Abstract
Chronic obstructive pulmonary disease (COPD) is a disorder of accelerated lung aging. Multiple pieces of evidence support that the aging biomarker short telomeres, which can be caused by mutations in telomerase reverse transcriptase (TERT), contribute to COPD pathogenesis. We hypothesized that short telomere risk-associated single nucleotide polymorphisms (SNPs) in TERT, while not able to drive COPD development, nonetheless modify the disease presentation. We set out to test the SNP carrying status in a longitudinal study of smokers with COPD and found that rapid decline of FEV1 in lung function was associated with the minor allele of rs61748181 (adjusted odds ratio 2.49, p = 0.038). Biochemical evaluation of ex vivo engineered human cell models revealed that primary cells expressing the minor allele of rs61748181 had suboptimal telomere length maintenance due to reduced telomerase catalytic activity, despite having comparable cell growth kinetics as WT-TERT expressing cells. This ex vivo observation translated clinically in that shorter telomeres were found in minor allele carriers in a sub-population of COPD patients with non-declining lung function, over the 5-year period of the longitudinal study. Collectively, our data suggest that functional TERT SNPs with mild catalytic defects are nonetheless implicated in the clinical presentation of COPD.
Similar content being viewed by others
Introduction
Telomere biology disorders consist of a spectrum of tissue degenerative disorders that have variable clinical manifestations and severities, but share the same disease etiology—short telomeres1. Telomeres are DNA–protein structures located at the ends of eukaryotic chromosomes2. Telomeres shorten with cell divisions and they limit the proliferation of human cells by inducing cellular senescence when a critical length is reached. Telomerase is a ribonucleoprotein complex that can copy the template within its RNA component, telomerase RNA (TER), to telomeric DNA by using its own polymerase, telomerase reverse transcriptase (TERT). De novo synthesis of telomeric repeats at chromosome ends by telomerase delays the depletion of telomeres and extends the proliferation lifespan3.
At the cellular level, mean telomere length is dynamically determined by the inherited starting telomere length, the rate of cell turnover, and the rate of telomere synthesis if telomerase is expressed in the cell. Individuals with the shortest telomeres (first percentile) in the general population are susceptible to a spectrum of premature aging disorders1, including idiopathic pulmonary fibrosis (IPF), even if they did not carry mutations in known telomere biology genes4. At the organismal level, telomere biology disorders, mostly caused by genetic deficiencies in telomerase function, affect tissue regenerative potential and present clinically as diverse disorders. Bone marrow is one tissue with a high proliferative demand that is particularly sensitive to short telomeres, as hematopoietic renewal failure occurs when the replicative potential and the stem cell pool of an affected individual is depleted. Consequently, bone marrow failure diseases, including dyskeratosis congenita and aplastic anemia, are frequently observed in patients with severe telomerase dysfunction.
On the other hand, the lung is a tissue with lower replicative demand but is also frequently affected by telomere dysfunction1,5. Patients with mutations in TERT suffer from IPF in an autosomal dominant pattern due to telomerase haploinsufficiency and the consequent short telomere-induced tissue renewal defects6. Furthermore, IPF is found in up to 20% of all patients with dyskeratosis congenita, the prototype and the first identified telomere biology disorder7. Disease presentation in the lung is driven by stem cell proliferative failure following telomere dysfunction in type 2 alveolar epithelial cells (AEC2)8.
Chronic obstructive pulmonary disease (COPD) is a multi-factorial trait where inflammation of the peripheral airways and/or loss of elastic recoil due to the destruction of alveoli (emphysema) cause irreversible progressive airflow limitation9. Patients with COPD are known to have short telomere length in their peripheral blood leukocytes10,11,12. Notably, the discovery of rare telomerase mutations in emphysema patients was previously reported13,14. Stanley et al. sequenced the telomerase genes (TERT and TER) in COPD patients across two large cohorts (COPDgene and the Lung Health Study (LHS)) and discovered that 1% of patients with early-onset severe emphysema had deleterious TERT mutations15. The prevalence of TERT driver mutations in COPD is comparable to alpha-1 antitrypsin deficiency—the only well-characterized genetic cause of emphysema15,16. Based on the findings of telomere dysfunction in COPD/emphysema, it has been proposed that stem cells in the lung (i.e., AEC2) were exhausted following cellular senescence during the pathogenesis and progression of COPD9.
Single nucleotide polymorphisms (SNPs) in the TERT region have been linked to telomere biology disorders, including, but not limited to, IPF17 and bone marrow failure18,19. Notably, a few non-coding (intronic) SNPs in the TERT gene were recently reported to associate with increased risks of COPD in Chinese and Japanese populations20,21. While rare, disease-causing TERT mutations precipitate the development of tissue regenerative deficiency such as emphysema22, in this study, we were interested in understanding whether common TERT SNP associated with mild telomere maintenance defects could modify the risk and clinical presentation of COPD.
The non-synonymous TERT SNP rs61748181 features a C > T nucleotide substitution, resulting in an amino acid change from alanine to threonine (TERT p.A279T). It is located at a linker domain of the N-terminus of TERT. Targeted deep sequencing has linked this SNP to bone marrow failure18, lung cancer23 and esophageal cancer24. Compared to other disease-associated TERT SNPs, it has a relatively high minor allele frequency (MAF) of 3.58%25 (Supplementary Table 1). Here, using a combination of human cell models, genetic analysis, and clinical data mining of the LHS cohort, we set out to investigate the effects of TERT SNP rs61748181 in the progression of COPD.
Results
rs61748181 carrying status does not affect the overall clinical progression of patients in the LHS cohort
The MAF of rs61748181 in the Caucasians from the LHS population was 3.29% (Table 1). The locus showed a slight deviation from Hardy–Weinberg equilibrium (p = 0.040), with a higher number of homozygotes for the minor allele than expected. The deviation is in agreement with a recent report on the excess of homozygotes for rs61748181 minor T allele in COPD patients than in control subjects26. 500 out of 3596 samples (13.90%) from Caucasian subjects in the LHS cohort failed the genotyping assay, likely due to DNA sample degradation, as we noted that there were unremarkable differences in baseline characteristics of the demographic profile between the typed and undetermined participants (Supplementary Table 2).
We sorted the Lung Health Study participants by annualized change (i.e., the slope) of FEV1 over 5 years (ΔFEV15yrs) and tested the association between rs61748181 and disease progression by a multivariate linear regression model. While age and smoking (both smoking history in pack-years and smoking status during the 5 years of the LHS study) affected disease progression, we did not observe any significant difference in ΔFEV15yrs between the rs61748181 minor allele carriers (CT and TT) (n = 197) and those with the CC genotype (n = 2899) (Supplementary Table 3). The lack of correlation between rs61748181 carrying status and ΔFEV15yrs suggested that COPD disease progression is not likely to be primarily driven by the carrier status of rs61748181.
Higher prevalence of rs61748181 carriers among rapid decliners of lung function than non-decliners in the LHS cohort
The lack of direct clinical correlation with telomerase genetic defects is not unexpected in telomere biology diseases, as physiological function and tissue renewal are protected by residual telomere length until a critical threshold for telomere dysfunction (unprotection) is reached. We reasoned that when carriers of TERT SNPs are subjected to an increased demand for telomere replenishment to maintain tissue integrity, the possible mild defects in telomere maintenance would be exacerbated. Over time, carriers of TERT SNPs should experience accelerated deterioration of existing pathological conditions, as a function of the sum total of all etiologic factors that contribute to the loss of tissue integrity, leading to accelerated progression of COPD pathologies9,27.
Based on this model, we explored the effect of this TERT SNP in patients with extremely rapid disease progression or no progression (Table 2). In a subset of 464 individuals (15% of the LHS population), carriers of rs61748181 TT or CT genotypes were more prevalent among rapid decliners (7.76%) than non-decliners (3.02%), with an adjusted OR of 2.49 (Table 3). In agreement with our model, restricting the comparator populations to patients with extremely rapid disease progression and no progression allowed us to tease out the accumulating biological effect of this TERT SNP.
rs61748181 in TERT can lead to compromised telomere length maintenance secondary to reduced telomerase activity
Next, we applied an ex vivo human cell model to assess the telomere maintenance capacity of the minor alleles of rs61748181 and two other TERT variants28. In addition to rs61748181, we include the telomere biology disorder-associated TERT SNP rs377639087, which features the deletion of codon 441 (TERT p.∆E441) and serves as a known telomerase activity defective control. rs112614087 (TERT p.A615T), which has a heterozygosity of 0.50029 and therefore is deemed unlikely to change TERT function, was chosen as the variant with normal telomerase activity control. The details of the TERT SNPs involved in the biochemical study are summarized in Supplementary Table 1.
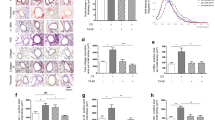
We used site-directed mutagenesis to introduce the sequences encoded by the minor alleles of the SNPs into the TERT open reading frame (Supplementary Table 4). Like other human fibroblasts, the human foreskin fibroblast BJ cell line silenced its endogenous TERT expression, whereas its TER expression is intact. Stable genomic integration of the coding sequences of TERT (WT and the SNP variants) through infection with retrovirus vectors (pBabe-Hygro-flag3B-TERT) in BJ cells restored telomerase activity. An examination of protein expression revealed that TERT protein levels in these reconstituted cell lines were comparable to each other (Fig. 1A). The population growth kinetics of the TERT-expressing BJ cell lines followed typical logarithmic curves (Fig. 1B). While the BJ vector control cell line (BJ-vector), which does not express telomerase activity, entered replicative senescence following 10 PDLs post-antibiotic selection, all BJ fibroblasts expressing recombinant telomerase (WT/SNP variants) were proliferating continuously in our ex vivo cell model system. Using this ex vivo system, we found the minor allele of SNP rs61748181, when translated into a variant version of TERT (expressed as TERT variant hereafter), was deficient in telomere length maintenance (Fig. 1C,D) while its TERT protein expressions were comparable (Fig. 1A) and growth kinetics remained the same as WT-TERT up to 50–60 PDLs (Fig. 1B).
Restoration of recombinant telomerase mediated by the TERT variants in BJ cell lines. (A) Recombinant expression of TERT in BJ fibroblasts. TERT expression levels (127 kDa) were normalized to those of vinculin (124 kDa) and the data are presented as mean ± SEM (n = 6). fluorescent & ECL ones). TERT and vinculin signals were differentiated with the use of differently labeled antibodies (chemiluminescent and fluorescent, respectively) and processed accordingly with different detection modalities. Full sets of data were available in the supplementary information (Supplementary Fig. 3) (B) Cellular growth kinetics of BJ cell lines expressing recombinant SNP-version of telomerase. (C) Telomere lengths in the cells at approximately every 10 PDLs were measured by TRF. Population doubling level (PDL) and telomere lengths (telomere restriction fragments, TRF) for each sample are illustrated. (D) Changes in telomere length over cell divisions are plotted.
To understand the mechanism behind defective telomere length maintenance by TERT variants, we applied the primer extension assay to check single nucleotide incorporation (nucleotide addition processivity, NAP) and repeat synthesis (repeat addition processivity, RAP) in WT-TERT and TERT SNP variant cell lines. The RAP activity of the rs61748181-TERT was 94.4 ± 5.5% compared to the WT-TERT (n = 6, p = 0.3313) and its NAP activity was 81.1 ± 7.3% of WT (n = 6, p = 0.0323) (Supplementary Fig. 1). The RAP and NAP activity of the telomerase activity defective control rs377639087-TERT was 99.41 ± 6.48% (p = 0.929) and 71.16 ± 11.22% (p = 0.022), respectively. We concluded that the TERT protein encoded by rs61748181 had similar repeat addition capacity as WT-TERT, but reduced nucleotide addition processivity. This minor difference in the polymerase’s processivity translated to the accumulated defects in telomere repeat synthesis overtime, as observed in the cell culture experiments.
Differences in median telomere length between CC homozygotes and rs61748181 minor allele carriers are evident in subgroups of LHS patients with extreme telomere synthesis demands
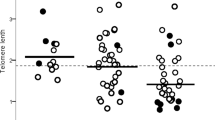
Considering the telomere maintenance defects observed in fibroblasts expressing the minor T allele of rs61748181 over the long term in our cell model, patients carrying the minor allele of rs61748181 should be at risk of increased telomere loss regardless of their COPD progression. We reasoned that we should be able to observe telomere length differences between CC homozygotes and rs61748181 minor allele carriers (genotypes CT and TT) in the LHS population. To test this, we investigated the association between rs61748181 minor allele carrying status and telomere lengths in 3050 genotyped Caucasian patients from the LHS with measurable telomeres. Our analyses did not reveal any statistical differences in median telomere length between rs61748181 minor allele carriers and CC homozygotes (Fig. 2A). Notably, the LHS cohort showed non-normal distribution of telomere length with several participants harboring leukocytes telomere measurements up to forty times longer than the median (Fig. 2B); perhaps not surprisingly, as it is well-known that telomere length regulation is multi-factorial, based on genetic, lifestyle and environmental influences.
Leukocyte telomere lengths for WT (CC) and rs61748181 carriers (CT/TT) (N = 3050). (A) Median telomere length for both WT and rs61748181 carriers showing no significant difference (Mann Whitney test). (B) Frequency distribution of relative telomere length among Caucasian members of the LHS cohort (median: 0.306, 95% CI 0.299–0.314). (C) Median telomere length of participants sorted based on rs61748181 status (Mann Whitney test). The cohort was divided into quartiles according to disease progression (ΔFEV15yrs); the first quartile represents participants with fastest disease progression, and the fourth quartile indicates those with non-declining lung function. Statistical analyses were performed using the JMP Statistics software package version 13 (SAS Institute Inc.) and R version 3.5.1.
We reasoned that the minor telomerase catalysis defects conferred by this SNP would be most evident in the subpopulation of patients with the highest demand for optimal telomere synthesis. Next, we accessed the leukocyte telomere length after dividing the cohort into four groups based on changes in their lung function over 5 years (ΔFEV15yrs). In each group, the leukocyte telomere length for the rs61748181 minor allele carriers was compared to their CC counterparts. In the fourth quartile (Fig. 2C), consisting of patients with no observable decline in their lung function over 5 years monitoring period (positive ΔFEV15yrs), carriers of rs6178181 minor allele (n = 43) had shorter leukocyte telomere lengths than CC patients (n = 719). This difference in telomere lengths based on SNP carrying status was statistically significant (p = 0.03). Similarly, rs6178181 minor allele carriers (n = 45) in the first quartile (where patients had the fastest disease progression and most negative ΔFEV15yr values) tended to have shorter leukocyte telomere lengths than CC patients (n = 717), though statistical significance was not reached. The difference in telomere length between rs6178181 minor allele carriers and CC individuals diminished in the middle subgroups (Fig. 2C), which may explain the lack of correlation between rs61748181 carrier status with telomere length, when the entire LHS cohort was considered (Fig. 2A).
Discussion
We investigated the biological and clinical effects of a TERT SNP rs61748181 which has previously been linked to telomere biology disorders18. With clinical samples collected in the LHS, we revealed that carrier status of the SNP rs61748181 aggravated existing disease conditions in COPD patients. Using a combination of molecular and cell biology tools in the laboratory, we further demonstrated that the non-synonymous SNP rs61748181 could lead to defective telomerase catalytic activity and deficient telomere length maintenance. Regression analysis showed that the association was independent of known risk factors such as aging and smoking history. These results implicated TERT as a biologically plausible candidate gene for lung function decline and COPD pathogenesis. Not only did our results agree with the previous observation that deleterious mutations in TERT contribute to COPD pathogenesis15, we also extended this observation to demonstrate that common variants in TERT with low penetrance may nonetheless modify disease presentations.
Our ex vivo results demonstrated in a laboratory setting that, when other confounding factors for telomere attrition were fixed, fibroblasts with recombinant expression of the disease-associating TERT variants were predetermined to suboptimal telomere maintenance. However, deficient telomere length maintenance in this context did not translate to a loss of proliferative capacity, as BJ cells expressing the minor allele of rs61748181 were kept in culture for up to 50–60 PDLs, the limit of fibroblast divisions during a lifetime30, and demonstrated the same growth rate as WT-TERT expressing cells. To our knowledge, this is the first study to investigate the long-term effects of common TERT variants on telomeres in primary cell lines. Our results confirmed that the short telomere risk-associated SNP rs61748181 encoded a TERT variant that was deficient in telomere length maintenance. We provided strong experimental evidence to support the previous clinical observations that in subjects with bone marrow failure, carriers (heterozygous and homozygous) of this SNP had shorter telomeres than their age-matched peers18. These biochemical results are also supported by reports in different cellular and experimental systems24,31.
This is the first study both to show the increased risk of rapid lung function decline in COPD patients carrying rs61748181 minor allele and to investigate the association of this SNP with short leukocyte telomeres in patient subsets from in a large cohort study of COPD. The current model of cellular senescence-based COPD pathogenicity9 stipulates that the carrier’s stem cell compartments would remain proliferative until their telomeres were too short and trigger stem cell exhaustion, which finally presented as rapid decline and poor lung performance at the end stage of the study. Telomeres shorten at similar rates in blood and somatic tissues32. Despite variation, telomere lengths correlate among most human tissue types, including lung tissues33,34. Notably, it is recently reported that carriers with rare variants in TERT, though not showing telomere biology disorders, have shorter telomere length across all tissues34. Therefore, our measurement of leukocyte telomere lengths in COPD patients should be representative of the lengths in their lung tissues.
We also considered the potential influence from a collider effect in our observed correlation between the rs61748181 minor allele carrier status with short leukocyte telomere length. For the collider effect to be true, two variables (telomere length and carrier status of rs61748181) are independently associated with an observation (COPD disease progression, in this case), but they should not be associated. Given that we have biological and empirical reasons to believe short telomere lengths are predicted by the rs61748181 minor allele, and we did not see an association between telomere length and COPD disease progression, we concluded that the collider effect is not relevant here. Finally, by stratifying our analyses to disease progression subgroups, the collider effect interference, if there is one, should be minimized.
An important limitation of our study is a lack of a validation cohort to confirm the clinical effects of rs61748181 carrying status. To reliably determine lung disease progression, longitudinal follow-up and repeated observations are necessary. The impediments from these resource-intensive follow-up procedures are compounded by the low allelic frequencies of rs61748181, requiring a large sample size to enroll in the study. While we do not have access to clinical samples from such a clinical study in lung function decline, we contend that the effects of rs61748181 should apply to other chronic tissue degenerative disorders. Our data should provide the impetus to include TERT non-synonymous SNPs in disease progression assessments.
An individual’s telomere length is determined by multiple factors, including genetics, inherited telomere length, lifestyle and environmental factors35. While the effect of environmental insults, such as smoking or infection, was highly variable and subject to huge inter-individual and temporal variations, the effects represented by the inherited telomere length and from the genotype of telomere-related genes are fixed. The comprehension of these variable and fixed effects could facilitate understanding of the pathogenesis and progression in complex disorders. Carriers with genetic variants of low penetrance in telomere biology genes could remain asymptomatic, while the telomere length maintenance defects keep accumulating as long as telomere length does not reach the threshold level. When these individuals encountered other fixed and/or variable factors that accelerate cellular turnover and telomere loss, such as exposure to cell death stimulants including smoking or recurrent infections, the defects in telomerase catalysis would be exacerbated. Over time, carriers of TERT SNPs should experience accelerated worsening of existing pathological conditions, as a function of the sum total of all etiologic factors that contributed to the loss of tissue integrity, leading to earlier onset or progression of multiple aging-related diseases, such as cancer27, IPF9, or COPD9 (Fig. 3).
The impact of genetic defects in telomere maintenance on lung function deterioration. Telomere Biology Diseases (TBD, right panel) are caused by rare genetic mutations that severely compromise the synthesis and/or the structural protection of the telomeres. TBD patients display extremely short telomeres at a young age, precipitating the onset of clinical symptoms. In contrast, carriers of common TERT genetic variants (SNPs) (middle panel) are not expected to display pathogenesis associated with short telomeres in the absence of additional factors (environmental, pathological or in combination with other genetic determinants) that accelerate telomere attritions. Even so, these carriers are still genetically predetermined to a decreased capacity to maintain their telomeres, when compared with noncarriers (left panel). These deficiencies, while silent phenotypically within the general population, could be unmasked by the massive increase in regenerative demand associated with tissue degenerative conditions, as seen in the case of COPD. Our work provides a proof-of-principle for novel epidemiological and clinical evaluations of the common SNPs in telomere biology genes on their effects as genetic modifiers rather than drivers of chronic tissue-degenerative disorders.
In contrast with our ex vivo model of rs61748181, where we observed telomere maintenance defects along the entire time course of the study, shorter telomeres were only observed in carriers of rs61748181 with COPD progression in the fourth quartile (non-decline in lung function group). We contend that while telomerase catalysis defects and accelerated telomere shortening should be consistent in carriers of rs61748181 within the LHS population, each individual’s variable factors (pathological factors and disease progression rate) could mask this direct correlation. Accordingly, we did not observe any cohort-wide association between rs61748181 with shorter telomere length. The moderate differences in telomere catalysis between CC homozygotes and rs6178181 minor allele carriers in a population could only be unmasked when the effects of these variable factors are removed, or approaching a fixed rate. We reasoned that maximum telomere attrition can be evident in the patient subgroup with the fastest disease progression profile. When we examined the subgroup of patients with the most negative ΔFEV15yrs, rapid lung tissue loss due to the end stage of the pathological processes indeed translated to a trend of shorter telomeres in carriers of rs61748181 minor allele than in CC patients, although statistical significance was not reached. Paradoxically, we demonstrated statistically significant difference in median telomere lengths between carriers of rs61748181 minor allele and CC-individuals, in a subgroup of patients with the most positive ΔFEV15yrs. Higher prevalence of smoking cessation (Supplementary Fig. 2) in this sub-group contributed to the stabilization of lung functions and reduced the lung tissue renewal demand that were associated with rapid telomere attrition. As well, reduction of direct oxidative damage on telomeres from cigarette smoke may also mitigate a significant environment variable in telomere length maintenance. We contend that the reduction in both (pathological and environmental) variables in telomere length regulation provided the opportunity to unmask the catalytic defects of rs61748181 minor allele at a population level.
Variants associated with mild penetrance in telomerase deficiency did not lead to cellular senescence in the absence of other confounding factors, and will not, on their own, drive the onset of chronic degenerative disorders. However, in the presence of accelerated cellular turnover, resulting from exposure to chronic environmental irritants and/or other genetic perturbations that affect tissue-specific functions, the presence of these genetic variants in the telomere biology pathway would be combined, resulting in faster telomere attrition. The effect of mildly deficient genetic polymorphisms in telomere biology genes can be seen as modifiers in accelerated COPD disease progression, when the pathogenetic process leads to excessive demand in lung tissue regeneration. Although the understanding of genetic profile in telomere biology genes does not unequivocally predict one’s disease susceptibility, we contend that a thorough understanding of all the determinants that affect cellular regenerative capacity is important in the prognosis and management of all existing chronic tissue degenerative disorders.
Methods
COPD patients
This study used clinical data and biological materials from the LHS. The LHS cohort initially recruited 5887 smokers, aged 35–60, with mild airway obstruction across 10 clinical centers in North America and collected their written informed consents36. Spirometry was performed annually over a period of 5 years for all study participants and in a subset after 11 years of study initiation as previously described37,38. Lung function was assessed as forced expiratory volume in one second (FEV1) % of predicted, i.e., FEV1 adjusted for age, height and sex39. We focused our data analyses on self-identified Caucasian patients in the LHS collection (n = 3596), as they constituted the major ethnicity group in the patient collection, and to avoid the confounding effects from haplotype differences in our analyses. In our study, FEV1 measurements over the 5-year follow-up period (reported as the slope from linear regression of FEV1 values plotted against time, ΔFEV15yrs) was used as a readout for changes in lung function and a measurement of COPD progression. For subgroup analyses based on disease progression, we restricted our study to 15% of the Caucasian subjects with confirmed genotype data, and included equal numbers of rapid- and slow decliners from each end of the linear regression of the rate of decline. This cutoff was used to capture extreme phenotypes that would be likely to be influenced by genetic factors, as we have done in previous studies. Using this cut-off, the rapid decliner group is restricted to LHS participants with ΔFEV1 lower than − 3.5, the characteristics of these selected subjects are shown in Table 2. All experiments were reviewed and performed in accordance to an Institutional Review Board-approved cohort study at the University of British Columbia (Providence Health Care Research Ethics Board certificate number H09-02042, ClinicalTrials.gov Identifier: NCT00000568).
Genotyping
A single polymorphism (rs61748181) from TERT was genotyped by TaqMan SNP Genotyping Assay (Applied Biosystems, Carlsbad, CA, USA). Template-free controls and known genotype controls were included in each experiment. In addition, for the purpose of assay quality control, 5% of the samples were typed again to check the repeatability of the genotyping assay. Genotypes were 100% reproducible in these samples. Herein, we refer WT as homozygous genotype for the reference allele (CC), while rs6178181 minor allele carriers are either homozygous or heterozygous carriers of the minor allele of rs61748181 (CT or TT).
Statistical analysis
Genotype distributions were assessed for Hardy–Weinberg equilibrium. Pearson’s χ2 test and student’s t-test or the Mann–Whitney test were used to assess the differences in dichotomous and continuous parameters between patients with different disease progression patterns, as well as participants with and without the minor allele of rs61748181. The frequencies of the alleles and genotypes between groups were analyzed by Pearson’s χ2 analysis for 2 × 2 contingency tables for crude odds ratios (ORs) and by multiple logistic regression for adjusted ORs. The ORs and their 95% confidence intervals (CI) were calculated. Multiple logistic regression for ΔFEV15yrs was performed to test the association with the TERT SNP. Confounding factors included age, gender, pack-years of smoking and smoking status. Statistical analyses were done using the JMP Statistics software package version 13 (SAS Institute Inc.) and R version 3.5.1. Figures were generated by GraphPad Prism 7.0 version (La Jolla, CA, US).
Cell culture
The primary human foreskin fibroblast BJ cell line was obtained from ATCC. All genetically-modified primary cell lines were cultured in DMEM high-glucose medium (Gibco, ThermoFisher Scientific, Waltham, MA, USA) with 15% fetal bovine serum (Gibco) and were maintained at 37 °C with 5% CO2. Population doubling levels (PDLs) were calculated based on the following equation: PDL = (log Xe − log Xb)/0.3 + S, where Xe was the cell harvest number, Xb was the inoculum number, and S was the starting PDL.
Retroviral gene delivery
The retroviral protein expression vector pBabe-hygro-Flag3B-WT-hTERT (a gift from Dr. Kathleen Collins) was used as the template for DpnI-mediated site-directed mutagenesis. Primers for the mutagenesis were synthesized by IDT DNA (Coralville, IA, USA) and are listed in Supplementary Table 4. The fidelity of positive clones was confirmed with Sanger sequencing of the entire TERT coding sequence (Genewiz, South Plainfield, NJ, USA). Silent mutations were introduced in each pair of primers to create a restriction site for easy identification of positive clones through restriction digestion with a selected endonuclease. The restriction enzymes of choice are also listed in Supplementary Table 4. Retroviral infection was performed by the 293 T cell-based system40 to integrate TERT (both WT and variant) coding sequences into the BJ genome. Hygromycin selection at 50 μg/ml was applied to select for positive clones for at least 14 days. Positive clones were pooled for continuous culture. Biological repeats of cell lines creation and long-term measurements for telomere maintenance dynamics were performed.
Telomere length measurement
The Southern blot-based terminal restriction fragment analysis (TRF) was applied to determine average telomere lengths in cell populations41. Data from TRF blots were analyzed by ImageQuant Software (GE Healthcare, Piscataway, NJ, USA) using weighted average analyses41. Data were reported as the nearest kilobase using the 1 kb DNA ladder (New England Biolabs, Beverly, MA, USA) as a reference.
Telomeres in peripheral blood leukocytes in the LHS cohort were measured by qPCR as previously described11.
Protein extraction
Cells were removed from culture flasks with trypsin/EDTA. Cell numbers in 100 μL of suspension were counted by Coulter Counter (Beckman Coulter, Brea, CA, USA) according to the manufacturer’s instruction and total cell numbers were calculated accordingly. Following the 2 × wash with ice-cold PBS, cells were resuspended in hypotonic lysis buffer (20 mM HEPES pH 8.0, 2 mM MgCl2, 0.2 mM EGTA, 10% glycerol, 1 mM DTT, and 0.1 mM PMSF). Cell suspensions were subjected to four consecutive freeze–thaw cycles from liquid nitrogen to a 37 °C water bath. NaCl was then added to the lysate in two parts to a final concentration of 400 mM. Cell suspensions were incubated on ice for 15 min before clearing the lysate by centrifugation at 13,200 rpm for 15 min. The supernatant (whole cell lysate, WCL) was transferred to a fresh tube, aliquoted, and stored in − 80 °C for further analysis. Protein concentrations were measured by Bradford protein assay.
Telomerase activity
Telomerase activity was measured by the classical radioactive primer extension assay42 using protein extracts from primary BJ cell lines expressing recombinant TERT isoforms. The quantifications of telomerase catalysis and repeat addition processivity were previously described43.
Immunoblot
100 μg WCL were resolved in 5% SDS-PAGE gel. A standard western blotting protocol was applied for the detection of TERT expression in the cell lysates. A sheep anti-human TERT polyclonal antibody (0.5 μg/ml, Abbexa, Cambridge, UK) and a Donkey anti-sheep horseradish peroxidase-conjugated antibody (0.2 μg/ml, Jackson ImmunoResearch, West Grove, PA, USA) were sequentially applied before detection by ECL (PerkinElmer, Waltham, MA, USA) on Kodak X-ray films. The cytoskeletal protein vinculin was used as a normalization control. It has a molecular weight of 124 kDa, which is similar to TERT (127 kDa). Rabbit anti-human vinculin monoclonal antibody (0.1 μg/ml, Cell Signaling, Danvers, MA, USA) and Alexa Fluor 680 Dye (ThermoFisher Scientific, Waltham, MA, USA) were applied before detection by Licor Odyssey CLx Infrared Imaging System (LI-COR Biotechnology, Lincoln, NE, USA) and quantification by ImageJ software (NIH, Bethesda, MD, USA).
Research ethics approval
This study utilized patient materials collected from an Institutional Review Board-approved cohort study at the University of British Columbia, with informed consent. All experiments in the study were reviewed and performed in accordance to the guidelines provided by the same Institutional Review Board at the University of British Columbia (Providence Health Care Research Ethics Board certificate number H09-02042, ClinicalTrials.gov Identifier: NCT00000568).
References
Armanios, M. & Blackburn, E. H. The telomere syndromes. Nat. Rev. Genet. 13, 693–704. https://doi.org/10.1038/nrg3246 (2012).
De Lange, T., Lundblad, V. & Blackburn, E. H. Telomeres 2nd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2006).
Blackburn, E. H. & Collins, K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a003558 (2011).
Alder, J. K. et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13051–13056. https://doi.org/10.1073/pnas.0804280105 (2008).
Alder, J. K. et al. Telomere length is a determinant of emphysema susceptibility. Am. J. Respir. Crit. Care Med. 184, 904–912. https://doi.org/10.1164/rccm.201103-0520OC (2011).
Armanios, M. Y. et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 356, 1317–1326. https://doi.org/10.1056/NEJMoa066157 (2007).
Ballew, B. J. & Savage, S. A. Updates on the biology and management of dyskeratosis congenita and related telomere biology disorders. Expert Rev. Hematol. 6, 327–337. https://doi.org/10.1586/ehm.13.23 (2013).
Alder, J. K. et al. Telomere dysfunction causes alveolar stem cell failure. Proc. Natl. Acad. Sci. U.S.A. 112, 5099–5104. https://doi.org/10.1073/pnas.1504780112 (2015).
Chilosi, M., Carloni, A., Rossi, A. & Poletti, V. Premature lung aging and cellular senescence in the pathogenesis of idiopathic pulmonary fibrosis and COPD/emphysema. Transl. Res. 162, 156–173. https://doi.org/10.1016/j.trsl.2013.06.004 (2013).
Savale, L. et al. Shortened telomeres in circulating leukocytes of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 179, 566–571. https://doi.org/10.1164/rccm.200809-1398OC (2009).
Lee, J. et al. The relationship between telomere length and mortality in chronic obstructive pulmonary disease (COPD). PLoS ONE 7, e35567. https://doi.org/10.1371/journal.pone.0035567 (2012).
Rode, L., Bojesen, S. E., Weischer, M., Vestbo, J. & Nordestgaard, B. G. Short telomere length, lung function and chronic obstructive pulmonary disease in 46,396 individuals. Thorax 68, 429–435. https://doi.org/10.1136/thoraxjnl-2012-202544 (2013).
Stanley, S. E. et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci. Transl. Med. 8, 35. https://doi.org/10.1126/scitranslmed.aaf7837 (2016).
Nunes, H. et al. Is telomeropathy the explanation for combined pulmonary fibrosis and emphysema syndrome? Report of a family with TERT mutation. Am. J. Respir. Crit. Care Med. 189, 753–754. https://doi.org/10.1164/rccm.201309-1724LE (2014).
Stanley, S. E. et al. Telomerase mutations in smokers with severe emphysema. J. Clin. Invest. 125, 563–570. https://doi.org/10.1172/JCI78554 (2015).
Zhou, J. J. et al. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am. J. Respir. Crit. Care Med. 188, 941–947. https://doi.org/10.1164/rccm.201302-0263OC (2013).
Mushiroda, T. et al. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J. Med. Genet. 45, 654–656. https://doi.org/10.1136/jmg.2008.057356 (2008).
Vulliamy, T. J. et al. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol. Dis. 34, 257–263. https://doi.org/10.1016/j.bcmd.2004.12.008 (2005).
Yamaguchi, H. et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 352, 1413–1424. https://doi.org/10.1056/NEJMoa042980 (2005).
Ding, Y. et al. TERT gene polymorphisms are associated with chronic obstructive pulmonary disease risk in the Chinese Li population. Mol. Genet. Genomic Med. 7, e773. https://doi.org/10.1002/mgg3.773 (2019).
Arimura-Omori, M. et al. Association between telomere-related polymorphisms and the risk of IPF and COPD as a precursor lesion of lung cancer: findings from the fukuoka tobacco-related lung disease (FOLD) registry. Asian Pac. J. Cancer Prev. (APJCP) 21, 667–673. https://doi.org/10.31557/apjcp.2020.21.3.667 (2020).
Tsuji, T., Aoshiba, K. & Nagai, A. Alveolar cell senescence in patients with pulmonary emphysema. Am. J. Respir. Crit. Care Med. 174, 886–893. https://doi.org/10.1164/rccm.200509-1374OC (2006).
Kachuri, L. et al. Fine mapping of chromosome 5p15.33 based on a targeted deep sequencing and high density genotyping identifies novel lung cancer susceptibility loci. Carcinogenesis 37, 96–105. https://doi.org/10.1093/carcin/bgv165 (2016).
Zhang, Y. et al. Telomerase variant A279T induces telomere dysfunction and inhibits non-canonical telomerase activity in esophageal carcinomas. PLoS ONE 9, e101010. https://doi.org/10.1371/journal.pone.0101010 (2014).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74. https://doi.org/10.1038/nature15393 (2015).
Qiao, D. et al. Whole exome sequencing analysis in severe chronic obstructive pulmonary disease. Hum. Mol. Genet. https://doi.org/10.1093/hmg/ddy269 (2018).
Shay, J. W. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593. https://doi.org/10.1158/2159-8290.CD-16-0062 (2016).
Calado, R. T. et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 106, 1187–1192. https://doi.org/10.1073/pnas.0807057106 (2009).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Shay, J. W. & Wright, W. E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 1, 72–76. https://doi.org/10.1038/35036093 (2000).
Zaug, A. J., Crary, S. M., Jesse Fioravanti, M., Campbell, K. & Cech, T. R. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 41, 8969–8978. https://doi.org/10.1093/nar/gkt653 (2013).
Daniali, L. et al. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun. 4, 1597. https://doi.org/10.1038/ncomms2602 (2013).
Saferali, A. et al. Longer telomere length in COPD patients with α1-antitrypsin deficiency independent of lung function. PLoS ONE 9, e95600. https://doi.org/10.1371/journal.pone.0095600 (2014).
Demanelis, K. et al. Determinants of telomere length across human tissues. Science https://doi.org/10.1126/science.aaz6876 (2020).
Trudeau, M. A. & Wong, J. M. Genetic variations in telomere maintenance, with implications on tissue renewal capacity and chronic disease pathologies. Curr. Pharmacogenomics Pers. Med. 8, 7–24. https://doi.org/10.2174/1875692111008010007 (2010).
Connett, J. E., Kusek, J. W., Bailey, W. C., O’Hara, P. & Wu, M. Design of the lung health study: a randomized clinical trial of early intervention for chronic obstructive pulmonary disease. Control Clin. Trials 14, 3S-19S (1993).
Anthonisen, N. R., Connett, J. E. & Murray, R. P. Smoking and lung function of Lung Health Study participants after 11 years. Am. J. Respir. Crit. Care Med. 166, 675–679. https://doi.org/10.1164/rccm.2112096 (2002).
Anthonisen, N. R. et al. Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1. The Lung Health Study. JAMA 272, 1497–1505 (1994).
Crapo, R. O., Morris, A. H. & Gardner, R. M. Reference spirometric values using techniques and equipment that meet ATS recommendations. Am. Rev. Respir. Dis. 123, 659–664. https://doi.org/10.1164/arrd.1981.123.6.659 (1981).
Swift, S., Lorens, J., Achacoso, P. & Nolan, G. P. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Protoc. Immunol. https://doi.org/10.1002/0471142735.im1017cs31 (2001).
Zeng, X. L. et al. The accumulation and not the specific activity of telomerase ribonucleoprotein determines telomere maintenance deficiency in X-linked dyskeratosis congenita. Hum. Mol. Genet. 21, 721–729. https://doi.org/10.1093/hmg/ddr504 (2012).
Hukezalie, K. R., Thumati, N. R., Côté, H. C. & Wong, J. M. In vitro and ex vivo inhibition of human telomerase by anti-HIV nucleoside reverse transcriptase inhibitors (NRTIs) but not by non-NRTIs. PLoS ONE 7, e47505. https://doi.org/10.1371/journal.pone.0047505 (2012).
Latrick, C. M. & Cech, T. R. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29, 924–933. https://doi.org/10.1038/emboj.2009.409 (2010).
Acknowledgements
We thank Sunny Yang for constructive comments on the manuscript and the patients and clinicians of the LHS for providing valuable clinical samples for our study. This work is supported by a grant to JMYW from the Natural Sciences and Engineering Research Council of Canada (RGPIN-05843).
Author information
Authors and Affiliations
Contributions
J.X., A.J.S. and J.M.Y.W. conceptualized the project and planned the experiments. J.X., M.A.T and J.C. performed the experiments and collected the data. J.X., D.Md.O. and Y.Y. analyzed the data, J.X., D.Md.O., A.J.S. and J.M.Y.W. co-wrote the manuscript. J.C., D.D.S. and A.J.S. performed the PCR-based telomere length measurements of COPD samples from the LHS population. All authors (J.X., D.Md.O., Y.Y., M.A.T., J.C., D.D.S., A.J.S. and J.M.Y.W.) contributed to the revisions and the editing of the final manuscript. JMWY is the corresponding author and guarantor of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Dr. Don Sin reports grants from Merck, personal fees from Sanofi-Aventis, personal fees from Regeneron, grants and personal fees from Boehringer Ingelheim, grants and personal fees from AstraZeneca and personal fees from Novartis. These financial interests bear no relation with the submitted work. The other authors declared no competing interests on this work.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, J., de Oliveira, D.M., Trudeau, M.A. et al. Mild catalytic defects of tert rs61748181 polymorphism affect the clinical presentation of chronic obstructive pulmonary disease. Sci Rep 11, 4333 (2021). https://doi.org/10.1038/s41598-021-83686-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-83686-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.