Abstract
The PvuII (rs2234693) Single Nucleotide Polymorphism (SNP) in the gene coding for the estrogen receptor-1 (ESR1), has been found associated with outcome in tamoxifen treated patients with early hormone-receptor positive breast cancer. However, it remains unclear whether this SNP is a predictive marker for tamoxifen efficacy or a prognostic marker for breast cancer outcome. The aim of this study was to examine the prognostic potential of this SNP in postmenopausal early breast cancer patients treated with adjuvant exemestane. Dutch postmenopausal patients randomised to 5 years of adjuvant exemestane of whom tissue was available (N = 807) were selected from the Tamoxifen Exemestane Adjuvant Multinational (TEAM) trial database. The SNP rs2234693 in the ESR1 gene was genotyped on DNA from formalin-fixed paraffin embedded (FFPE) tumor tissue using Taqman assays and related to the primary endpoint disease-free survival (DFS) and secondary endpoint overall survival (OS). Survival analyses were performed using Cox regression analysis. In total 805 patients were included in the analyses (median follow up of 5.22 years) and genotypes were obtained in 97% of the samples. The variant T allele of PvuII in ESR1 (rs2234693) was associated with a better DFS (hazard ratio (HR) 0.689, 95% confidence interval (CI) 0.480–0.989, P = 0.044) in univariate analysis only, and a better OS in both univariate (HR 0.616, 95%, CI 0.411–0.923, P = 0.019) and multivariate analyses (HR 0.571, 95% CI 0.380–0.856, P = 0.007), consistent with a prognostic rather than a predictive drug response effect. Variation of PvuII in the ESR1 gene is related to OS in postmenopausal, early HR + breast cancer patients treated with exemestane in the TEAM study. Variation in the ESR1 gene may therefore be a prognostic marker of early breast cancer survival, and warrants further research.
Similar content being viewed by others
Background
Endocrine treatment is the cornerstone of treatment of hormone receptor positive breast cancer1, and for many years, tamoxifen has been the gold standard adjuvant endocrine therapy2. Over the last decades, however, in postmenopausal women third generation aromatase inhibitors have consistently demonstrated superior efficacy over tamoxifen, and optimal treatment is sequential use of tamoxifen followed by an aromatase inhibitor or an aromatase inhibitor alone1,3,4,5. Unfortunately, still 40% of women using optimal endocrine therapy experience a relapse6. To date, only the presence of hormone receptors are predictive markers for outcome7. Identification of more accurate biomarkers remains crucial to predict which women are responsive to endocrine treatment and the optimal therapy6,8. In a pharmacogenetic analysis of the Tamoxifen Exemestane Adjuvant Multinational (TEAM) study an association between tamoxifen efficacy and the rs2234693 SNP in the estrogen receptor-1 (ESR1) gene has been found in postmenopausal hormone receptor positive breast cancer patients, whereas patients with an increasing number of C alleles of PvuII in ESR1 had a decreased disease free survival (DFS)9. Moreover, in a case–control study patients carrying the CC or CT genotype had a 4.14-fold increased relative risk to develop breast cancer10. Results from other studies on breast cancer risk or survival after breast cancer are conflicting, which may be explained by differences in hormone receptor expression, menopausal status or ethnicity11,12,13,14. Therefore, it remains unclear if this biomarker is predictive or prognostic, i.e. the latter being independent of treatment.
In order to investigate the value of this biomarker further, the aim of this TEAM substudy was to examine the prognostic potential of this SNP in postmenopausal, early breast cancer patients treated with adjuvant exemestane alone.
Methods
Study population
For the current pharmacogenetic substudy, data and tumor tissue of Dutch patients treated with exemestane alone enrolled in the Tamoxifen Exemestane Adjuvant Multinational (TEAM) trial were used. From January 2001 until January 2006 women participated in the multinational TEAM trial, randomizing between 5 years adjuvant exemestane (25 mg once a day, orally) or 2.5–3 years tamoxifen (20 mg once a day, orally) followed by exemestane for a total of 5 years6. Eligible patients had histologically confirmed hormone receptor positive early breast cancer, postmenopausal status and completed local treatment with curative intent. Other inclusion and exclusion criteria have been described elsewhere6. Informed consent was obtained from all individual participants included in the study. The international TEAM study was conducted in accordance with the Declaration of Helsinki and was approved by the respective institutional Ethics Committees whereof the Dutch part was approved by the central medical ethics review board of the Erasmus University Medical Center in Rotterdam. The current pharmacogenetic study was separately approved by the central medical ethics review board of the Erasmus University Medical Center in Rotterdam, The Netherlands in accordance with the Declaration of Helsinki.
Endpoints
The primary endpoint of the current substudy was DFS, defined as time from date of randomization until the date of the earliest documented locoregional or distant recurrence, ipsilateral or contralateral breast cancer excluding ductal carcinoma in situ, or death from any cause. A secondary endpoint was overall survival (OS), defined as time from randomization until date of death from any cause.
Genotyping
For genotyping tumor-negative tissue was used, however, when this was unavailable tissue from tumor-containing blocks were used. DNA was extracted from the FFPE tumor tissue as described earlier15. Briefly, three slides of 20 μm were used to extract DNA from with the Maxwell forensic DNA isolation kit (Promega, Leiden, The Netherlands) according to manufacturer’s protocol. Pre-amplification was accomplished for enrichment of the target DNA15. Genotypes of PvuII in ESR1 (rs2234693) were established using commercially available pre-designed Taqman assays obtained from Applied Biosystems (Nieuwerkerk aan den IJssel, the Netherlands). Endpoint genotyping was performed on the LightCycler 480 Real Time PCR System (Roche, Almere, The Netherlands) according to standard procedures.
Statistical analysis
A SNP call rate > 85% was found acceptable, as FFPE samples were used and therefore a low call rate could be expected. Genotype distributions were tested for the Hardy–Weinberg equilibrium (HWE) and chi-squared test were performed. Cox regression models were used to evaluate the effect of the SNP on covariates and DFS and OS. Hazard ratios (HRs) and 95% confidence intervals (95% CIs) were given. If the SNP was found to be (borderline) significant with a p-value of less than 0.1 in univariate Cox regression models it carried forward to a multivariate model. Risk factors found to have a p-value of less than 0.1 in univariate analyses were carried forward in the multivariate model as well. The median follow-up time was calculated by the reverse Kaplan–Meier methodology16. All tests were two tailed and p-values of less than 0.05 were considered significant. The data was analyzed using Statistical Package for Social Sciences (SPSS) software 23.0 (IBM Corp., Armonk, NY, USA).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the central medical ethics review board of the Erasmus University Medical Center in Rotterdam, The Netherlands and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Results
Patient characteristics
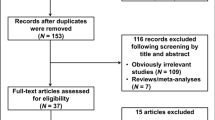
From the 9779 patients enrolled in the international TEAM study, 2753 patients were included in the Netherlands, whereof 1374 were randomized to exemestane alone. FFPE tissue was available of 807 (58.7%) of the Dutch exemestane patients. Two patients were ineligible because of metastatic disease before they started treatment and were therefore excluded from analyses as summarized in the consort diagram (Fig. 1). No genotypes could be called due to insufficient DNA quality for 74 patients.
Genotyping
The genotyping call rate for the rs2234693 SNP was 97%, as shown in Table 1. The genotyped samples (N = 731) were not significantly different from the whole group (N = 805) concerning age, BMI, TNM stage, tumor size, nodal status, histological grade of the tumor, progesterone status, type of operation (wide excision or mastectomy) and received adjuvant therapy, as determined by visually comparing continuous distributions and comparing frequencies for categorical variables, making informative drop-out unlikely (data not shown). Genotype frequencies were similar to those observed in a publicly available dataset from European subjects, with a Minor Allele Frequency (MAF) of the C allele of 0.464.
Survival analysis
The median follow-up for the genotyped samples of Dutch Exemestane patients was 5.22 years (95% CI 5.10–5.35). Kaplan–Meier curves are shown in Fig. 2. The estimated HRs and associated 95% CIs for univariate and multivariate Cox model analyses for DFS and OS, respectively, are shown in Tables 2 and 3. The risk factors T stage, PR status, histological grade and type of operation were associated with DFS, while T stage and type of surgery were associated with OS. The multivariate Cox models were adjusted for these covariates. The variant T allele of PvuII (rs2234693) in ESR1 was associated with a better DFS (HR 0.689, 95% CI 0.480–0.989, P = 0.044) in univariate analysis, but not anymore in multivariate analysis (HR 0.696, 95% CI 0.469–1.031, P = 0.071). Moreover, the variant T allele of this SNP was associated with improved OS (HR 0.616, 95% CI 0.411–0.923, P = 0.019) in both univariate and multivariate analyses (HR 0.571, 95% CI 0.380–0.856, P = 0.007).
Discussion
We found that the T genotype of PvuII (rs2234693) in ESR1 was associated with a better OS and a trend for better DFS than the C genotype in postmenopausal, hormone receptor positive early breast cancer patients treated with adjuvant exemestane alone in the TEAM study. We previously described the association of the TT and TC genotypes (versus CC genotype) of PvuII in ESR1 and a better survival in a comparable subgroup of Dutch TEAM patients treated with tamoxifen9. Therefore, our results are more consistent with a prognostic factor, rather than a predictive drug response effect, as this SNP is related to OS in early breast cancer patients for both types of endocrine treatment (exemestane or tamoxifen). Particularly, because exemestane and tamoxifen differ in working mechanism17.
Our results are in line with those of Gabrinski et al. who found an association between the ESR1 PvuII genotype and DFS regardless of type of endocrine treatment, however final data are not yet published18.
The PvuII restriction site is localized in intron 1 of the ESR1, 400 basepairs upstream of exon 213. Loss of the PvuII restriction site due to T- > C transition, may result in a binding site for transcription factor B-Myb, which amplifies transcription of the estrogen receptor19,20. This upregulation of the estrogen receptor possibly explains the association of the C-allele with worse survival. Alternatively, this SNP may be in linkage disequilibrium with causal synonymous polymorphisms elsewhere in the ESR1 or another gene19.
A limitation of our study is that DNA from FFPE samples is not ideal for genotyping and samples with intact genomic DNA as blood or frozen tissue would be preferred, however, this was not available in our study, and earlier research showed that FFPE tumor tissue and normal tissue was highly concordant21,22. Although there are discordant results described in the literature, for example the deletion of the CYP2D6 gene in breast tumor tissue is reported to cause departures from HWE23, it seems unlikely that the ESR1 gene is deleted in our study as HWE is not violated (Table 1).
As the C allele of rs2234693 is a frequently observed allele with a MAF of 0.464 in our study, this SNP is of interest and may be validated in other large studies. Moreover, to personalize endocrine therapy in breast cancer in the future a 18F‐fluoroestradiol (FES) positron emission tomography (PET) may be used for whole‐body imaging for receptor status assessment to research the phenotype-genotype of the PvuII in ESR124.
Conclusions
The T allele of PvuII (rs2234693) in the ESR1gene is associated with improved overall survival in postmenopausal, hormone receptor positive early breast cancer patients and may be considered as a prognostic marker in early breast cancer. Further studies into the prognostic value of this biomarker are warranted.
Abbreviations
- DFS:
-
Disease-free survival
- ESR1:
-
Estrogen receptor-1
- FES:
-
18F‐fluoroestradiol
- FFPE:
-
Formalin-fixed paraffin embedded
- HR:
-
Hazard ratio
- HWE:
-
Hardy–Weinberg equilibrium
- MAF:
-
Minor Allele Frequency
- OS:
-
Overall survival
- PET:
-
Positron emission tomography
- SNP:
-
Single nucleotide polymorphism
- SPSS:
-
Statistical package for social sciences
- TEAM:
-
Tamoxifen exemestane adjuvant multinational
References
Blok, E. J., Derks, M. G., van der Hoeven, J. J., van de Velde, C. J. & Kroep, J. R. Extended adjuvant endocrine therapy in hormone-receptor positive early breast cancer: current and future evidence. Cancer Treat Rev. 41, 271–276. https://doi.org/10.1016/j.ctrv.2015.02.004 (2015).
Osborne, C. K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 339, 1609–1618. https://doi.org/10.1056/NEJM199811263392207 (1998).
Derks, M. G. M. et al. Adjuvant tamoxifen and exemestane in women with postmenopausal early breast cancer (TEAM): 10-year follow-up of a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 18, 1211–1220. https://doi.org/10.1016/S1470-2045(17)30419-9 (2017).
Howell, A. et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet 365, 60–62. https://doi.org/10.1016/S0140-6736(04)17666-6 (2005).
Coates, A. S. et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: update of study BIG 1–98. J. Clin. Oncol. 25, 486–492. https://doi.org/10.1200/JCO.2006.08.8617 (2007).
van de Velde, C. J. et al. Adjuvant tamoxifen and exemestane in early breast cancer (TEAM): a randomised phase 3 trial. Lancet 377, 321–331. https://doi.org/10.1016/S0140-6736(10)62312-4 (2011).
Noordhoek, I. et al. Higher ER load is not associated with better outcome in stage 1–3 breast cancer: a descriptive overview of quantitative HR analysis in operable breast cancer. Breast Cancer Res. Treat. 176, 27–36. https://doi.org/10.1007/s10549-019-05233-9 (2019).
Ayoub, N., Lucas, C. & Kaddoumi, A. Genomics and pharmacogenomics of breast cancer: current knowledge and trends. Asian Pac. J. Cancer Prev. 12, 1127–1140 (2011).
Dezentje, V. O. et al. CYP2D6 genotype in relation to tamoxifen efficacy in a Dutch cohort of the tamoxifenexemestane adjuvant multinational (TEAM) trial. Breast Cancer Res. Treat. 140, 363–373. https://doi.org/10.1007/s10549-013-2619-6 (2013).
Madeira, K. P. et al. Estrogen receptor alpha (ERS1) SNPs c454–397T>C (PvuII) and c454–351A>G (XbaI) are risk biomarkers for breast cancer development. Mol. Biol. Rep. 41, 5459–5466. https://doi.org/10.1007/s11033-014-3419-8 (2014).
Boyapati, S. M. et al. Polymorphisms in ER-alpha gene interact with estrogen receptor status in breast cancer survival. Clin. Cancer Res. 11, 1093–1098 (2005).
Ghali, R. M. et al. Differential association of ESR1 and ESR2 gene variants with the risk of breast cancer and associated features: a case-control study. Gene 651, 194–199. https://doi.org/10.1016/j.gene.2018.02.011 (2018).
Modugno, F. et al. Association of estrogen receptor alpha polymorphisms with breast cancer risk in older Caucasian women. Int. J. Cancer 116, 984–991. https://doi.org/10.1002/ijc.21105 (2005).
Zhang, Y. et al. Association between ESR1 PvuII, XbaI, and P325P polymorphisms and breast cancer susceptibility: a meta-analysis. Med. Sci. Monit. 21, 2986–2996. https://doi.org/10.12659/MSM.894010 (2015).
Baak-Pablo, R., Dezentje, V., Guchelaar, H. J. & van der Straaten, T. Genotyping of DNA samples isolated from formalin-fixed paraffin-embedded tissues using preamplification. J. Mol. Diagn. 12, 746–749. https://doi.org/10.2353/jmoldx.2010.100047 (2010).
Schemper, M. & Smith, T. L. A note on quantifying follow-up in studies of failure time. Control Clin. Trials 17, 343–346. https://doi.org/10.1016/0197-2456(96)00075-x (1996).
Lonning, P. E. The potency and clinical efficacy of aromatase inhibitors across the breast cancer continuum. Ann. Oncol. 22, 503–514. https://doi.org/10.1093/annonc/mdq337 (2011).
Grabinski, J. L. et al. ER alpha genotypes and breast cancer recurrence. J. Clin. Oncol. https://doi.org/10.1200/jco.2008.26.15_suppl.501 (2008).
Anghel, A. et al. Estrogen receptor alpha polymorphisms: correlation with clinicopathological parameters in breast cancer. Neoplasma 57, 306–315 (2010).
Herrington, D. M. et al. Common estrogen receptor polymorphism augments effects of hormone replacement therapy on E-selectin but not C-reactive protein. Circulation 105, 1879–1882 (2002).
van Huis-Tanja, L. et al. Concordance of genotype for polymorphisms in DNA isolated from peripheral blood and colorectal cancer tumor samples. Pharmacogenomics 14, 2005–2012. https://doi.org/10.2217/pgs.13.169 (2013).
Pander, J. et al. Correlation between germline polymorphisms and the efficacy of cetuximab in metastatic colorectal cancer. Eur. J. Cancer 46(10), 1829–1834 (2010).
Goetz, M. P. et al. Re: concordance between CYP2D6 genotypes obtained from tumor-derived and germline DNA. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/dju063 (2014).
Venema, C. M. et al. Androgen and estrogen receptor imaging in metastatic breast cancer patients as a surrogate for tissue biopsies. J. Nucl. Med. 58, 1906–1912. https://doi.org/10.2967/jnumed.117.193649 (2017).
Acknowledgements
We thank all the participating hospitals and patients for participation in the trial. Furthermore, we thank the Datacenter Heelkunde for data collection and preparation of the datasets. The long-term TEAM study datamanagement was supported by a Dutch Cancer Society grant.
Funding
This TEAM trial was supported by unrestricted Grants from Pfizer and KWF.
Author information
Authors and Affiliations
Contributions
E.M.K.K., C.M.S., C.J.H.vdV. participated in the data acquisition and coordination of the TEAM study. R.B.P., D.H., H.J.G., H.G. initiated and designed the pharmacogenetic study. S.G., D.H. and R.B.P. designed the experiments and interpreted the data. S.B. gave advice on the statistical analysis and wrote the statistical section of the manuscript. S.G. and D.H. performed statistical analysis, wrote the manuscript and was responsible for editing, and preparation of the manuscript for submission. H.J.G., H.G., J.R.K., E.M.K.K. critical revised the manuscript. All authors read and approved the final article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Houtsma, D., de Groot, S., Baak-Pablo, R. et al. The variant T allele of PvuII in ESR1 gene is a prognostic marker in early breast cancer survival. Sci Rep 11, 3249 (2021). https://doi.org/10.1038/s41598-021-82002-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82002-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.