (33/70) (Supplementary Table S1). All LNM-upregulated and top10 LNM-downregulated genes are depicted in Fig. 2.
Distribution of normalized LNM/PT ratios of LNM-upregulated and -downregulated genes. Genes were classified as LNM-upregulated (red) and LNM-downregulated (blue; top10 out of 33 genes are depicted) based on the median normalized LNM/PT ratio and opposite LYMPHNODE/BREAST and median matched LNM/PT ratios. Grey dashed lines represent the cut-off for LNM-enrichment (1) and LNM-depletion (−1).
Considering their biological role, 3 LNM-upregulated genes (ATG10, GATA3 and S100B) are potential markers of aggressive phenotype and increased metastatic potential of cancer cells. On the other hand, 33 LNM-downregulated genes are mainly associated with innate immunity, in particular, with the complement cascade, as revealed by the functional annotation analysis (Fig. 3, Supplementary Table S2).
LNM-downregulated genes are associated with the complement system. (A) ConsensusPathDB over-representation analysis [pathways ordered according to − log 10(FDR adjusted p value), complement-related pathways marked in blue] and (B) STRING protein interaction analysis (complement-related protein products marked in purple).
Complement component 3 (C3) protein expression decreases during metastasis—IHC validation
Downregulation of transcripts related to complement cascade in LNM, when compared to corresponding PT, indicates that local expression of complement might play a distinct role in PT and LNM. For that reason we decided to validate our transcriptional findings at the protein level, focusing on the central element of all activation cascades—complement component 3 (C3). Using IHC staining, we evaluated C3 expression in the larger cohort of luminal BCa patient (n = 79) with both positive (N+, n = 43) and negative (N−, n = 36) nodal involvement status, including the samples analysed with NanoString technology.
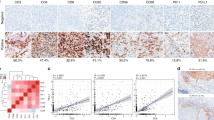
The C3 staining was informative for 36 of 36 N− PT, 42 of 43 N+ PT and 36 of 43 LNM specimens, resulting in 35 matched PT-LNM pairs. We evaluated stromal, tumoral and overall (i.e. combined C3 expression in both tumour cells and stroma) C3 expression status and dichotomized it as negative or positive based on staining intensity. In 11 matched PT-LNM pairs for which both NanoString and IHC data were available, mRNA levels and protein status tended to correlate when C3 protein was evaluated in tumour cells, however, did not reach the statistical significance due to the low number of samples (Supplementary Figure S2). In addition, in N+ patients, tumoral C3 was significantly decreased in LNM in comparison to PT (Fig. 4A), while no difference in C3 presence was noted in the stromal compartment (Fig. 4B). Combined stromal and tumoral assessment (overall status) showed a trend toward reduced C3 positivity in LNM (Fig. 4C). Similar tendencies were also observed when N+ and N− patients were analysed together (Fig. 4D–F). Representative images of tumoral C3 staining are presented in Fig. 4G. Of note, C3 expression in tumour cells of PT did not correlate with any clinical features, including stage, T status, grade and CTC status (data not shown). Intriguingly, loss of C3 expression in LNM tumour cells was associated with shorter 3-year overall survival (Supplementary Figure S3).
C3 protein expression is reduced during the lymphatic spread of BCa. N+ group evaluated for tumoral (A), stromal (B) and overall (C) C3 expression in PT and LNM; whole cohort evaluated for tumoral (D), stromal (E) and overall (F) C3 expression in PT and LNM; number of informative specimens is presented in Supplementary Figure S4; distribution was estimated with Fisher’s exact test. Representative images of C3 immunohistochemical staining in PT and LNM (G); presented tumoral C3-positive tissues also show a weak stromal C3 expression.
Discussion
Interactions between tumour cells and TME remain still unexplored, particularly in the context of cancer progression. In the current study, we focused on the most common molecular subtype of BCa—luminal cancers, where we demonstrate that the immune-related transcriptome changes occurring between primary tumour (PT) and lymph node metastasis (LNM) are mainly related to innate immunity, i.e. complement pathway.
In the exploratory group consisting of matched PT and LNM from 11 non-metastatic luminal BCa patients, 9% of transcripts were downregulated, whereas only 0.8% of transcripts were upregulated in LNM when compared to PT. According to the literature, all 3 identified LNM-upregulated genes, i.e. ATG10, GATA3 and S100B, are considered markers of breast cancer cells and their invasive potential9,10,11,12,13. ATG10 encodes a protein involved in autophagy cascade known to support or even induce metastatic process9,10. GATA3 codes for a transcription factor that is particularly abundant in luminal epithelial cells and directly correlates with ESR1 expression, thus is considered a marker of luminal breast cancer11,12, whereas S100B belongs to a family of genes coding for calcium-binding inflammatory proteins, regulators of p53, and might be involved in acid-induced EMT of ER-positive breast cancer cells in vitro14. The above-described enrichment in breast cancer and metastasis-associated transcripts proves that our algorithm is capable of adjusting the detected changes to the original characteristics of compared tissues, presumably due to the appropriate application of healthy tissue normalization step. On the other hand, the genes that were downregulated in LNM in comparison to PT were mainly associated with innate immunity, in particular complement system pathways. Interestingly, the identified complement-related genes are implicated in the early steps of the cascade, including all 3 pathways of its activation—C1QB, C1S, C1R, C4B, CFB and C3, regulation—SERPING1, as well as its effector mechanisms—C3AR1 (coding for receptor expressed on the surface of a variety of immune cells).
The complement system is known to play a dual role in cancer. As a fundamental part of the innate immunity, it is capable of targeting cancer cells and managing the immune response against the tumour. On the other hand, as a potent pro-inflammatory mechanism, the complement system is thought to substantially contribute to tumour growth by generating chronic inflammation state that facilitates mobilization of immune suppressor cells15 and supports angiogenesis16. In general, proteins comprising the complement system are synthesized in the liver and then released into plasma, resulting in the extracellular body compartments being the main environment for their interaction and cascade activation17,18,19,20. Nonetheless, a growing body of evidence suggests that the individual components of the complement pathway may also be produced within a tumour by both cancer and stromal cells. This is also supported by our data on high expression of C3 protein in tumour cells in BCa. Still, the locally produced complement proteins are thought to have a non-canonical function and act in a context-dependent manner, which merits further exploration18,21,22.
The association between the complement system and tumour dissemination has been broadly studied at the mRNA level. Several reports show that in the absence of EMT-promoting signals, C3 could enhance metastasis of epithelial cancer cells by loosening cellular junctions23, modulating vascularization and endothelial cells function24 or promoting survival of cancer cells via increasing their interactions with platelets25,26. On the other hand, the ovarian cancer mouse model provided evidence on the correlation between mRNA C3 expression and lymphatic vasculature24. Finally, the complement system also appeared to facilitate early stages of metastasis via modulation of cell adherence in several cancer types27.
In breast cancer, local expression of complement inhibitors was reported and perceived as a mechanism of evading immune response and cytocidal complement function. Namely, in primary breast tumours the expression of factor I and CD46 correlated with larger tumour size, lower differentiation index, and poor prognosis28,29. Moreover, in animal models of BCa, two groups reported the role of complement in premetastatic niche formation in the lungs30,31. Nevertheless, there is little clinical material-derived evidence on the exact role of the complement system in breast cancer dissemination. Still, a recent report by Chatterjee et al.32 has comprehensively summarized gene expression changes from healthy to metastatic (sentinel and non-sentinel) lymph node.
In our study, the reduction in local mRNA expression of C3—the central node of all complement activation cascades—in lymph node metastases was confirmed at the protein level. To the best of our knowledge, this is the first evidence for the association between C3 protein expression in breast cancer and metastatic colonization of lymph nodes. By far only one study by Chen and colleagues examined the relationship between the C3 protein and TNM staging and nodal involvement in pancreatic cancer, providing negative results33. The report by Vadrevu et al. also demonstrated that in BCa patients the complement proteins, including C3, are produced in both metastatic and metastasis-free nodes, yet the expression level in colonized lymph nodes was substantially higher. Still, the authors conclude that the complement system affects the metastatic process in a context-dependent manner30. This is in line with the outcomes of a comprehensive TCGA data analysis by Roumenina et al., investigating the complement system, TME and their prognostic properties in 30 cancer types. Based on survival analysis according to the level of complement-related transcriptome, BCa was classified as a cancer type with uncertain complement significance18, perhaps due to the distinct biology of the molecular subtypes of breast tumours. This provides a rationale for further exploration of the complement system role in BCa biology.
One of the limitations of our study was the small size of the exploration group (n = 11) in NanoString analysis. To compensate for that we performed an immunohistochemical validation of selected results on a larger cohort of patients (n = 79), proving the accuracy of our transcriptomic results. Another limitation was the application of NGS (RNA-seq) data on healthy tissues transcriptome for the healthy background normalization step. Since healthy tissue material is rarely included in high-throughput gene expression studies due to both its limited availability and high cost of the analysis, no compatible NanoString data set was available for use in public databases. A growing number of studies proves that NanoString and RNA-seq are compatible for gene expression analysis, both at the single gene and pathway level34,35,36,37. According to Zhang et al., NanoString and RNA-seq show the highest correlation coefficient among all available transcriptomic methods38. Nonetheless, aware of the limitations of comparing data from two different platforms, we did not normalize our NanoString cancer data using the NGS results for healthy tissues; instead, to minimize platform-specific differences, normalization was conducted within the same platform (Fig. 1).
To conclude, based on our findings we propose that the complement system potentially contributes to breast cancer lymphatic spread. We observed reduced mRNA expression of complement system genes in colonized lymph nodes when compared to corresponding primary tumours. Importantly, we also demonstrate that protein expression of complement C3 in cancer cells decreases in the course of metastatic spread and its further decrease in lymph node metastases is linked with patients poor prognosis. Consequently, we hypothesize that in non-metastatic luminal breast cancer patients the local PT expression of complement system-related genes may facilitate their invasion and metastasis, and eventually become decreased to alleviate immune system reaction when the cells reach the lymph node. Due to its potential role in shaping the aggressive phenotype of luminal breast tumours, the complement system appears to be a potential target for cancer treatment and thus merit further studies.
Methods
Patients
The study group consisted of 79 non-metastatic luminal BCa patients staged I–III, who underwent surgical treatment at the Medical University Hospital in Gdansk between 2011 and 2013. The study was approved by the Ethical Committee of the Medical University of Gdansk (NKBBN 94/2017) and informed consent was collected from all participants. All experiments were conducted in accordance with the Declaration of Helsinki, REMARK39 and STROBE40. Patients were characterized by different clinicopathological parameters, including nodal involvement and CTC status, as described previously41 and summarised in Table 1. Transcriptome analysis covered archival FFPE samples of PT and 11 LNM from 11 selected N+ patients for whom matched PT-LNM pairs were available. The algorithm of patients/sample selection is depicted in Supplementary Figure S4.
nCounter transcriptome profiling of primary breast cancer and corresponding lymph node metastasis fragments
Total RNA was isolated from archival FFPE blocks using RNeasy Mini Kit (Qiagen) and the expression of 730 target genes was evaluated with nCounter PanCancer Immune Profiling Panel (NanoString Technologies), as reported previously42.
For each analysed sample, background correction and normalization against global mean were performed as described42, using nSolver 4.0 software (NanoString Technologies). PT and LNM samples were normalized together. In brief, the background level was estimated by thresholding over the mean plus 2 standard deviations of the negative control counts. Subsequently, the data were normalized according to the global mean of the counts of positive controls and 4 most stably expressed housekeeping genes—ABCF1, EDC3, HDAC3, and CNOT4. The negative and positive control probes were included in the assay. Following normalization, low-expression genes (log2 mean count in all samples < 6) were excluded, leaving 593 target genes for analysis.
Healthy tissues transcriptome profiles from GeneCards database
Illumina Body Map expression data for normal lymph node (LYMPH NODE) and normal breast tissue (BREAST) generated within the Genotype-Tissue Expression (GTEx) project8 and deposited in the GeneCards database43 were obtained as a courtesy of Weizmann Institute of Science. 578 out of 593 genes expressed in the NanoString data were present in the Illumina Body Map dataset. Low-expression genes (log2 FPKM in each normal tissues < 5) were excluded, leaving 360 target genes for analysis.
Transcriptome data analysis
Analysis was performed for 360 genes expressed in both the NanoString and Illumina Body Map datasets (as depicted in Fig. 1, genes listed in Supplementary Table S3). The NanoString dataset comprised 11 matched LNM-PT pairs of BCa samples, whereas the Illumina Body Map dataset included the healthy control tissues—LYMPH NODE and BREAST. In brief, all data were log2 transformed and a log2 LYMPH NODE/BREAST (HEALTHY) ratio was calculated for each gene using Illumina Body Map data. In parallel, a matched log2 LNM/PT (CANCER) ratio was calculated for each patient using NanoString data. Subsequently, the HEALTHY ratio was subtracted from the matched CANCER ratio, giving a healthy background-normalized log2 LNM/PT ratio (further referred to as normalized LNM/PT ratio) for each patient. Eventually, the median normalized LNM/PT ratio was calculated for each gene based on the data of the whole cohort.
Genes with median normalized LNM/PT ratio > 1 were considered LNM-enriched compared to healthy lymph node (Supplementary Figure S5A). Genes with median normalized LNM/PT ratio < −1 were considered LNM-depleted compared to healthy lymph node (Supplementary Figure S5B). LNM-enriched/depleted genes were then analysed for changes in gene expression level from PT to LNM in reference to the healthy background. Genes with LYMPH NODE < BREAST and median normalized LNM > PT expression tendencies were considered LNM-upregulated in comparison to PT (Supplementary Figure S5C), while genes with LYMPH NODE > BREAST and median normalized LNM < PT expression tendencies were considered LNM-downregulated in comparison to PT (Supplementary Figure S5D).
Immunohistochemical evaluation of complement C3 protein
Tissue microarrays (TMA) comprising five 1-mm diameter tumour samples per each patient were prepared as previously described44. In brief, to detect C3, TMA sections were deparaffinised and treated with citrate buffer (pH 6, Dako) for 10 min and Peroxidase-Blocking Solution (Dako) for 5 min. The sections were incubated for 1 h at RT with polyclonal rabbit anti-C3 antibody (NBP1-32080, NOVUS Biologicals) diluted 1:250, envisioned by EnVision Kit, Rabbit/Mouse (Dako) and counterstained with haematoxylin (Sigma Aldrich). All tumour samples were evaluated in both tumour cells and surrounding stroma. Intensity of the staining and its semi-quantitative presence was documented. Staining was categorized based on the intensity as negative (i.e. no or weak expression), or positive (i.e. moderate to strong expression), and the tumour samples were scored for stromal, tumoral and overall (i.e. tumour cells and stroma) C3 expression. For each specimen, maximum record out of all examined and informative tumour samples was assigned for further analysis.
Statistical analysis
Data were analysed and visualized using R computing environment (3.6.1)45 and GraphPad Prism 8 (GraphPad Software) licensed for Medical University of Gdańsk. Differences in the distribution of C3 expression status were estimated with Fisher’s exact test. Association between tumoral C3 and overall survival was evaluated using log-rank test. Statistical significance was inferred for p values ≤ 0.05.
Selected genes were functionally annotated with Reactome pathways using over-representation analysis tool by ConsensusPathDB46. Interactions between protein products of selected genes were visualized using STRING v1147.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on request.
References
Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging biological principles of metastasis. Cell 168, 670–691 (2017).
Brown, M. et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 359, 1408–1411 (2018).
Pereira, E. R. et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 359, 1403–1407 (2018).
Kodama, T., Mori, S. & Nose, M. Tumor cell invasion from the marginal sinus into extranodal veins during early-stage lymph node metastasis can be a starting point for hematogenous metastasis. J. Cancer Metastasis. Treat. 4, 56 (2018).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011).
Zhu, B. et al. Immune gene expression profiling reveals heterogeneity in luminal breast tumors. Breast Cancer Res. 21, 147 (2019).
Netanely, D., Avraham, A., Ben-Baruch, A., Evron, E. & Shamir, R. Expression and methylation patterns partition luminal-A breast tumors into distinct prognostic subgroups. Breast Cancer Res. 18, 74 (2016).
Lonsdale, J. et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Mowers, E. E., Sharifi, M. N. & Macleod, K. F. Autophagy in cancer metastasis. Oncogene 36, 1619–1630 (2017).
Jo, Y. K. et al. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS ONE 7, e52705 (2012).
Perue, C. et al. Molecular portraits of human breast tumors. Nature 406, 747–752 (2000).
Hoch, R. V., Thompson, D. A., Baker, R. J. & Weigel, R. J. GATA-3 is expressed in association with estrogen receptor in breast cancer. Int. J. Cancer 84, 122–128 (1999).
Yen, M.-C. et al. S100B expression in breast cancer as a predictive marker for cancer metastasis. Int. J. Oncol. 52, 433–440 (2018).
Sadeghi, M. et al. Integrative analysis of breast cancer cells reveals an epithelial-mesenchymal transition role in adaptation to acidic microenvironment. Front. Oncol. 10, 304 (2020).
Cisse, B. Nature reviews. Immunology 8, 6 (2008).
Corrales, L. et al. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J. Immunol. 189, 4674–4683 (2012).
Zhang, R., Liu, Q., Li, T., Liao, Q. & Zhao, Y. Role of the complement system in the tumor microenvironment. Cancer Cell Int. 19, 300 (2019).
Roumenina, L. T., Daugan, M. V., Petitprez, F., Sautès-Fridman, C. & Fridman, W. H. Context-dependent roles of complement in cancer. Nat. Rev. Cancer 66, 1–18 (2019).
Afshar-Kharghan, V. et al. The role of the complement system in cancer. J. Clin. Investig. 127, 780–789 (2017).
Kourtzelis, I. & Rafail, S. The dual role of complement in cancer and its implication in anti-tumor therapy. Ann. Transl. Med. 4, 66 (2016).
Reis, E. S., Mastellos, D. C., Ricklin, D., Mantovani, A. & Lambris, J. D. Complement in cancer: Untangling an intricate relationship. Nat. Rev. Immunol. 18, 5 (2018).
Kolev, M., Le Friec, G. & Kemper, C. Complement—Tapping into new sites and effector systems. Nat. Rev. Immunol. 14, 811–820 (2014).
Conyers, G., Milks, L., Conklyn, M., Showell, H. & Cramer, E. A factor in serum lowers resistance and opens tight junctions of MDCK cells. Am. J. Physiol. Cell Physiol. 259, C577–C585 (1990).
Nunez-Cruz, S. et al. Genetic and pharmacologic inhibition of complement impairs endothelial cell function and ablates ovarian cancer neovascularization. Neoplasia 14, 994-IN1 (2012).
Labelle, M., Begum, S. & Hynes, R. O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 20, 576–590 (2011).
Polley, M. & Nachman, R. Human platelet activation by C3a and C3a des-arg. J. Exp. Med. 158, 603–615 (1983).
Ajona, D., Ortiz-Espinosa, S., Pio, R. & Lecanda, F. Complement in metastasis: A comp in the camp. Front. Immunol. 10, 669 (2019).
Okroj, M. et al. Local expression of complement factor I in breast cancer cells correlates with poor survival and recurrence. Cancer Immunol. Immunother. 64, 467–478 (2015).
Maciejczyk, A. et al. CD46 Expression is an unfavorable prognostic factor in breast cancer cases. Appl. Immunohistochem. Mol. Morphol. 19, 540–546 (2011).
Vadrevu, S. K. et al. Complement c5a receptor facilitates cancer metastasis by altering T-cell responses in the metastatic niche. Can. Res. 74, 3454–3465 (2014).
Sharma, S. K. et al. Pulmonary alveolar macrophages contribute to the premetastatic niche by suppressing antitumor T cell responses in the lungs. J. Immunol. 194, 5529–5538 (2015).
Chatterjee, G. et al. Molecular patterns of cancer colonisation in lymph nodes of breast cancer patients. Breast Cancer Res. 20, 1–12 (2018).
Chen, J. et al. Expression and clinical significance of complement C3, complement C4b1 and apolipoprotein E in pancreatic cancer. Oncol. Lett. 6, 43–48 (2013).
Speranza, E. et al. Comparison of transcriptomic platforms for analysis of whole blood from Ebola-infected cynomolgus macaques. Sci. Rep. 7, 1–15 (2017).
Bondar, G. et al. Comparing NGS and NanoString platforms in peripheral blood mononuclear cell transcriptome profiling for advanced heart failure biomarker development. J. Biol. Methods 7, 66 (2020).
Kwong, L. N. et al. Biological validation of RNA sequencing data from formalin-fixed paraffin-embedded primary melanomas. JCO Precis. Oncol. 2, 1–19 (2018).
Picornell, A. et al. Breast cancer PAM50 signature: correlation and concordance between RNA-Seq and digital multiplexed gene expression technologies in a triple negative breast cancer series. BMC Genomics 20, 1–11 (2019).
Zhang, W. et al. A large-scale comparative study of isoform expressions measured on four platforms. BMC Genomics 21, 1–14 (2020).
McShane, L. M. et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res. Treat. 100, 229–235 (2006).
Von Elm, E. et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: Guidelines for reporting observational studies. Int. J. Surg. 12, 1495–1499 (2014).
Markiewicz, A. et al. Mesenchymal phenotype of CTC-enriched blood fraction and lymph node metastasis formation potential. PLoS ONE 9, e93901 (2014).
Popeda, M. et al. NF-kappa B signaling-related signatures are connected with the mesenchymal phenotype of circulating tumor cells in non-metastatic breast cancer. Cancers 11, 1961 (2019).
Stelzer, G. et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 54, 1.30.1-1.30.33 (2016).
Markiewicz, A. et al. Epithelial-mesenchymal transition markers in lymph node metastases and primary breast tumors-relation to dissemination and proliferation. Am. J. Transl. Res. 6, 793 (2014).
Team, R. C. & others. R: A language and environment for statistical computing (2013).
Kamburov, A. et al. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 39, D712–D717 (2011).
Szklarczyk, D. et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Acknowledgements
The authors would like to acknowledge Marcin Okrój, PhD, DSc for critical review of the manuscript. This research was funded by the National Science Centre (Poland), grant number 2016/22/E/NZ4/00664.
Author information
Authors and Affiliations
Contributions
Conceptualization—N.B.K., A.M., M.P. and A.J.Z.; Methodology—N.B.K., A.K., M.N., M.P., T.S. and J.S.; Formal analysis—A.M., M.P. and T.S.; Investigation—N.B.K., A.M., M.N. and M.P.; Resources—N.B.K., A.K., M.N. and A.J.Z.; Data curation—M.P. and T.S.; Original draft preparation—A.M. and M.P.; Draft review and editing—all authors; Visualization—M.P. and T.S.; Supervision—N.B.K., A.M. and A.J.Z.; Project administration—A.J.Z.; Funding acquisition—A.J.Z.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Popeda, M., Markiewicz, A., Stokowy, T. et al. Reduced expression of innate immunity-related genes in lymph node metastases of luminal breast cancer patients. Sci Rep 11, 5097 (2021). https://doi.org/10.1038/s41598-021-84568-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84568-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.