Abstract
The native marmoset of the Southeastern Atlantic Forest in Brazil is among the 25 most endangered primates of the world. Hybridization with alien species is one of its main threats registered since the early 2000s based on phenotype, so far, without genetic confirmation. Using uniparental molecular markers, we analyzed 18 putative hybrids, captured from 2004 to 2013 in different localities of the Atlantic Forest. A nine base pair deletion in the SRY gene of C. aurita was used to investigate paternal ancestry. Maternal ancestry was assessed by DNA sequencing of ca. 455 bp from the COX2 gene. Hybridization was confirmed for 16 out of the 18 marmosets since they inherited COX2 haplotypes of the alien C. penicillata or C. jacchus and the SRY deletion specific to C. aurita. Two individuals inherited both parental lineages of C. aurita, which is probably related to backcrossing or hybrid interbreeding. The direction of hybridization of females with the matrilineal lineage of invasive species with males descending from the native lineage was predominant in our sampling. This is the first time that hybridization between C. aurita and invasive species has been confirmed through genetic analysis.
Similar content being viewed by others
Introduction
The buffy-tufted-ear marmoset, Callithrix aurita is endemic to the Brazilian Atlantic Forest, occurring in the states of Rio de Janeiro, eastern and northeastern São Paulo and adjacent parts of Minas Gerais. These areas, together with the states of Espírito Santo and southern Bahia, have long been recognized as a ‘core area’ for primates1, since they host 19 of the 22 species found in the Atlantic rainforest biome, being the number of species updated2.
The small distribution range, habitat destruction and in-situ hybridization with invasive species are the major threats to the conservation of C. aurita, however, the extent of the latter remains obscure3,4. The species once considered extinct in Rio de Janeiro state during the 1980s1,5,6 is currently among the 25 most endangered primates of the world7. It is also configured in the national and international lists of threatened species4,8,9 as endangered.
The Callithrix genus comprises six taxa, originally parapatric10. Among them, two species with the most extensive distribution, the common marmoset C. jacchus and the black-tufted-ear marmoset C. penicillata, emerged as invaders to the southeastern and southern regions of Brazil1,11,12,13,14. Callithrix jacchus is native to Bahia, Tocantins, Maranhão, Piauí, Ceará and Rio Grande do Norte, while C. penicillata is original to Central Brazil in the states of Mato Grosso do Sul, Goiás, Tocantins, Bahia, part of São Paulo and part of Minas Gerais10. In contrast to C. aurita, both species are generalists and inhabitants of secondary forests, which putatively enable them to have a greater degree of environmental plasticity to adapt to degraded areas and expand their territories15,16. These two species hybridize naturally in a contact zone of their original range in the northeast and by human-mediated introduction in Rio de Janeiro state13. There are no natural hybrid zones registered between C. aurita and the invasive species5.
Interspecific hybridization occurs when parental individuals are from genetically distinct populations, subspecies or species17. At the species taxonomic level, it results in progeny that carry a mixture of previously isolated gene pools. It occurs naturally at distribution boundaries between closely related species in hybrid zones that act as a "filter testing new genotypic combinations through natural selection”18. Thus, gene introgression may or may not occur and is considered a dynamic evolutionary mechanism capable of contributing to the genetic enrichment of species while preserving distinct biological entities19. New genotypic combinations can also allow for the exploration of new niches resulting in speciation. It is estimated to occur naturally in more than 10% of primate species18.
However, human accidentally induced hybridization has been a concern in conservation. The direct or indirect introduction of nonnative taxa by alteration of the habitat allows the expansion of territories of allopatric or parapatric species, and sometimes the suppression of reproductive isolation between sympatric species facilitates interbreeding20. Hybridization can be especially problematic for rare species contacting more abundant species17. Among the most relevant outcomes of hybridization is genetic introgression, i.e. the movement of a gene from one species to the genetic pool of another through repeated backcrosses between a hybrid and its original parent generation17. If hybridization events are rare, introgression may not occur since F1 individuals are subject to the same selective forces as others or even when common, because the offspring are sterile or less adapted21. The consequence may also be less severe for the rare species if there are pure stocks in isolated populations not affected by hybridization. If introgression is widespread over the entire distribution of the species, the outcome in general is the disintegration of the genome of the rare species in hybrid swarms and its assimilation by the most abundant species21.
According to distribution modeling, C. penicillata is the species with the highest potential of impact over C. aurita due to similarities in the pattern of environmental fitness between both species. Areas with more pronounced seasonality, higher altitudes and mild temperatures that adjust to C. penicillata are less suitable for the dispersion of C. jacchus22. However, this scenario can be changed because of global warming, where an expansion of the distribution of C. jacchus is predicted and the opposite for C. penicillata and C. aurita23.
Illegal pet trafficking is considered the main factor in the introduction of exotic Callithrix species in the southeastern region12,24, with habitat invasion probably favored by long-term Atlantic Forest disturbance25. The loss of biodiversity, both by deforestation and by climate change, reduces habitat heterogeneity, relaxes divergent selection, and promotes interspecific hybridization26.
Hybridization has been considered a threat to C. aurita since 20083 when the first putative hybrids were described in the Serra dos Órgãos National Park (Parque Nacional da Serra dos Órgãos-PARNASO) municipality of Teresópolis in Rio de Janeiro state27, based on morphological features, followed by other publications based on the same grounds27,28,29. The appearance of the ear tufts, the pattern of the face and the coat coloration of the back are the main phenotypic features considered as a diagnosis for the three species in question, and thus of hybridization among them. Accordingly, with some authors, observed hybrids presented an intermediate phenotype to that of the parental species28,29,30,31,32. Vocalization is also intermediate between the pattern of the parental species33.
However, the detection of hybrids only assuming that they will be phenotypically intermediate to parental species can represent an inaccuracy, considering that there is greater morphological variation within and among populations than is recognized and, in general, the hybrids can express a mosaic of parental phenotypes34. For example, an unexpected finding was registered in four putative male hybrids with an intermediate phenotype between C. jacchus and C. penicillata, since they carried an acrocentric Y chromosome described only for C. aurita35. Once F1 has been generated and if the hybrids are viable and fertile, the backcross with one of the parental species is facilitated, making it impossible to distinguish them phenotypically from the parental species36. The outcome is that the frequency of backcrosses in nature is probably underestimated.
A factor that can contribute for the variety of phenotypes in marmoset hybrids is the successful interbreeding, probably due to the same diploid number, 2n = 46, and the morphological similarity among the autosomes of five Callithrix species (C. flaviceps was not studied). The five studied species differ only in terms of a small polymorphic uni- or biarmed Y chromosome and its Ag-NOR banding pattern35,37,38. Ex situ hybridization experiments during the 1970s and the beginning of the 1980s proved a degree of fertility of the hybrids among the six species, but with apparent reduced fertility in hybrids between C. aurita and C. jacchus5. Once fertile, backcrossing to one or both parental taxa, as well as mating among the hybrids, can generate hybrid swarms21.
Interspecific breeding has been suggested by cytogenetic analyses based on the finding of a small acrocentric Y chromosome exclusive to C. aurita in three male marmosets with a mixed phenotype31,39. Additionally, by DNA sequencing of the cytochrome c oxidase subunit II gene (COX2), the matrilineal lineage of C. aurita was identified in hybrids in whose group the dominant male was C. aurita with a female descendant of C. jacchus30. Nevertheless, neither record genetically confirmed hybridization among the native species and the invasive species.
A genetic polymorphism that can distinguish a male of C. aurita from the other marmosets is a deletion of nine base pairs (bp) in the SRY (sex-determining region of the Y chromosome) gene sequence, which is found exclusively in this species40. This polymorphism can be useful to determine whether the patrilineal lineage of the free-living putative hybrids comes from native or invasive species. In turn, specific haplotypes of COX2 can represent a molecular marker30 to identify the matrilineal lineage.
Our main goal was to investigate the occurrence of in situ hybridization using uniparental markers amongst a sample of marmosets with a mixed phenotype, captured over nine years from different places of the Atlantic Forest and to assess whether there is a pattern in mating.
Results
Sample data
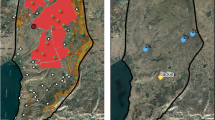
Eighteen marmosets considered putative C. aurita hybrids (Csp) were captured in three different localities in the state of Rio de Janeiro and one place in the state of São Paulo, Brazil, from 2004 to 2013 (Fig. 1, Table 1).
Place of origin and sampling of the 18 individuals of Callithrix sp. studied. (A). Map of South America; (B). Southeastern Brazil; (C). Sampling locations at Rio de Janeiro state: Guapimirim, Teresópolis and Petrópolis. In São Paulo state, only Biritiba Mirim. The size of the circles represents the sample number. Half of the circle in black represents the SRY haplotype with the deletion of nine base pairs exclusive to C. aurita (SRYaur), identified in all hybrids. In the other half, the COX2 haplotypes identified among the samples are represented. Two individuals from Guapimirim carried the haplotype of C. aurita (HCa). In all the others, the haplotypes of invasive C. jacchus (HCj) and C. penicillata (HCp and HCp2) were identified. (Map created by R. S. Carvalho using QGIS 3.16. QGIS Geographic Information System. QGIS Association. http://www.qgis.org).
Among the putative hybrids, there were 15 males and three females. Fifteen wild-born individuals of Callithrix aurita (six), C. jacchus (five) and C. penicillata (four) were sampled as positive controls (Table 1).
In general, the putative hybrids exhibited short ear tufts, in some cases fan-shaped with darker color. Commonly, the loss of the yellowish supra-cranial line peculiar to C. aurita was observed. The color of the body was lighter with eventual grooves (Fig. 2). These features confer to the individuals sampled a mixed pelage pattern of the tufts and the back, suggesting that they are hybrids between C. aurita and C. penicillata or C. jacchus29,32,41 (since both alien species are observed at the sampling locations).
Callithrix aurita (above) and an example of hybrid phenotypes (below) of the sampled individuals of Callithrix sp.: (a, d) loss of the yellowish supracranial line peculiar of C. aurita; (b, e) short ear tufts, in some cases fan-shaped with darker color; c, f) lighter color of the body with eventual grooves; (g–i) C. jacchus phenotype; (j–l) C. penicillata phenotype (Photographs by R. S. Carvalho).
SRY analysis
In our preliminary analysis to test the efficacy of the SRY primer set developed in this research to ascertain the patrilineal lineage of the putative hybrids, we sequenced the PCR products of one pure individual of each C. aurita (Ca), C. jacchus (Cj) and C. penicillata (Cp) used as a positive control and five putative hybrids of Callithrix sp. (Csp3, Csp4, Csp5, Csp6 and Csp7). We obtained the SRY nine base pair deletion 117_125delTAAGTATCG exclusive to C. aurita in the Ca positive control and in the five Csp samples tested. At the same position is the insertion in the Cj and Cp positive controls, which presented identical DNA sequences (Fig. 3).
Alignment sequence of 210 base pairs (bp) of the SRY (sex-determining region Y) gene in five Callithrix sp. males (Csp3, Csp4, Csp5, Csp6 and Csp7) and one male of each C. penicillata (Cp), C. jacchus (Cj) and C. aurita (Ca). The dashes indicate the nine base pair deletion 117_125delTAAGTATCG observed in C. aurita and in the five Callithrix sp.
Afterward, using the same primer set labelled with 6-FAM fluorescent dye, an amplicon of the SRY of 196 bp was observed for all six Ca positive controls and 18 putative hybrids samples attesting to the patrilineal lineage of the native threatened species. For all nine positive controls of Cj and Cp an electropherogram peak of 205 bp was generated (Table 1 provides an example of the electropherogram for each subset of obtained peaks. The image of the 33 peaks can be found in Supplementary Fig. S1 and S2 online).
The amplification of the SRY fragment in the eight female samples among the positive controls (Ca2, Cj3, Cj4, Cp1 and Cp3) and the putative hybrids (Csp1, Csp11, and Csp18) was possible due to the occurrence of placental anastomosis which allows hematopoietic chimerism resulting from the high incidence of twin gestation in callitrichids42.
COX2 analysis
PCRs with the COX2 primer set amplified a single product of the expected size of ca. 445 bp for all the Callithrix positive controls and for the 18 Csp samples analyzed. Through the alignment of DNA sequences of the positive controls and GenBank® sequences of each species, one COX2 haplotype was found for Callithrix jacchus and C. aurita, identified as HCj and HCa, respectively (Fig. 4). Among the 18 sequences obtained from the putative hybrids we found five individuals with HCj (Csp1, Csp2, Csp9, Csp10 and Csp11) and two with HCa (Csp6 and Csp8). Regarding C. penicillata, three haplotypes were detected (Figs. 4 and 5). One of them corresponds to the GenBank® sequence Cpe1_AY118194.1, which clustered with the C. geoffroyi Cge_AY 118,192.1 haplotype and did not cluster with any of the Callithrix sp. neither with our positive control samples. The second haplotype identified as HCp was shared by four of our positive controls, Cpe3_AY118196.1 from GenBank® and seven Callithrix sp. (Fig. 5). The third haplotype, HCp2, was shared by Cpe2_AY118195.1, and four of our Callithrix sp. samples (Fig. 5). For C. kuhli, one COX2 haplotype, AY118193.1, represented a specific branch (Fig. 5).
Alignment of the DNA sequencing of 445 base pairs of the cytochrome c oxidase subunit II (COX2) gene. One haplotype was detected for Callithrix aurita (Ca), one for C. jacchus (Cj), one for C. geoffroyi, one for C. kuhlii and three for C. penicillata (Cp). The 18 sequences from the Callitrix sp. (Csp) free-living individuals aligned with Ca, Cj, and two Cp haplotypes. The gray color represents similarities among the nucleotide sequences of all individuals. The blue colored bar represents cytosine in that position, yellow for guanine, green for thymine and red for adenine. The sequences identified by AY were obtained from GenBank®.
Phylogenetic Bayesian tree was built with MRBAYES 3.2.1 based on alignment of 445 base pairs of the cytochrome c oxidase subunit II gene alignment from Callithrix aurita (Ca, orange), C. jacchus (Cj, blue), and C. penicillata (Cp 1–3, brown, green and black). Sequences of C. kuhli (Cku) and C. geoffroyi (Cge) were also included. The 18 Callithrix sp. (Csp) clustered with Ca, Cj, Cp and Cp2 haplotypes. GenBank® sequences are designated by AY. Numbers on branches indicate posterior probabilities.
Each of the five haplotypes identified was separated by at least one variable of a total of 37 polymorphic sites. Haplotype diversity was 0.8205 ± 0,0356, and nucleotide diversity (π) was 0,0313 ± 0,0159. Among the haplotypes identified, there were 22 diagnostic variable sites for C. aurita, nine for C. penicillata and one for C. jacchus (see Supplementary Table S1 online). Most nucleotide changes among the GenBank® sequences of pure individuals and those mitochondrial haplotypes found in our positive controls and the hybrids are transitions that resulted in synonymous substitutions. Only four nonsynonymous substitutions were detected. One transition at m.84T > C specific to Cja1_AY118190.1 resulted in p.L84P, which clustered with another GenBank® sequence, those from our positive controls and five hybrids. Specific of Cpe2_AY118195.1, a transition m.126C > T resulted in the exchange of p.T42M. Four hybrids clustered with this haplotype identified as HCp2. Two transversions, one detected in all C. aurita samples m.429A > G, led to the replacement of p.V143M and the other m.489G > A changing p.A165T was present in Cp3_AY118196.1, where all our positive controls of C. penicillata and seven hybrids clustered, representing the HCp haplotype (see the nucleotide and amino acid alignment in Supplementary Fig. S3a, S3b, S3c and S4 online).
The unrooted phylogenetic tree, generated by the alignment of the COX2 sequences, including those from GenBank®, shows the greatest genetic variability of C. penicillata forming three well-defined clusters with posterior probabilities over 0.83 of node support (Fig. 5). One cluster is closer to that of C. geoffroyi (Cge_AY118119.1). Our Cp positive controls and seven Csp (12–18) clustered with Cpe3_AY118196. Four Csp (3–5 and 7) clustered with Cpe2_AY118.195.1, phylogenetically closer to C. jacchus. The sequences of none of the positive controls grouped with C. geoffroyi or C. kuhlii.
Discussion
Hybridization was undoubtedly confirmed for 16 out of the 18 free-living marmosets studied since all of them inherited the SRY deletion 117_125delTAAGTATCG specific to C. aurita and COX2 haplotypes of C. penicillata or C. jacchus. The molecular markers of patrilineal and matrilineal ancestry combined here were suitable to attest hybridization in almost 90% of the sampled individuals. Two male individuals, identified as Csp6 and Csp8, from PARNASO in the Teresópolis municipality of Rio de Janeiro state, although exhibiting a mixed phenotype, inherited both parental lineages exclusive to C. aurita which is probably related to backcrossing or hybrid interbreeding. These two individuals were members of a group of C. aurita and C. penicillata, where the only adult male belonged to the native species27.
The hybrids sampled were distributed throughout the original area of occurrence of C. aurita, with one locality in São Paulo state at Biritiba Mirim and three localities of Rio de Janeiro state, in the municipalities of Teresópolis, Guapimirim and Petrópolis. At these localities, C. penicillata is present in almost 60% of the interspecific breeding, C. jacchus in 30% and less than10% of backcrossing to C. aurita was registered. Despite being a small sampling by location, the greater participation of C. penicillata in hybridization with C. aurita corroborates the expectation due to the similarities in the pattern of environmental fitness between both species22 and also with the greatest ex situ reproductive success observed between these species when compared to C. jacchus5.
Although the fertility of the hybrids had already been confirmed experimentally through many directional matings through the six species of Callithrix5 and in situ among C. penicillata and C. jacchus24, nothing is known about the fertility of free-living hybrids of C. aurita and still less about the patterns of in situ mating selection. The divergence of the Atlantic Forest marmosets occurred lightly more than 5 Ma in the Pliocene–Pleistocene scenario43,44,45. In birds, a typical young pair of sister species has only 1–2 Ma, and fertile hybrids still occur when species have nearly 7–17 Ma. Complete hybrid unfeasibility occurs when species are separated by approximately 11–55 Ma46. Therefore, considering only the phylogenetic proximity among the marmoset species, several kinds of crossings could result in viable and fertile hybrids representing an additional competitor to the threatened C. aurita.
We drew attention to the fact that most individuals inherited the nuclear DNA marker of C. aurita and that the mitochondrial DNA marker of one of the invaders cannot ensure that they represent the F1 generation without previous life history knowledge. The same limitation exists to confirm backcrossing or interbreeding among hybrids as observed from samples of Csp6 and Csp8 individuals who presented patrilineal and matrilineal DNA sequences of C. aurita but with phenotypes that correspond neither to the native species nor to the invasive ones (see Supplementary Fig. S5 online). Multiple nuclear markers are needed to attest to which generation they belong to and for a more complete understanding of the extent of introgression. Microsatellite markers47,48,49 and a panel of SNPs developed from C. jacchus and C. penicillata50 are already available and should be tested on C. aurita for this purpose.
The suspicion of hybridization based on the morphology of the Y chromosome and mixed phenotype in five samples39 analyzed here (Csp3, Csp4, Csp5, Csp6 and Csp7) was now confirmed with the SRY and COX2 primer sets. Due to the subtle difference in the morphology among the Y chromosome of the native species and the invasive species and the polymorphism of this chromosome observed for the invasive species35,38, it is advisable to confirm through molecular genetics the inheritance of the C. aurita Y chromosome in hybrids based on cytogenetics31. Since SRY 117_125delTAAGTATCG is specific to C. aurita, the SRY primer set is suitable for this purpose.
According to COX2, we found greater genetic variability in C. penicillata than in C. jacchus and C. aurita. Our C. penicillata samples, engendered three haplotypes forming well-defined clusters with above 83% node support. Considering that Cge_AY118119.1 (representing C. geoffroyi) and Cpe1_AY118194.1 (representing C. penicillata) are from GenBank® and that there is no other sample matching them, we will not discuss their proximity. However, the Cpe2_AY118195.1 and Cpe3_AY118196.1 haplotypes, which matched our samples and generated two distinct groups, showed greater distances between each other than the one shown by the Cpe2_AY118195.1 and the C. jacchus cluster. This unprecedented result may suggest hidden phylogenetic diversity that may be revealed through extensive phenotype-genetic research along the C. penicillata home range.
Apparent directional mating was observed between males carrying the C. aurita Y chromosome with invasive females or at least female descendants of matrilineal lineage of the invaders. All 18 hybrids sampled over nine years in different places in the Atlantic Forest of RJ and SP carry the Y chromosome of C. aurita and 16, the COX2 haplotype of C. jacchus or C. penicillata. The occurrence of the small isolated populations in contact with the closely related invasive species in a fragmented habitat is much more likely to hybridize because of the difficulty of finding mates of the same species17, the same scenario experienced by C. aurita in the Atlantic forest. The population decline and the consequent reduction in the number of native females for breeding may lead to hybridization as an alternative strategy to carry on reproduction. Considering that in most primates, females are philopatric and males disperse, male-mediated asymmetric introgression is the most likely outcome18. The prevalence of males of one population/species mating with heterospecific females, instead of reciprocal crosses, has already been observed for other mammalian taxa. For example, the introduced American mink (Mustela vison) is more numerous, and males are larger than the European threatened native mink (Mustela lutreola) which favors them to mate with European mink females51. As male of C. aurita is larger than the invaders, it could be one factor favoring their success with congeneric females. Added to the reduction of encounters, habitat devastation is recognized for favoring hybridization, since the modification of the environment can hinder communication and recognition of conspecifics52. Hybridization in São Paulo and Rio de Janeiro states represented in our sample can be boosted by deforestation, which increased just this year by up to 400% in São Paulo and more than doubled in Rio de Janeiro53. The Atlantic Forest retains only 12.4% of its original vegetation54, which is highly fragmented, and the largest fragments generally do not exceed 50 hectares53. It is urgent to seek information in the field on populations of C. aurita that inhabit more extensive and preserved forest fragments to ascertain whether the establishment of exotic species occurs and what kind of interaction takes place between them. The pure populations of the native species can also act as a source of parental native genome that can mitigate hybridization effects19. In the municipality of Sapucaia, Rio de Janeiro, one of the few records of the occurrence of populations of C. aurita was made without the presence of aliens in the vicinity55.
This is the first genetic confirmation that hybridization takes place among C. aurita and the invasive species C. jacchus and C. penicillata over time, although based on sampling in a small extension of the species distribution. The uniparental molecular markers used, despite the limitations that we have already noticed, allowed basic aspects of hybridization to be raised based on newly generated data.
In the end, we highlight the fact that there are possibly among the hybrids studied here, individuals that carry approximately 50% and maybe more of the genomic composition of the threatened native species C. aurita. This percentage is among what is currently recommended by several researchers in this field as a reasonable limit for protection56,57 representing an advance in relation to the minimum of 75% proposed in the intercross policy of 199657,58. In Brazil, the recent creation of a captive breeding program12 can provide the necessary support to deepen the genetic, reproductive and physiological research regarding C. aurita hybrids. However, to a coherent hybrid policy, ecological features and historical process that resulted in admixture individuals should be incorporated57. As already highlighted, we must bear in mind that each case of hybridization is peculiar and to be effective, conservation general rules must be reassessed21,57. Considering that the southeastern region of Brazil presents different scenarios where habitat loss and forest fragmentation coexist with some remaining but significantly large and continuous Atlantic Rain forested areas, we emphasize that relative to the hybridization among Callithrix aurita and invasive marmosets, we must think about it regionally.
Methods
Sampling location
Sampling of the pure individuals for positive controls occurred in areas of the Cerrado, Caatinga and of the southeastern region of the Atlantic Forest, the natural areas of occurrence of C. penicillata, C. jacchus and C. aurita, respectively. The positive controls were represented by six individuals of C. aurita, five C. jacchus and four C. penicillata, among which there were five females. All of them were wild born out of one C. jacchus (Cj1) born in captivity at Centro de Primatologia do Estado do Rio de Janeiro/CPRJ in 2009 (microchip number 030000007075), whose parents were wild born. Two individuals of C. jacchus were in captivity at the time of sample collection at the Wildlife Screening Center (Centro de Triagem de Animais Silvestres/CETAS/IBAMA) in Recife city in the state of Pernambuco, Northeast Brazil (original habitat of this species) and one C. penicillata in CETAS/IBAMA of Brasília city in the Midwest region of Brazil (original habitat of this species). They are identified by the respective initials of the pure species (Ca, Cj and Cp) followed by an Arabic numeral (Table 1).
The putative hybrid marmosets were sampled in areas of the Atlantic Forest, a highly threatened ecosystem, classified as a global hotspot of biodiversity, due to its exceptional concentration of endemic species and loss of more than 90% of its primary vegetation59 and is the natural area of occurrence of C. aurita. The putative hybrids were identified with the acronym Csp (Callitrix sp.) followed by an Arabic numeral (Table 1).
Five males out of the eighteen putative hybrids studied here (Csp3, Csp4 and Csp5 from Guapimirim and Csp6 and Csp7 from PARNASO) have already been analyzed by cytogenetics and according to the morphology of their Y chromosome they were supposed to be hybrids fathered by a C. aurita male or a hybrid with the Y chromosome of this species39. According to the subtle difference among the morphology of the Y chromosome of the Callithrix species, it is difficult to attest patrilineal lineage solely based on cytogenetic data. For this reason, those samples were included for analysis using nuclear and mitochondrial markers.
The free-living animals studied were captured in Tomahawk traps, baited with fruits, and sedated with ketamine hydrochloride (10 mg/kg) via intramuscular injection for morphological evaluation and blood sampling.
All research conducted followed guidelines for the ethical treatment of nonhuman primates approved by the Ethical Commission of the Instituto de Biologia Roberto Alcantara Gomes of Universidade do Estado do Rio de Janeiro (UERJ) for the Care and Use of Experimental Animals and adhered to Brazilian legislation number CEUA035/2014. Research permits were approved by the Brazilian Ministry of Environment, MMA/IBAMA/SISBIO number 31570-5.
The study was carried out in compliance with the ARRIVE guidelines (https://arriveguidelines.org).
SRY and COX2 analysis
To investigate the paternal lineage we used the sex-determining region of the Y chromosome (SRY) gene based on GenBank® sequences (AF338377, AF338381 and AF338379) of Callithrix40 to design consensus primers FSRY (5’-TACAGGCCATGCACAGAGAG-3’) and RSRY (5´-CTAGCGGGTGTTCCATTGTT-3’) for amplification through polymerase chain reaction (PCR), of an amplicon of ca. 200 bp, which includes the nine base pair deletion exclusive of C. aurita40. The organization of the mitogenome of the five Callithrix species was assumed to be free of NUMTS according to the high-quality base calls with no ambiguous nucleotides that could indicate a population of nuclear DNA of mitochondrial origin50,60.
To analyze the matrilineal lineage we used the primer set which flanks mitochondrial cytochrome c oxidase subunit II (COX2) gene following the advised PCR protocol30.
DNA was extracted from blood samples through phenol–chloroform protocol61. PCRs to amplify the SRY fragment contained 20–25 ng of DNA, 1–2 µM of each primer, 0.25–1 U of Taq DNA polymerase Platinum (Invitrogen®), 1X Taq buffer, 1.5 mM (for SRY)—2.5 mM (for COX2) MgCl2, 0.2 mM dNTPs and ultrapure water for a total reaction volume of 25 µL. The amplifications were carried out in a Veriti® 96-well Thermal cycler (Applied Biosystems, Co.) under the following conditions: a denaturation step of 94 °C for 5 min, followed by 30 cycles at 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min, and a final extension step of 72 °C for 10 min.
PCR products were checked by electrophoresis in 2% agarose gel dyed with ethidium bromide (10 mg/ml). The amplicons of SRY and COX2 were purified with Sephadex Resin and sequenced in an ABI PRISM 3500 (Applied Biosystem, Co).
Sequencing of the SRY amplicon was performed for three positive controls, one of each Callithrix species and five putative hybrids to check for the presence of the nine base pair insertions/deletions described in the pure species. Once it was confirmed, the forward primer was labelled with 6-FAM fluorescent dye, and all the PCR products were analysed in a ABI PRISM 3500 system (Applied Biosystem, Co) to identify the size of the amplicons using Geneious software. Our purpose was to make the analysis of putative hybrids more agile, eliminating the DNA sequencing step.
All samples were sequenced for the COX2 amplicon. The DNA sequences generated were aligned with Clustal W using the Megalign DNASTAR/Lasergene program, Inc. The GenBank® sequences of C. aurita (AY118189.1; AY118188.1), C. jacchus (AY118190.1; AY118191.1), C. penicillata (AY118194.1; AY118195.1; AY118196.1), C. geoffroyi (AY118192.1) and C. kuhlii (AY118193.1) were aligned with the haplotypes identified here to verify nucleotide and amino acid similarities using MEGA-X62. DNAsp663 was used to identify parsimony informative sites for each species and to estimate haplotype and nucleotide diversity. MRBAYES 3.2.164 was used to build the Bayesian tree to analyze phylogenetic relationships under an HKY85 substitution model using a Metropolis-coupled, Markov Chain Monte Carlo (MCMC). Four chains were run simultaneously for 500,000 generations in two independent runs, sampling trees every 100 generations. A burn-in of 1000 trees was carried out for each independent run. A consensus tree with nodal posterior probability support was obtained for five Callithrix species and Callithrix sp. samples.
References
Mittermeier, R. A., Coimbra-Filho, A. F., Constable, I. D., Rylands, A. B. & Valle, C. Conservation of primates in the Atlantic forest region of eastern Brazil. Int. Zoo Yearb. 22, 2–17 (1982).
Mittermeier, R. A., Rylands, A. B. & Wilson, D. E. W. Handbook of the Mammals of the World, vol. 3, Primates. Lynx, Barcelona (2013).
Rylands, A. B., Kierulff, M. C. M., Mendes, S. L. & Oliveira, M. M. Callithrix aurita. The IUCN Red List of Threatened Species 2008. IUCN Red List Threat. Species 8235, 1–7 (2008).
IUCN. The IUCN Red List of Threatened Species. Version 2020–2, Vol. 8235 (2020).
Coimbra-Filho, A., Pissinatti, A. & Rylands, A. B. Experimental mutiple hybridism and natural hybrids among Callithrix species from easterna Brazil. In Marmosets and Tamarins Systematics Behaviour and Ecology (ed. Rylands, A. B.) 93–120 (Oxford University Press, 1993).
Mittermeier, R. A., Coimbra-Filho, A. F., Rylands, A. B. & Constable, I. D. Atlantic Forest region of eastern Brazil a top primate of conservation priority. IUCN / SSC Primate Spec. Gr. Newsl. 1, 9–11 (1981).
Carvalho, R. S. et al. Buffy-tufted-year marmoset Callithrix aurita É. Geoffroy Saint-Hilaire, 1812 Brazil. In Primates in Peril: The World’s 25 Most Endangered Primates 2018–2020 (eds. Schwitzer, C. et al.) 136 (IUCN SSC Primate Specialist Group, International Primatological Society, Global Wildlife Conservation, Bristol Zoological Society, 2019).
ICMBio. Instituto Chico Mendes de Conservação da Biodiversidade. Livro Vermelho da Fauna Brasileira Ameaçada de Extinção. (2018).
SMA-SP. Decreto no63.853, de 27 de novembro de 2018. (2018).
Rylands, A. B. et al. An assessment of the diversity of New World Primates. Neotrop. Primates 8, 61–93 (2000).
Brandão, L. D. & Develey, P. F. Distribution and conservation of the buffy-tufted-ear marmoset, Callithrix aurita, in lowland coastal, Atlantic Forest, Southeast Brazil. Neotrop. Primates 6, 86–88 (1998).
Carvalho, R. S. et al. Callithrix aurita: a marmoset species on its way to extinctionin the Brazilian Atlantic Forest. Neotrop. Primates 24, 1–8 (2018).
Malukiewicz, J. et al. Natural and anthropogenic hybridization in two species of eastern Brazilian marmosets (Callithrix jacchus and C. penicillata). PLoS ONE 10, 1–22 (2015).
Ruiz-Miranda, C. R., Affonso, A. G., Martins, A. & Beck, B. Distribuição do sagui (Callithrix jacchus) nas áreas de ocorrência do mico-leão-dourado (Leontopithecus rosalia) no Estado do Rio de Janeiro. Neotrop. Primates 8, 98–101 (2000).
Mendes, S. L. Hybridization in free-ranging Callithrix flaviceps and the taxonomy of the Atlantic forest marmosets. Neotrop. Primates 5, 6–8 (1997).
Santos, C. V. et al. Ecologia, comportamento e manejo de primatas invasores e populações-problema. A Primatologia no Brasil 10, 101–118 (2007).
Rhymer, J. M. & Simberloff, D. Extinction by hybridization and introgression. Annu. Rev. Ecolology Evol. Syst. 27, 83–109 (1996).
Zinner, D., Arnold, M. L. & Roos, C. The strange blood: Natural hybridization in primates. Evol. Anthropol. 20, 96–103 (2011).
Brumfield, R. T. Speciation genetics of biological invasions with hybridization. Mol. Ecol. 19, 5079–5083 (2010).
Steeves, T. E., Maloney, R. F., Hale, M. L., Tylianakis, J. M. & Gemmell, N. J. Genetic analyses reveal hybridization but no hybrid swarm in one of the world’s rarest birds. Mol. Ecol. 19, 5090–5100 (2010).
Allendorf, F. W., Leary, R. F., Spruell, P. & Wenburg, J. K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 16, 613–622 (2001).
Bechara, I. M. Abordagens metodológicas em Biogeografia da Conservação para avaliar risco de extinção de espécies: um estudo de caso com Callithrix aurita (Primates: Callitrichidae). (UFRJ, 2012).
Braz, A. G., Lorini, M. L. & Vale, M. M. Climate change is likely to affect the distribution but not parapatry of the Brazilian marmoset monkeys (Callithrix spp.). Divers. Distrib. 25, 536–550 (2019).
Malukiewicz, J. A review of experimental, natural, and anthropogenic hybridization in Callithrix marmosets. Int. J. Primatol. 40, 72–98 (2019).
Pinto, L. F. G. & Voivodic, M. Reverse the tipping point of the Atlantic Forest for mitigation. Nat. Clim. Chang. 11, 364–365 (2021).
Seehausen, O., Takimoto, G., Roy, D. & Jokela, J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol. Ecol. 17, 30–44 (2008).
Pereira, D. G., De Oliveira, M. E. A. & Ruiz-Miranda, C. R. Interações entre calitriquídeos exóticos e nativos no Parque Nacional da Serra dos Órgãos - RJ. Espaço e Geogr. 11, 87–114 (2008).
Aximoff, I., Soares, H. M., Pissinatti, A. & Bueno, C. Registros de Callithrix aurita (Primates, Callitrichidae) e seus híbridos no Parque Nacional do Itatiaia. Oecologia Aust. 20, 520–525 (2016).
Detogne, N. et al. Spatial distribution of buffy-tufted-ear (Callithrix aurita) and invasive marmosets (Callithrix spp.) in a tropical rainforest reserve in southeastern Brazil. Am. J. Primatol. 79, 1–11 (2017).
Carvalho, R. S. et al. Molecular identification of a buffy-tufted-ear marmoset (Callithrix aurita) incorporated in a group of invasive marmosets in the Serra dos Órgãos National Park, Rio de Janeiro - Brazil. Forensic Sci. Int. Genet. Suppl. Ser. 4, e230–e231 (2013).
Novaes, C. M. et al. Karyotypic characteristics of hybrid marmosets of the genus Callithrix (Erxeleben, 1777) suggest the participation of three parental species. Bol. do Mus. Biol. Mello Leitão 39, 11–21 (2017).
Pereira, D. G. Densidade, genética e saúde populacional como ferramentas para propor um plano de controle e erradicação de invasão biológica: o caso de Callithrix aurita (Primates) no Parque Nacional da Serra dos Órgãos, RJ, Brasil (Universidade do Estado do Rio de Janeiro, 2010).
Veracini, C., Galeni, L. & Forti, M. The concept of species and the foundations of biology, a case study: The Callithrix jacchus group (Primates-Platyrrhini). Riv. Biol. 95, 75–100 (2002).
Campton, D. E. Natural hybridization and introgression in fishes: methods of detection and genetic interpretations. In Population Genetics and Fishery Management (eds Ryman, N. & Utter, F.) 16–192 (University of Washington Press, 1987).
Nagamachi, C. Y., Pieczarka, J. C., Schwarz, M., Barros, R. M. S. & Mattevi, M. S. Comparative chromosomal study of five taxa of genus Callithrix, group jacchus (Platyrrhini, Primates). Am. J. Primatol. 41, 53–60 (1997).
Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (2005).
Ardito, G., Lamberti, L., Bigatti, P., Stanyon, R. & Govone, D. NOR distribution and satellite associations in Callithrix jacchus. Caryologia 40, 185–194 (1987).
Nagamashi, C. & Ferrari, I. Cytogenetic studies of Callithrix jacchus (Callitrichidae, Platyrrhini) from two different sites in Brazil. I. Morphological variability of Y chromosome. Rev. Bras. Genética 7, 497–507 (1984).
Nogueira, D. M. et al. Cytogenetic study in natural hybrids of Callithrix (Callitrichidae: Primates) in the Atlantic forest of the state of Rio de Janeiro, Brazil. Iheringia, Série Zool. 101, 156–160 (2011).
Moreira, M. A. M. SRY evolution in Cebidae (Platyrrhini: Primates). J. Mol. Evol. 55, 92–103 (2002).
de Morais, M. M. OS SAGUIS (Callithrix spp., ERXLEBEN, 1777) Exóticos Invasores na bacia do Rio São João, Rio de Janeiro: Biologia populacional e padrão de distribuição em uma paisagem fragmentada. Uenf.Br (2010).
Sweeney, C. G., Curran, E., Westmoreland, S. V., Mansfield, K. G. & Vallender, E. J. Quantitative molecular assessment of chimerism across tissues in marmosets and tamarins. BMC Genomics 13, 98. https://doi.org/10.1186/1471-2164-13-98 (2012).
Buckner, J. C., Lynch Alfaro, J. W., Rylands, A. B. & Alfaro, M. E. Biogeography of the marmosets and tamarins (Callitrichidae). Mol. Phylogenet. Evol. 82, 413–425 (2015).
Schneider, H. et al. A molecular analysis of the evolutionary relationships in the Callitrichinae, with emphasis on the position of the dwarf marmoset. Zool. Scr. 41, 1–10 (2012).
Perelman, P. et al. A molecular phylogeny of living primates. PLoS Genet. 7, 1–17 (2011).
Price, T. D. & Bouvier, M. M. The evolution of F1 postzygotic incompatibilities in birds. Evolution (N. Y). 56, 2083–2089 (2002).
Nievergelt, C. M., Mundy, N. I. & Woodruff, D. S. Microsatellite primers for genotyping common marmosets (Callithrix jacchus) and other callitrichids. Mol. Ecol. 7, 1432–1434 (1998).
Raveendran, M. et al. Polymorphic microsatellite loci for the common marmoset (Callithrix jacchus) designed using a cost- and time-efficient method. Am. J. Primatol. 70, 906–910 (2008).
Takabayashi, S. & Katoh, H. Noninvasive genotyping of common marmoset (Callithrix jacchus) by fingernail PCR. Primates 56, 235–240 (2015).
Malukiewicz, J. et al. Mitogenomic phylogeny of Callithrix with special focus on human transferred taxa. BMC Genomics 22, 1–14 (2021).
Rozhnov, V. V. Extinction of the European mink: ecological catastrophe or a natural process?. Lutreola 1, 10–16 (1993).
Rosenthal, G. G. Individual mating decisions and hybridization. J. Evol. Biol. 26, 252–255 (2013).
Fundação SOS Mata Atlântica. Notícias. www.sosma.org.br/noticias/desmatamento-da-mata-atlantica-cresce-em-dez-estados/ (2021).
Banks-Leite, C. et al. Using ecological thresholds to evaluate the costs and benefits of set-asides in a biodiversity hotspot. Science 345, 1041–1045 (2014).
Oliveira, A. B. L. Presença ou Ausência do Callithrix aurita em Fragmentos de Mata Atlântica (Instituto Superior de Agronomia - Universidade Técnica de Lisboa, 2012).
Allendorf, F. W. et al. Intercrosses and the U.S. endangered species act: Should hybridized populations be included as westslope cutthroat trout?. Conserv. Biol. 18, 1203–1213 (2004).
Wayne, R. K. & Shaffer, H. B. Hybridization and endangered species protection in the molecular era. Mol. Ecol. 25, 2680–2689 (2016).
U.S. Fish and Wildlife Service and National Oceanic and Atmospheric Administration. Endangered and threatened wildlife and plants: Proposed policy and proposed rule on the treatment of intercrosses and intercross progeny (the issue of “hybridization"). Fed. Regist. 67, 4710–4713 (1996).
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Fonseca, G. A. B. & Kent, J. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000).
Malukiewicz, J., Hepp, C. M., Guschanski, K. & Stone, A. C. Phylogeny of the jacchus group of Callithrix marmosets based on complete mitochondrial genomes. Am. J. Phys. Anthropol. 162, 157–169 (2016).
Sambrook, J., Maniatis, T. & Fritsch, E. F. Molecular Cloning. A Laboratory Manual. Vol. 1 (Cold Spring Harbor Laboratory Press, 2001).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Rozas, J. et al. DnaSP6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Ronquist, F., van der Mark, P. & Huelsenbeck, J. P. Bayesian phylogenetic analysis using MRBAYES. Phylogenetic Handb. https://doi.org/10.1017/cbo9780511819049.009 (2012).
Acknowledgements
We thank the Brazilian Institute of Natural Resources/IBAMA for the capture license, and Centro de Primatologia do Rio de Janeiro/INEA for the maintenance of the hybrids in captivity. We thank the manager of Serra dos Órgãos National Park for allowing the capture of the hybrids and Gisele Lôbo-Hajdu for technical support. This work was supported by Grants from Fundação Carlos Chagas de Amparo à Pesquisa do Estado do Rio de Janeiro/FAPERJ (Proc. E-26/171.271/2006 e E-26/111.569/2010). HGB thanks CNPq, FAPERJ and Sr2/UERJ for the research fellowships. DGP thanks the PhD scholarship from CAPES. DGP also thanks the Pos-Doc scholarship and DMN for the research fellowship both from FAPERJ.
Author information
Authors and Affiliations
Contributions
D.M.N. designed the study; D.M.N. and R.S.C. wrote the manuscript; R.S.C., D.G.P. and H.G.B. conducted field work; R.S.C. prepared most of the figures; D.G.P. provided the images of the Csp6 and Csp8 hybrids; A.P. provided biological samples, D.M.N., A.M.O. and T.S.P. designed the primers; D.A.S. supervised laboratory work; D.M.N., R.S.C., A.M.O. and S.O.L. performed genetic analysis; E.F.C. and A.M.R.F. provided the laboratory infrastructure for performing the analyses. All the authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nogueira, D.M., de Carvalho, R.S., de Oliveira, A.M. et al. Uniparental genetic markers to investigate hybridization in wild-born marmosets with a mixed phenotype among Callithrix aurita and invasive species. Sci Rep 12, 1487 (2022). https://doi.org/10.1038/s41598-021-04276-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-04276-7
This article is cited by
-
Morphological and Genetic Variation Among Callithrix Hybrids in Rio de Janeiro, Brazil
Evolutionary Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.