Abstract
The architectural features of cellular life and its ecologies at larger scales are built upon foundational networks of reactions between molecules that avoid a collapse to equilibrium. The search for life’s origins is, in some respects, a search for biotic network attributes in abiotic chemical systems. Radiation chemistry has long been employed to model prebiotic reaction networks, and here we report network-level analyses carried out on a compiled database of radiolysis reactions, acquired by the scientific community over decades of research. The resulting network shows robust connections between abundant geochemical reservoirs and the production of carboxylic acids, amino acids, and ribonucleotide precursors—the chemistry of which is predominantly dependent on radicals. Moreover, the network exhibits the following measurable attributes associated with biological systems: (1) the species connectivity histogram exhibits a heterogeneous (heavy-tailed) distribution, (2) overlapping families of closed-loop cycles, and (3) a hierarchical arrangement of chemical species with a bottom-heavy energy-size spectrum. The latter attribute is implicated with stability and entropy production in complex systems, notably in ecology where it is known as a trophic pyramid. Radiolysis is implicated as a driver of abiotic chemical organization and could provide insights about the complex and perhaps radical-dependent mechanisms associated with life’s origins.
Similar content being viewed by others
Introduction
The origins of life may be conceptualised as a chronology that occurred within a completely enclosed box. Initially, energy and molecules are present within the box as abiotic, geochemically plausible forms of photons, gases, liquids and rocks. After some period of time, the chemical composition of the box includes biomolecules, living cells, molecular waste products, waste heat and any remaining initial substrates and side products. The basic assumption is that life and its organizational properties emerged from the conditions and materials present within the box, and was not delivered from elsewhere. At what point in this chronology did life’s complex organizational attributes arise? Are there abiotic settings that exhibit ‘life-like’ behaviours or chemical relationships near the very beginning of this prebiotic chronology?
Radiation chemistry has long been employed to model prebiotic chemical reaction networks, starting with Calvin’s 1951 report that radiolysis of aqueous CO2 results in its reduction1. Radical chemistry is responsible for a large fraction of the products observed after radiolysis of dilute aqueous solutions containing various solutes. The relatively promiscuous nature of radicals, generated by radiolysis or other sources of energy, may be a possible means of driving the de novo emergence of chemical systems with complex attributes. Radicals (species with unpaired outer electrons) afford diverse sets of products starting from simple initial inputs and conditions, and can be generated by a variety of plausible mechanisms including ionizing radiation2, UV light3, spark discharge4 and mineral corrosion5, all of which have been implicated in proposed early Earth scenarios6. Radical reactions can proceed via low barriers without requiring further activation by a catalyst7. Radicals can exhibit powerful electrochemical potentials (far greater than even the most potent combinations of geochemical substrates8) and can be readily sourced from both organic and inorganic substrates to drive a wide array of organic redox reactions9,10,11. Indeed, a number of enzyme-facilitated biochemical redox potentials exceed tabulated geochemical potentials, indicating that non-enzymatic predecessors of these reactions, for kinetic reasons alone, may have required radiolysis, photolysis or extremely powerful phosphorylating intermediates to drive protometabolism12. For all of these reasons, radical-driven chemical networks may serve as insightful model systems for investigating the production of prebiotic molecules.
The chemical reactions that produce key reactive prebiotic molecules can be analysed at the network level much in the same ways as ecosystems and metabolic pathways13,14,15,16. A network is constructed by counting the object types (e.g., distinct molecules, atoms or photons) within a system and graphing each as a node; interactions (e.g., chemical reactions) between objects are depicted with lines, which are called edges. Mapped network interactions offer clues to how the laws of physics and chemistry can give rise to emergent features17. By describing the relationships between chemical objects that are synthesised alongside one another18,19, elements of hierarchical organization, feedback and order can be grasped—features that are missed at the level of studying individual reactions20,21,22. Through the analysis of reaction network maps, it is possible to assess whether an abiotic chemical system exhibits complex organizational attributes associated with life.
Living systems exhibit persistent network attributes that reflect a balance between generic operating principles (e.g., persistence, robustness or redundancy) and the functional constraints associated with exhibiting these principles (e.g., cost, resource limitation or transport efficiency)23. We focus on three such attributes that are also implicated with broader examples of complex systems behaviour—states of matter capable of dynamic response as a result of state-dependent feedback mechanisms that regulate internal relationships between objects24,25,26,27. As in many different examples of complex systems, biological objects interact with a heterogeneous distribution that appears as a gradually decreasing slope (i.e., a ‘heavy tail’) on a connectivity histogram28,29. An observable is heavy-tailed if the probability of observing extremely large values is more likely than it would be for an exponentially-distributed variable30. Biological objects are also formed and interconverted within a topology of overlapping cycles of energy and matter31,32,33,34. Cycles are critical relationships that afford dynamic attributes such as buffering against supply fluctuations35, persistence36,37 and homeostatic maintenance38,39. Finally, biological objects exist within a nested hierarchy40,41 in which the production of numerous, smaller-sized objects supports the production of fewer, larger-sized objects. This arrangement is also known as a bottom-heavy energy-size spectrum42 which in the field of ecosystem ecology is more commonly referred to as a trophic pyramid. Bottom-heavy population distributions are intrinsically correlated with energy flow and entropy production across hierarchical tiers42,43, as objects at each level are inefficiently cycled through formation and degradation in closed loops to maintain far-from-equilibrium configurations44,45,46,47.
It was recently demonstrated that radiolysis, by means of a continuous reaction network, generates a complex mixture of products that includes precursors for nucleotide synthesis48. These findings add to a growing body of evidence that radicals provide particularly efficient means of connecting geochemical substrates to macromolecular precursors with minimal experimental intervention2,12,49,50,51,52,53,54. Here we analyse the structure of a reaction network compiled from hundreds of experiments conducted over 60 years of research that links common geochemical substrates (CO2, H2O, N2, NaCl, chlorapatite and pyrite) to radiolytically-produced prebiotic precursors, and compare this network to life and its ecologies55,56,57.
Results
A chemical reaction network of solid-, liquid-, and gas-phase reactions was assembled from 44 peer-reviewed publications spanning 31 journals over a period of time from 1961 to 2020 (Datafile S1). The network includes 782 reactions and 386 distinct atomic, molecular, and photonic species. To aid visualisation of the network, reactions were assigned to chemical reaction categories based on their reactive substrates (e.g., free radical reactions, physicochemical reactions, nitrile radical reactions, chloride reactions, geochemical reactions, etc.). Atoms are initially present in the system as their most geochemically abundant molecular species (H2O, CO2, N2, NaCl, a proxy for the apatite mineral group Ca10Cl2(PO4)6, and pyrite FeS2). The electromagnetic spectrum is binned into gamma, X-ray, UV, visible and infrared photons to account for reactions across key atomic and molecular energetic thresholds (i.e., strong nuclear binding force, inner valence electrons, outer valence electrons and ambient system temperature). Radiolytic energy was incorporated from natural modes of decay of radiogenic uranium oxide, which serves as a generic geochemical proxy for naturally-occurring radiolytic energy sources such as galactic cosmic radiation58, solar flares59 and planetary radiation belts60. A complete list of these reactions and source manuscripts is provided in Supplementary Datafile S1 (Excel spreadsheet) and Datafile S2 (.graphml network file).
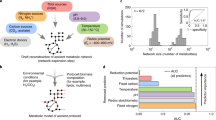
The radiolysis network is depicted in Fig. 1a using a ForceAtlas2 layout. Species (green) have radii weighted according to the total number of connections. The network has a diameter of 28, with an average directed path length of about 5.4 steps. Radical reactions (shaded in red) contain hubs that dominate the network and link together inputs and outputs from each of the reaction categories. The most connected network hub is water (234 total degree connections; Supplementary Table S1), but the next largest hubs are the low-mass, highly reactive reducing H (165 connections) and oxidising OH (122 connections) radicals, which are readily produced by radiolysis or photolysis of water. The most powerful forms of energy in the system (gamma rays, alpha particles, beta particles, X rays and UV light) are also major hubs within the network. Carboxylic acids are produced via radical reactions53. Amino acids are produced through a combination of radiolysis and acid-hydrolysis of uncharacterised polymers2,61. Nucleotide precursors that lead to cytidine and uridine cyclic phosphates are all present within the network48,49,62.
Visualisation and statistical analysis of radiolysis network. (a) Bi-partite network with molecular species (green circles) size-weighted according to degree of connectivity to reactions (red circles). Groupings of chemical reaction categories are individually shaded and labelled. (b) Statistical comparison of species node degree connectivity distribution indicates that an exponential fall-off fit is poor compared to a power law or lognormal fit of the empirical data. (c) Statistical comparison of the cycle size distribution indicates that an exponential fall-off fit is not reliably distinguishable from a power law, lognormal or exponentially truncated power law fit of the empirical data.
Heavy-tailed connectivity distribution
A list of the most connected chemical species is provided in Supplementary Table S1. Scaling analysis and, specifically, evaluation of the relevance of the power law scaling, were conducted as described in the “Methods” section. The analysed dataset includes 384 degree values, corresponding to the number of nodes in the partition of the bi-partite network (Fig. 1a, green nodes) comprising reaction participants such as molecules, radicals, ions, etc. Fitting a power law to the observed degree distribution yields α = − 1.56 with standard error σ = 0.03. The cumulative probability of the node degree for the reaction components found in the system was assessed using a combination of significance value (p) and loglikelihood ratio (R) analyses to determine whether it more closely follows a heavy-tailed distribution, such as a power law, than a distribution with an exponential fall-off (Fig. 1b; please see “Methods” section for specific details of the statistical tools employed). The positive sign of log-likelihood ratio R = 198.3 and significance value of p < 0.05 (p = 2e−8) indicated that a heavy-tailed distribution fits the data better (see Refs.63,97 for more detailed discussion regarding the interpretation of R and p values). A similar comparison of heavy-tailed distribution types between power law and lognormal distributions (Fig. 1b) indicated that a lognormal distribution was a better fit than a power law distribution (negative sign of R = − 13.1, p = 2e−5).
Enumerated closed-loop cycling of chemical species
Reactions that compose the network create at least 2618 possible closed cycles of molecules and photons. Chemical species ranked by frequency of presence within closed cycle subnetworks is provided in Supplementary Table S2. Water was explicitly excluded as an intermediate in enumerated cycles owing to its frequent presence as an unreactive bystander (solvent) molecule. Statistical analyses of cycle size distribution (number of nodes in each cycle, inclusive of participating chemical species and reactions) were conducted as described in the preceding paragraph and the Methods section. Each comparison had p values that exceeded a p < 0.05 threshold for statistically distinguishing between better fits63. This indicates that an exponential fall-off fit is not strongly distinguishable from heavy-tailed distributions such as power law (p = 0.90), lognormal (p = 0.37) or exponentially truncated power law (p = 0.29) fits (Fig. 1c).
Most cycles of reactions include low mass radicals directly produced from radiolysis and recombination of feedstock molecules (e.g., N2, CO2, H2O, etc.) such as H, N, O, OH, CN, NO, HNO, etc. H, OH and UV photons are each present in over 1000 of the total number of 2618 enumerated cycles. Three examples of overlapping cycles (IDX8, IDX1763, IDX916; IDX denotes the software-assigned cycle identification number) that include the most common species found in all cycles are illustrated in Fig. 2. The coupling of simple radicals to one another and to the initial substrates, and then to higher mass molecules produced over time, makes possible more complex cycles as molecular species diversify in the system. The cycle enumeration analysis indicates that as energy is input into the system, a facile return to the initial state (or to any particularly simple final state) becomes unlikely over time. As the number of organic species diversifies, and the total number of possible cyclical connections multiply accordingly, species that were not initially present within the system may persist through stability afforded by cyclic relationships37,39.
Hierarchical stratification
A potential for hierarchical stratification within the radiolytic network is indicated by a histogram of compound connectivity plotted as a function of chemical species size (Fig. 3a). The connectivity distribution indicates that smaller-mass compounds are more frequently connected than larger compounds, and therefore that greater numbers of smaller-mass compounds should be produced and converted into other compounds at greater frequency within a radiolytic system than larger-mass compounds. However, these network description data lack estimates of relative product yield, or of the dominant synthetic pathways found in actual chemical systems where all of the reactions that make up the network are co-occurring with one another.
Indicators of hierarchical stratification in small molecule radiolytic chemical systems. (a) Connectivity degree per compound in the assembled radiolysis network, plotted versus chemical species mass. (b) Experimentally obtained radiolytic abundance (G values), plotted versus chemical species mass. Most plotted values are based on radiolysis of aqueous HCN/-CN. The plotted G value for hydrogen cyanide is based on irradiation of nitrogen and methane. See Supplementary Table S3 for all
A more insightful physical parameter known as radiolytic yield (G value, typically reported in units of number of molecules produced per 100 eV of input absorbed; see notes in Supplemental Table S3) takes into account the complex balance of all production and degradation reactions that are co-occurring for each compound in a radiolysis experiment. G values can vary depending on conditions such as pH, temperature, presence of radical scavengers, and concentrations of starting materials or intermediate substrates formed. A species with a high G value (for a given array of experimental parameters) will be produced more frequently and probably be found in greater abundance than a species with a low G value. In experimental systems where radiolytic consumption outstrips production, G values can be negative. G values can therefore indicate dominant radiolytic pathways of consumption and production that tend to occur in actual chemical systems, and can be used to distinguish more frequently-occurring species and pathways from all possible such species and pathways mapped within the chemical reaction network. Radiolytic yield values depicted in this way reflect dominant modes of energy flow in irradiated chemical systems, analogous to the way time-integrated abundances of species offer a representation of energy flow in complex ecosystems that correlate with biomass or trophic level.
The plots of network connectivity distribution versus mass (Fig. 3a) and the radiolytic G values versus mass (Fig. 3b) depict a bottom-heavy energy-size spectrum of products where the time-averaged numbers of smaller objects greatly exceed those of larger objects. Bottom-heavy energy-size spectra are interpreted as indicators of hierarchical tiering and systemic stability, and are pervasive across ecosystem ecology (with a few, well-understood exceptions)42. The link between lower mass species heterogeneity leading to higher mass species heterogeneity, and the trend of decreasing abundance with increasing mass, implicates a relationship in which fewer, more massive molecules emerge from reactions between many, less massive input molecules and initial radicals. This hierarchical relationship is corroborated by direct observations from radiolytic experiments for which detailed quantitation data are available2,48,54 and the atmospheric chemistry of numerous solar system bodies15.
Robustness is a feature of living systems that is difficult to quantify compared to enumerable traits like number of cycles or connectivity distribution. Vector representations of graphs and relational structures, such as chemical reaction networks, enable the application of numerical data analysis to these complex structures64. We employed the representation learning framework Node2vec65 to conduct a pairwise comparison of the topological similarity of all reactions to one another, to assess whether disparate chemical reactions within the network occur in similar contexts; the identification of multiple, dissimilar reactions occurring in similar contexts would provide a measure of robustness. Node2vec has been shown to effectively learn features from diverse graph structures by employing a random walk exploration algorithm, which reduces connectivity paths around a node to vector representations66,67,68. The vector representations between any two reactions may be numerically compared to one another using cosine similarity, defined to equal the cosine of the angle between them, yielding an absolute value between 0 and 1 (indicating extrema of low and high similarity, respectively). Any two reactions in the assembled radiolysis network with a high cosine similarity (defined for this analysis as exceeding a numerical cut-off of 0.85) would be considered ‘synonymous’ with one another, in the sense of occurring within similar chemical contexts. The analysis uncovered 16 synonymous pairings with a cosine similarity of embedded reactions that exceeded the numerical cut-off (Supplementary Table S4). Excluding three placeholder equations (i.e., equations connecting known inputs and outputs but unknown intermediate species) and one mirrored forward/reverse reaction pairing, seven of these pairs involve radical species, and ten pairs involve either nitriles or carboxylic acids. This analysis suggests that nitriles and carboxylic acids form robust radical-driven subnetworks that connect geochemical substrates to organic compound production.
Discussion
The topological attributes of this radiolytic network, i.e., (1) heterogeneous connectivity, (2) closed-loop cycling, and (3) hierarchy and a bottom-heavy energy-size spectrum, outline chemical conditions that have been sought for plausible prebiotic settings13. These attributes are intimately associated with generic capabilities of evolvability27 and self-organization29 that also underpin cell biology69,70 and ecosystem ecology. Two general observations are apparent based on the layout of the network. The first is that the subnetwork of radical interactions (Fig. 1a, shaded in red) dominates the core of the network and represents an efficient way to connect the production of different compounds to one another. The second is that water, beyond its long-recognized role as a solvent for prebiotic chemistry, can serve as an essentially inexhaustible source of powerful reagents that can drive reactions throughout the network. Water-derived radicals (namely, H/eaq– and OH) are the most highly connected species, and their reactions yield some organic compounds in greater abundance than others of comparable carbon number or molecular mass. A notable example is simple hydroxyaldehyde production from hydrogen cyanide via Kiliani-Fischer homologation49,50,71, from which the synthesis of 2-aminooxazole62 and 2-aminoimidazole72 by the reaction of glycolaldehyde with cyanamide is afforded48.
A fit of the degree distribution of the reaction network participants to a power law yields an exponent value close to − 1.5, a value which is correlated with self-organizational behaviours in the vicinity of a phase transition between stable and chaotic states for many different complex systems such as earthquakes, avalanches, biological interactomes, neural activity, solar flares and cosmological structures25,73,74,75,76,77,78,79. Claims of ubiquitous power law scaling in biological and other complex networks17,29,70 have been reconsidered in light of more rigorous statistical treatments80,81; however, knowledge of whether a distribution is heavy-tailed or not is more important than whether it specifically fits a power law82,83. There is no universal prescription for the generative mechanisms that underlie such heavy-tailed distributions, nor for the possible self-organizational behaviours they are capable of exhibiting82; object properties that give rise to such distributions must be experimentally enumerated for each unique system25.
Radiolysis is generally considered to be a degradative chemical process, so how can it lead to the production and net accumulation of successively larger molecules? The patterns of compound synthesis observed in radiolytic systems may derive, in part, from the chemical kinetics of radicals. Lower mass radicals can more easily attain higher average speeds (which are proportional to m‒0.5) in the process of thermalization. These species may therefore be more likely to escape back-reaction than larger radicals at a given temperature. Small radicals may also yield progressively larger molecules over time because mon- and di-atomic species such as H, N, O, OH, CN, etc. possess relatively few internal degrees of freedom with which to distribute the energy released during radical recombination. This contrasts with larger molecules, where excess recombination energy can be quickly redistributed across many vibrational and rotational degrees of freedom84. This difference in numbers of degrees of freedom increases the probability that a recombination reaction that terminates a radical chain may lead to larger molecule production. It is important to note, however, that in the gas phase, excited CO and the CN radical have been observed to be exceptionally stable and tend to dominate initially, at least in the context of spark-discharge and laser-induced plasmas generated from a range of gaseous mixtures all containing sources of C, H, O and N51; differences between gas-phase and aqueous-phase radical reaction dynamics will be carefully considered in future network descriptions. The kinetic attributes of radicals, combined with their electrochemical extrema and ready sourcing from geochemical substrates, indicate that compound- and reaction-level radical attributes can play critical roles in producing complex network-level properties within abiotic chemical systems15,85. More specifically, the underlying requirements that can generate scale-free networks (continuous addition of new nodes, and increased likelihood of reactivity of new nodes with existing, highly-linked nodes)28,86 are consistent with the chemical properties of radicals such as H, OH, CN and the solvated electron.
The emergence of many closed cycles from only a handful of initial substrates is one of the most significant general attributes of radiolytic systems that finds parallels in the trophic dynamics of ecosystem networks87. Orgel88 observed: “The demonstration of the existence of a complex, nonenzymatic metabolic cycle, such as the reverse citric acid, would be a major step in research on the origin of life, while demonstration of an evolving family of such cycles would transform the subject…The recognition of sequences of plausible reactions that could close a cycle is an essential first step toward the discovery of new cycles, but experimental proof that such cycles are stable against the challenge of side reactions is even more important”. Families of radical cycles are known to play key roles in the atmospheric composition of different planetary bodies89,90, offering real examples that closed loops of radical reactions can be stable against side reactions. It remains to be seen whether radiolytically driven cycles meet the formal definition of autocatalytic sets as outlined by previous workers91, but they do implicate a potential for self-organization and dynamic feedback through cyclic ensembles prior to the emergence of catalytic macromolecules.
Alternative interpretations of the network topology stem from sources of potential observational bias. In some cases, the experiments that form the basis for the reaction network were designed to search for targeted species, or conducted with reference to specific type localities or prebiotic scenarios. In others, the analytical methods employed were limited to the technology available at the time, which improved in the years that followed. These potential sources of bias, though real, are nevertheless likely to exert a minimal impact on the observed network topology. The underlying experiments cover a wide range of parameters (i.e., total dose, dose rate, initial reactant selections and concentrations, energy sources) that span what would most likely be found in a natural system. Additionally, the chemical reactions inferred to be occurring in the experimental systems are evaluated within a context of accounting for the relative abundances of multiple products. By employing conservative criteria for network inclusion (Supplementary Information), further experimentation is likely to add to the existing network reactions, rather than alter or eliminate them in any statistically significant manner. Conservative criteria reduce the possibility that the inferred topological attributes described in this manuscript (connectivity distribution, hierarchical arrangement, etc.) will change as a result of greater experimental scrutiny. Finally, although this effort focused on the role that radicals play in shaping the network topology, future iterations should also evaluate the effects that ions may exert on expected network topology and attributes.
It remains an open question as to whether living systems can only emerge from non-living settings that already possess the network organizational features observed in current biology prior to the appearance of the first cell, or whether these attributes more likely arise as features at intermediate or even terminal points throughout a prebiotic chronology. The radiolytic network described here indicates that the network attributes that characterize forms of life on Earth and its ecologies may be tractably derived from abiotic network parallels found in relatively common atmospheric and geochemical conditions. Whether or not radiolysis was directly implicated in the historical origins of life on our planet requires more detailed study. From a broader perspective, though, the methods employed here to describe each of these reaction network attributes as parallels for higher-level ecosystem organization can be used to test diverse hypotheses about prebiotic chronologies such as the RNA World92, for evaluating the relative systemic complexities of terrestrial surface93 or hydrothermal94 chemosynthetic paradigms, or for testing theories about the likelihood of de novo emergence of pre-metabolic, autocatalytic cycles88. Considered separately from the question of life’s origins, this network may also serve as a basis for investigating whether radiolysis underpins a universal potential for organosynthetic self-organization or natural computation carried out within discrete chemical reaction networks95.
Methods
Chemical reaction network assembly
Chemical reaction network data are chemical reactions from radiolytic and polar chemical experiments that include key atomic species (CHONPS) of living systems. All reactions were assigned to chemical reaction categories (i.e., free radical reactions, physicochemical reactions, nitrile radical reactions, chloride reactions, etc.) based on their reactive substrates. Reactions that include solid, liquid and gaseous phases are combined into a single network, as it is assumed that (1) all phases and species within the ‘enclosed box’ of reactions can occur alongside one another; and (2) all reactants are within diffusive range of one another. These assumptions are consistent with plausible natural conditions that exist near a planetary surface:atmosphere interface, and with experimental radiolysis setups (many of which incorporate mixed phases) that form the basis of the compiled network.
A complete list of network reactions is provided as Supplementary Datafile S1 (Excel spreadsheet), and reaction criteria and source materials are described in the attached Supplementary Information file. Atoms are initially present in the system as their most geochemically abundant molecular species (H2O, CO2, N2, NaCl, a proxy for the apatite mineral group Ca10Cl2(PO4)6, and pyrite FeS2). The electromagnetic spectrum was binned into gamma, X-ray, UV, visible and infrared photons to account for reactions across key atomic and molecular energetic thresholds (i.e., strong nuclear binding force, inner valence electrons, outer valence electrons and ambient system temperature). Hydrated species complexed with water and reactions contingent upon an aqueous medium include water as a bystander species. Amino acid source polymers are chemically uncharacterised and are binned into polymer placeholder species that can be thermally hydrolysed to yield amino acids. Reaction rates and kinetic coefficients were omitted from this study.
The energy sources that could drive network structure and chemosynthesis were forms of radiation sourced from radiogenic uranium oxide. Uranium is a geochemical proxy for naturally-occurring energy sources such as galactic cosmic radiation58, solar flares59 and planetary radiation belts60. The assembled database includes 782 reactions and 386 distinct atomic, molecular, and photonic species.
Chemical reaction network visualisation
The acquired reaction equations were used to construct a reaction network where nodes are divided into two sets, i.e. partitions, and connections are allowed only between nodes in different partitions. Networks of this type are called bi-partite networks. The first partition in the constructed network includes reaction participants, such as atomic, molecular, and photonic species; the second partition includes reactions themselves. The network is constructed as a directed network where directionality reflects the form of the stoichiometric equation: the entities on the left-hand side of the equation are connected to the node representing the reaction, and the reaction node is connected to the entities on the right-hand side of the equation. This definition of directionality simplifies network analytics, such as cycle search. It is related to, but not definitive of, reversibility of the chemical reactions. The resulting network was graphed and analysed using Gephi 0.9.296. The network file is included as Datafile S2 (.graphml format).
Statistical analyses
Analysis of the degree distribution was performed following the methodology introduced and extensively discussed in Abbot et al. using a python library powerlaw97. A log-likelihood test is employed to evaluate pairs of alternative distributions as possible sources of the observed data. For example, the test elucidates whether the dataset exhibits a heavy-tailed distribution by comparing a power law fit (the simplest approximation of a heavy-tailed distribution on a finite interval) to one with an exponential fall-off. The test also helps to check if alternative distributions, such as a lognormal distribution, exponentially truncated power law, etc. offer better fits than a power law. Per convention used in powerlaw library implementation, the computed loglikelihood ratio R for the pair of distributions (d1, d2) is positive if data are more likely to fit the first distribution d1, and negative if the data are more likely to fit the second distribution d2. Loglikelihood ratio is accompanied by the significance value p; following Clauset et al., we consider p > 0.05 as an indicator that neither distribution is a stronger fit63. Detailed discussion of these tools is beyond the scope of our contribution; for more details, we refer the inquisitive reader to the original papers describing these tools and implementation63,97.
Closed loop cycle enumeration
Closed loops of chemical species in the directed bi-partite network were identified through exhaustive enumeration between pairs of nodes by finding the shortest paths from the first node to the second and all the shortest paths from the second node to the first. Pair-wise combinations of the forward/backward paths would constitute distinguishable cycles. Enumerated cycles were expressed as sub-networks formed by these forward–backward paths; each sub-network was labelled with an ordered sequence of the indices of the included nodes to identify unique subnetworks. Water, as a frequent bystander molecule, was omitted as a possible link to prevent overcounting of trivial closed cycles. All cycles were assigned a numerical identifier according to the order of discovery within the network, with a prefix IDX. The order of discovery within the network, and thus the enumeration of the cycles, carries no particular physical or chemical significance. Each IDX cycle was saved as an individual Graphml network file, which are available upon request. Summary information about all of the IDX cycles (species composition of the cycles and cycle size) are included as Supplementary Datafile S3 (Excel spreadsheet).
Assessing systemic robustness with a machine-learning algorithm
The framework for representation learning on graphs Node2vec65 was used to learn continuous features of molecules and reactions, and assess whether disparate chemical reactions within the network occur within similar contexts regardless of their reaction category assignments. Network-based chemical semantic similarity was evaluated by carrying out random walks within the network, embedding the nodes visited by each walk into a vector space, and evaluating cosine similarity of the embedded reactions with a cut-off for similar reactions set at 0.85.
Data availability
Data files generated and analysed during the current study, but not included as supplementary information files, are available from the corresponding author on request.
References
Garrison, W. M., Morrison, D. C., Hamilton, J. G., Benson, A. A. & Calvin, M. Reduction of carbon dioxide in aqueous solutions by ionizing radiation. Science 114, 416–418 (1951).
Draganić, Z. D., Draganić, I. G. & Borovičanin, M. The radiation chemistry of aqueous solutions of hydrogen cyanide in the megarad dose range. Radiat. Res. 66, 42–53 (1976).
Bar-Nun, A. & Hartman, H. Synthesis of organic compounds from carbon monoxide and water by UV photolysis. Origins Life 9, 93–101 (1978).
Miller, S. L. & Urey, H. C. Organic compound synthesis on the primitive earth. Science 130, 245–251 (1959).
Pasek, M. A., Dworkin, J. P. & Lauretta, D. S. A radical pathway for organic phosphorylation during schreibersite corrosion with implications for the origin of life. Geochim. Cosmochim. Acta 71, 1721–1736 (2007).
Lim, R. W. J. & Fahrenbach, A. C. Radicals in prebiotic chemistry. Pure Appl. Chem. 92, 1971–1986 (2020).
Studer, A. & Curran, D. P. Catalysis of radical reactions: A radical chemistry perspective. Angew. Chem. Int. Ed. 55, 58–102 (2016).
Shock, E. L. et al. Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, USA. Geochim. Cosmochim. Acta 74, 4005–4043 (2010).
Bím, D., Maldonado-Domínguez, M., Rulíšek, L. & Srnec, M. Beyond the classical thermodynamic contributions to hydrogen atom abstraction reactivity. Proc. Natl. Acad. Sci. USA 115, E10287–E10294 (2018).
Mayer, J. M. Hydrogen atom abstraction by metal–oxo complexes: Understanding the analogy with organic radical reactions. Acc. Chem. Res. 31, 441–450 (1998).
Gutowski, M. & Kowalczyk, S. A study of free radical chemistry: Their role and pathophysiological significance. Acta Biochim. Pol. 60, 1–16 (2013).
Moran, J. & Rauscher, S. Energy and self-organization at the origin of metabolism. Commun. Chem. (in rev.).
Nghe, P. et al. Prebiotic network evolution: Six key parameters. Mol. BioSyst. 11, 3206–3217 (2015).
Jolley, C. & Douglas, T. Topological biosignatures: Large-scale structure of chemical networks from biology and astrochemistry. Astrobiology 12, 29–39 (2012).
Solé, R. V. & Munteanu, A. The large-scale organization of chemical reaction networks in astrophysics. Europhys. Lett. 68, 170–176 (2004).
Shenhav, B., Solomon, A., Lancet, D. & Kafri, R. in Transactions on Computational Systems Biology I (ed. Priami, C.) 14–27 (Springer, Berlin, 2005).
Brown, J. H. et al. The fractal nature of nature: Power laws, ecological complexity and biodiversity. Philos. Trans. R. Soc. Lond. B 357, 619–626 (2002).
Walker, S. I. & Mathis, C. in Prebiotic Chemistry and Chemical Evolution of Nucleic Acids (ed. Menor-Salvár, C.) 263–291 (Springer, Berlin, 2018).
Hordijk, W., Hein, J. & Steel, M. Autocatalytic sets and the origin of life. Entropy 12, 1733–1742 (2010).
Albert, R. Scale-free networks in cell biology. J. Cell. Sci. 118, 4947–4957 (2005).
Liu, R., Mao, G. & Zhang, N. Research of chemical elements and chemical bonds from the view of complex network. Found. Chem. 21, 193–206 (2019).
Estrada, E. The complex networks of earth minerals and chemical elements. MATCH Commun. Math. Comput. Chem. 59, 605–624 (2008).
Fricker, M. D., Boddy, L., Nakagaki, T. & Bebber, D. P. In Adaptive Biological Networks (eds. Gross, T. & Sayama, H.) 51–70 (Springer, Berlin, 2009).
Nicolis, G. Chemical chaos and self-organization. J. Phys. Condens. Matter 2, SA47–SA62 (1990).
Pérez-Mercader, J. In Astrobiology (eds. Horneck, G. & Baumstark-Khan, C.) 337–360 (Springer, Berlin, 2002).
Li, W. Expansion-modification systems: A model for spatial 1/f spectra. Phys. Rev. A 43, 5240–5260 (1991).
Albert, R. & Barabási, A.-L. Topology of evolving networks: Local events and universality. Phys. Rev. Lett. 85, 5234–5237 (2000).
Barabási, A.-L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Marković, D. & Gros, C. Power laws and self-organized criticality in theory and nature. Phys. Rep. 536, 41–74 (2014).
Adler, R., Feldman, R. & Taqqu, M. (eds.) A Practical Guide to Heavy Tails: Statistical Techniques and Applications (Springer, Berlin, 1998).
Patten, B. C. & Higashi, M. Modified cycling index for ecological applications. Ecol. Modell. 25, 69–83 (1984).
Essington, T. E. & Carpenter, S. R. Nutrient cycling in lakes and streams: Insights from a comparative analysis. Ecosystems 3, 131–143 (2000).
Christian, R. R. & Thomas, C. R. Network analysis of nitrogen inputs and cycling in the Neuse River estuary, North Carolina, USA. Estuaries 26, 815–828 (2003).
Allesina, S. & Ulanowicz, R. E. Cycling in ecological networks: Finn’s index revisited. Comput. Biol. Chem. 28, 227–233 (2004).
Loreau, M. Material cycling and the stability of ecosystems. Am. Nat. 143, 508–513 (1994).
DeAngelis, D. L. et al. Nutrient dynamics and food-web stability. Annu. Rev. Ecol. Syst. 20, 71–95 (1989).
Artzy-Randrup, Y. & Stone, L. Connectivity, cycles, and persistence thresholds in metapopulation networks. PLoS Comput. Biol. 6, e1000876 (2010).
Newsholme, E. A. & Crabtree, B. Substrate cycles in metabolic regulation and in heat generation. Biochem. Soc. Symp. 41, 61–109 (1976).
Kritz, M. V., dos Santos, M. T., Urrutia, S. & Schwartz, J.-M. Organising metabolic networks: Cycles in flux distributions. J. Theor. Biol. 265, 250–260 (2010).
Valentine, J. W. & May, C. L. Hierarchies in biology and paleontology. Paleobiology 22, 23–33 (1996).
McShea, D. W. The hierarchical structure of organisms: A scale and documentation of a trend in the maximum. Paleobiology 27, 405–423 (2001).
Trebilco, R., Baum, J. K., Salomon, A. K. & Dulvy, N. K. Ecosystem ecology: Size-based constraints on the pyramids of life. Trends Ecol. Evol. 28, 423–431 (2013).
Lindeman, R. L. The trophic-dynamic aspect of ecology. Bull. Math. Biol. 53, 167–191 (1991).
Kleidon, A. & Lorenz, R. D. (eds.) Non-equilibrium Thermodynamics and the Production of Entropy: Life, Earth, and Beyond (Springer, Berlin, 2005).
Goldenfeld, N. & Woese, C. Life is physics: Evolution as a collective phenomenon far from equilibrium. Annu. Rev. Condens. Matter Phys. 2, 375–399 (2011).
Braakman, R. & Smith, E. The compositional and evolutionary logic of metabolism. Phys. Biol. 10, 011001 (2013).
Ji, S. Molecular Theory of the Living Cell: Concepts, Molecular Mechanisms, and Biomedical Applications (Springer, Berlin, 2012).
Yi, R. et al. A continuous reaction network that produces RNA precursors. Proc. Natl. Acad. Sci. USA 117, 13267–13274 (2020).
Yi, R., Hongo, Y., Yoda, I., Adam, Z. R. & Fahrenbach, A. C. Radiolytic synthesis of cyanogen chloride, cyanamide and simple sugar precursors. ChemistrySelect 3, 10169–10174 (2018).
Ritson, D. & Sutherland, J. D. Prebiotic synthesis of simple sugars by photoredox systems chemistry. Nat. Chem. 4, 895–899 (2012).
Ferus, M. et al. High energy radical chemistry formation of HCN-rich atmospheres on early Earth. Sci. Rep. 7, 6275 (2017).
Getoff, N. Significance of solvated electrons (eaq−) as promoters of life on Earth. In Vivo 28, 61–66 (2014).
Negrón-Mendoza, A., Draganić, Z. D., Navarro-González, R. & Draganić, I. G. Aldehydes, ketones, and carboxylic acids formed radiolytically in aqueous solutions of cyanides and simple nitriles. Radiat. Res. 95, 248–261 (1983).
Adam, Z. R. et al. Estimating the capacity for production of formamide by radioactive minerals on the prebiotic Earth. Sci. Rep. 8, 265 (2018).
Bedau, M. A. et al. Open problems in artificial life. Artif. Life 6, 363–376 (2000).
Grassberger, P. in Information Dynamics NATO ASI Series (Series B: Physics) (eds. Atmanspacher, H. & Scheingraber, H.) 15–33 (Springer, Berlin, 1991).
Kaneko, K. Chaos as a source of complexity and diversity in evolution. Artif. Life 1, 163–177 (1993).
Buhl, D. & Ponnamperuma, C. Interstellar molecules and the origin of life. Sp. Life Sci. 3, 157–164 (1971).
Airapetian, V. S., Glocer, A., Gronoff, G., Hébrard, E. & Danchi, W. Prebiotic chemistry and atmospheric warming of early Earth by an active young Sun. Nat. Geosci. 9, 452–455 (2016).
Paranicas, C., Cooper, J. F., Garrett, H. B., Johnson, R. E. & Sturner, S. J. in Europa (eds. Pappalardo, R. T. et al.) 529–544 (University of Arizona Press, Tucson, 2009).
Takano, Y., Masuda, H., Kaneko, T. & Kobayashi, K. Formation of amino acids from possible interstellar media by γ-rays and UV irradiation. Chem. Lett. 31, 986–987 (2002).
Powner, M. W., Gerland, B. & Sutherland, J. D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459, 239–242 (2009).
Clauset, A., Shalizi, C. R. & Newman, M. E. J. Power-law distributions in empirical data. SIAM Rev. 51, 661–703 (2009).
Grohe, M. in Proceedings of the 39th ACM SIGMOD-SIGACT-SIGAI Symposium on Principles of Database Systems 1–16 (Portland, OR, USA, 2020).
Grover, A. & Leskovec, J. in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 855–864 (San Francisco, CA, USA, 2016).
Palumbo, E. et al. in The Semantic Web: European Semantic Web Conference Vol. 11155, 117–120 (Springer, Crete, Greece, 2018).
Kim, M., Baek, S. H. & Song, M. Relation extraction for biological pathway construction using node2vec. BMC Bioinform. 19, 206 (2018).
Shen, Z., Chen, F., Yang, L. & Wu, J. Node2vec representation for clustering journals and as a possible measure of diversity. J. Data Inf. Sci. 4, 79–92 (2019).
Barabási, A.-L. & Oltvai, Z. N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 (2004).
Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabási, A.-L. The large-scale organization of metabolic networks. Nature 407, 651–654 (2000).
Ritson, D. J. & Sutherland, J. D. Synthesis of aldehydic ribonucleotide and amino acid precursors by photoredox chemistry. Angew. Chem. Int. Ed. 52, 5845–5847 (2013).
Fahrenbach, A. C. et al. Common and potentially prebiotic origin for precursors of nucleotide synthesis and activation. J. Am. Chem. Soc. 139, 8780–8783 (2017).
Muñoz, M. A. Colloquium: Criticality and dynamical scaling in living systems. Rev. Mod. Phys. 90, 031001 (2018).
Langton, C. G. Computation at the edge of chaos: Phase transitions and emergent computation. Phys. D Nonlinear Phenom. 42, 12–37 (1990).
Bak, P., Tang, C. & Wiesenfeld, K. Self-organized criticality: An explanation of the 1/f noise. Phys. Rev. Lett. 59, 381–384 (1987).
Gaveau, B., Moreau, M. & Toth, J. Scenarios for self-organized criticality in dynamical systems. Open Syst. Inf. Dyn. 7, 297–308 (2000).
Bak, P. & Paczuski, M. Complexity, contingency, and criticality. Proc. Natl. Acad. Sci. USA 92, 6689–6696 (1995).
Hoffmann, H. & Payton, D. W. Optimization by self-organized criticality. Sci. Rep. 8, 2358 (2018).
Lovecchio, E., Allegrini, P., Geneston, E., West, B. J. & Grigolini, P. From self-organized to extended criticality. Front. Physiol. 3, 98 (2012).
Lima-Mendez, G. & van Helden, J. The powerful law of the power law and other myths in network biology. Mol. BioSyst. 5, 1482–1493 (2009).
Broido, A. D. & Clauset, A. Scale-free networks are rare. Nat. Commun. 10, 1017 (2019).
Stumpf, M. P. H. & Porter, M. A. Critical truths about power laws. Science 335, 665–666 (2012).
Mitzenmacher, M. A brief history of generative models for power law and lognormal distributions. Internet Math. 1, 226–251 (2003).
Glassman, I., Yetter, R. A. & Glumac, N. G. Combustion 41–69 (Elsevier, New York, 2015).
Gleiss, P. M., Stadler, P. F., Wagner, A. & Fell, D. A. Relevant cycles in chemical reaction networks. Adv. Complex Syst. 4, 207–226 (2001).
Dančík, V., Basu, A. & Clemons, P. in Systems Biology (eds. Prokop, A. & Csukas, B.) 129–178 (Springer, Berlin, 2013).
Patten, B. C., Higashi, M. & Burns, T. P. Trophic dynamics in ecosystem networks: Significance of cycles and storage. Ecol. Modell. 51, 1–28 (1990).
Orgel, L. E. The implausibility of metabolic cycles on the prebiotic Earth. PLoS Biol. 6, e18 (2008).
Monks, P. S. Gas-phase radical chemistry in the troposphere. Chem. Soc. Rev. 34, 376–395 (2005).
Platt, U. et al. in Tropospheric Chemistry: Results of the German Tropospheric Chemistry Programme (eds. Seiler, W. et al.) 359–394 (Springer, Berlin, 2002).
Vasas, V., Fernando, C., Santos, M., Kauffman, S. & Szathmáry, E. Evolution before genes. Biol. Direct 7, 1 (2012).
Robertson, M. P. & Joyce, G. F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 4, a003608 (2012).
Damer, B. & Deamer, D. The hot spring hypothesis for an origin of life. Astrobiology 20, 429–452 (2020).
Martin, W., Baross, J., Kelley, D. & Russell, M. J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 6, 805–814 (2008).
Soloveichik, D., Cook, M., Winfree, E. & Bruck, J. Computation with finite stochastic chemical reaction networks. Nat. Comput. 7, 615–633 (2008).
Bastian, M., Heymann, S. & Jacomy, M. in Proceedings of the Third International AAAI Conference on Weblogs and Social Media 361–362 (2009).
Alstott, J., Bullmore, E. & Plenz, D. powerlaw: A Python package for analysis of heavy-tailed distributions. PLoS One 9, e85777 (2014).
Acknowledgements
This work was supported by a Grant from the Simons Foundation’s Simons Collaboration on the Origins of Life (494291, Z.R.A.). B.K. acknowledges support from the National Science Foundation (#1724090) and the John Templeton Foundation (#61926). A.C.F acknowledges support from the University of New South Wales Strategic Hires and Retention Pathways (SHARP). We wish to thank H.J. Cleaves and A.H. Knoll for helpful comments on an earlier drafts of this manuscript.
Author information
Authors and Affiliations
Contributions
Z.R.A., B.K., A.C.F. and D.Z. designed the study; Z.R.A., S.M.J. and D.Z. collected data; Z.R.A., B.K., A.C.F. and D.Z. analysed and interpreted data; Z.R.A., B.K., A.C.F. and D.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adam, Z.R., Fahrenbach, A.C., Jacobson, S.M. et al. Radiolysis generates a complex organosynthetic chemical network. Sci Rep 11, 1743 (2021). https://doi.org/10.1038/s41598-021-81293-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-81293-6
This article is cited by
-
Physical non-equilibria for prebiotic nucleic acid chemistry
Nature Reviews Physics (2023)
-
The curious consistency of carbon biosignatures over billions of years of Earth-life coevolution
The ISME Journal (2021)
-
Nanoscale neural network using non-linear spin-wave interference
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.