Abstract
CUX1, encoding a homeodomain-containing transcription factor, is recurrently deleted or mutated in multiple tumor types. In myeloid neoplasms, CUX1 deletion or mutation carries a poor prognosis. We have previously established that CUX1 functions as a tumor suppressor in hematopoietic cells across multiple organisms. Others, however, have described oncogenic functions of CUX1 in solid tumors, often attributed to truncated CUX1 isoforms, p75 and p110, generated by an alternative transcriptional start site or post-translational cleavage, respectively. Given the clinical relevance, it is imperative to clarify these discrepant activities. Herein, we sought to determine the CUX1 isoforms expressed in hematopoietic cells and find that they express the full-length p200 isoform. Through the course of this analysis, we found no evidence of the p75 alternative transcript in any cell type examined. Using an array of orthogonal approaches, including biochemistry, proteomics, CRISPR/Cas9 genomic editing, and analysis of functional genomics datasets across a spectrum of normal and malignant tissue types, we found no data to support the existence of the CUX1 p75 isoform as previously described. Based on these results, prior studies of p75 require reevaluation, including the interpretation of oncogenic roles attributed to CUX1.
Similar content being viewed by others
Introduction
Protein isoforms and splice variants often have important and distinct biological functions. Since protein isoforms, by definition, have considerable sequence homology and may be expressed at different levels within the cell, it can be challenging to accurately differentiate between and functionally characterize such isoforms1. One such protein reported to have multiple isoforms is CUT-like homeobox 1 (CUX1), a HOX-family transcription factor with critical roles in development and tumorigenesis. In vertebrates, the CUX1 locus contains two distinct genes that partially share exons: CUX1, which encodes a transcription factor localized to the nucleus, and CASP, which encodes a golgi-associated transmembrane protein involved in retrograde transport2,3,4. Altered levels and mutations of the CUX1 transcription factor have been implicated in cancer across several tumor types and species5,6. CASP, on the other hand, has not been implicated in human disease7,8. The RefSeq database documents seven mRNA isoforms for the human CUX1 locus; five of these are CASP transcripts and two are CUX1 (Fig. S1). Due to its relevance to human health, we focus our attention herein on CUX1. For the sake of simplicity, our subsequent references to the CUX1 gene or mRNA allude to those isoforms that encode CUX1, unless stated otherwise.
CUX1 is highly conserved, ubiquitously expressed, and essential for survival in mice and Drosophila9. CUX1 controls many cellular processes including determination of cell identity, cell cycle progression, cell–cell communication, and cell motility9. In cancer, however, there are conflicting reports of CUX1 acting alternately as an oncogene or tumor suppressor gene6. To resolve this discrepancy, we hypothesized that distinct CUX1 protein isoforms explain these disparate functions.
The two RefSeq-annotated CUX1 mRNA transcripts vary only by alternative first exons and encode a full-length protein of 1505 amino acids length, described in the literature as p200 (Figs. 1a, S1). p200 CUX1 has four DNA-binding domains, comprised of three CUT-repeat domains and one homeodomain (Fig. 1a). A truncated p110 CUX1 isoform is generated by post-translational proteolytic processing of full-length p200 CUX1 by cathepsin L (Fig. 1a)10. This cleavage occurs during the S phase in normal cells, and can become constitutive in transformed cells10,11. p110 CUX1 lacks one CUT-repeat domain and the N-terminal region but retains the three C-terminal DNA-binding domains. A third isoform, p75 CUX1, is reported to arise from an alternative transcription start site (TSS) embedded within intron 20 and retains one CUT-repeat and the homeodomain (Fig. 1a)6,12. p75 has been identified in human breast cancer cell lines and mouse thymocytes12. Despite fewer DNA binding domains, p75 and p110 bind DNA more stably than p20011,13. Rarer CUX1 isoforms have been described to be generated by post-translational proteolytic processing; p80, p90 and p150 CUX113,14,15. However, these isoforms are less well characterized and it is unclear if they bind DNA and exert transcriptional activity13,14,15.
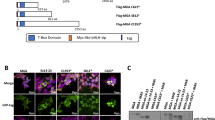
Human hematopoietic cells only express the p200 CUX1 isoform. (a) Schematic representation of the CUX1 mRNA. There are two CUX1 mRNA transcripts that vary only by the alternative first exons (1a and 1b). CUX1 encodes a full-length protein of 1505 amino acids which runs at 200 kDa (p200). A truncated p110 CUX1 protein is reported to be generated by proteolytic cleavage by cathepsin L. The p75 CUX1 isoform is reported to arise from an alternative transcription site embedded within intron 20. (b) Schematic representation of the predominant CUX1 protein isoforms, with protein domains indicated, and the CUX1 antibodies used in this study. (c) Immunoblot of CUX1 in the indicated human AML cell lines, using the B-10 antibody (n = 3). 10 μg of protein was loaded for the K562 and Kasumi-1 cell line, and 15 μg of protein was loaded for all other cell lines. (d) Immunoblot of CUX1 in primary human CD34 + HSPCs using the B-10-HRP antibody (n = 3). (e) Immunoblot of CUX1 in the NIH-3T3 fibroblast line and several human breast cancer cell lines previously reported to express p75 CUX1 using the B-10-HRP antibody (n = 3). (f) Immunoblot of CUX1 in indicated human AML cell lines, using the PUC antibody (n = 3). (g) Immunoblot of CUX1 in indicated human AML cell lines, using the ABE217 antibody (n = 3). (h) Immunoblot of GFP in a KG-1 cell line where endogenous CUX1 is C-terminally tagged with GFP. Protein from unedited KG-1 cells is also included (n = 3). Blot is cropped from the same gel to remove an intervening irrelevant lane.
By some reports, CUX1 is thought to be oncogenic in cancer. Over-expression of short p75 or p110 CUX1 isoforms in fibroblasts and breast cancer cells causes increased proliferation, cell cycle progression, and tumor formation in vivo11,12,15,16,17,18,19. p75 CUX1 transgenic mice engendered a higher proportion of adenosquamous mammary carcinomas and lung metastases compared to p110 or p200 transgenic mice18. On the other hand, large scale cancer genome resequencing efforts demonstrate that patterns of CUX1 inactivating mutations or deletions are more characteristic of a tumor suppressor20. CUX1 deletions and inactivating mutations are prevalent across cancer types20,21,22. In myeloid malignancies, CUX1 falls within the commonly deleted region of chromosome 7q2223. Consistent with a tumor suppressor role, CUX1 knockdown in mouse models leads to MDS/MPN that is reminiscent of human disease24,25. CUX1 knockdown in human hematopoietic stem cells provides an engraftment advantage in immunodeficient mice23. Even in Drosophila models, the CUX1 orthologue, cut, exerts tumor suppressive activity21,23.
Given the clinical significance, it is critical to parse out the putative oncogenic and tumor suppressive roles of CUX1. We reasoned that uncovering isoform-specific properties would reveal therapeutic strategies for inhibiting oncogenic CUX1 isoforms or promoting the expression of tumor suppressive isoforms to treat malignancies with CUX1 alterations. To this end, we characterized the CUX1 isoforms in a panel of human cell types and leveraged publicly available functional genomic datasets across a spectrum of tissue types. To our surprise, we identified no evidence supporting the existence of the p75 mRNA isoform in any cell type examined and demonstrate that p75 is likely a western blotting artefact. Focusing on hematopoietic cells, we only identify the p200 isoform. Our data indicate that in hematopoietic cells, the tumor suppressive role of CUX1 is attributable to the full-length protein. In addition, the lack of evidence for a p75 transcript calls into question prior interpretations of studies using exogenously overexpressed short isoforms that ascribe an oncogenic role for CUX1.
Results
Human hematopoietic cells only express the p200 CUX1 isoform
Given the relevance of CUX1 to myeloid malignancies, we first sought to identify the CUX1 isoforms expressed in human acute myeloid leukemia (AML) cells. Immunoblotting with an antibody that recognizes an epitope shared across all CUX1 isoforms (clone B-10, Fig. 1b) reveals six of eight AML cell lines express a dominant p200 CUX1 band (Fig. 1c). We also observed a less prominent 75 kDa band in five cell lines (Fig. 1c). p200 was also the predominant isoform in primary human CD34+ hematopoietic stem and progenitor cells (HSPCs), the normal counterpart thought to give rise to myeloid malignancies (Fig. 1d). To discern if the p75 band we observed corresponds to that described previously, we assessed cell lines reported to express p75 or p110: murine NIH-3T3 fibroblasts (p110), and MCF7, T47D, and MDA-MB-231 human breast cancer cell lines (p75)12,26. In contrast to prior findings, we did not observe a short isoform protein band in any of these cell lines, using both the B-10 and the PUC antibodies (Fig. 1e, S2). The absence of p110 in NIH-3T3 cells was also previously observed12.
We next blotted AML cell lines to determine if we could detect the p75 band with other CUX1 antibodies. We used a polyclonal antibody we previously generated, PUC, that recognizes amino acids 1223–1242 of CUX127, and ABE217, a polyclonal antibody raised against an epitope spanning amino acid 861 of CUX1 (Fig. 1b). There were several background bands observed with PUC, but no dominant p75 band (Fig. 1f). The faint band at 75 kDa seen with the ABE217 antibody cannot be the p75 isoform, as the antibody recognizes an epitope of CUX1 upstream of the p75 protein sequence (Fig. 1b, g). Thus, p200 is the predominant CUX1 isoform in human hematopoietic cells, and we did not detect p75 or p110 in cells previously reported to express short isoforms.
To circumvent potential antibody artefacts, we took an alternative approach to determine if the p75 band is in fact CUX1, by tagging the endogenous CUX1 allele with an in-frame C-terminal GFP tag by CRISPR/Cas9 homology-mediated repair. We used KG-1 cells, which express both p75 and p200 bands. KG-1 cells have a partial deletion of chromosome arm 7q that includes CUX1, thus they are mono-allelic for CUX1, enabling facile CRISPR/Cas9 editing (Fig. S3). GFP-tagged CUX1 migrated at a higher molecular weight than endogenous CUX1, as expected (Fig. S4). Probing these cells with an anti-GFP antibody only identified a single unique p200 CUX1 band in the tagged cell line (Fig. 1h). Together, these data indicate that human AML and primary HSPCs express p200 and not shorter CUX1 isoforms.
CRISPR/Cas9 genomic editing precludes the existence of a CUX1 p75 isoform
To further interrogate if the p75 band is encoded by the CUX1 locus or is a non-specific western blotting artefact, we devised several CRISPR/Cas9 strategies to selectively target KG-1 genomic DNA encoding p75, p200, or both (Figs. 2a, d, 3a). As illustrated in Fig. 2a, we designed a gRNA targeting exon 4 of CUX1 which is only expressed in the genomic region encoding the p200 isoform. This would be expected to selectively introduce a frameshift mutation in the p200 protein only and thereby induce nonsense-mediated decay of only p200, leaving p75 intact. We identified two single-cell CRISPR-edited clones that had a complete loss of p200 CUX1 (C9 and H9), best appreciated with the ABE217 antibody (Fig. 2b). The B-10 antibody also shows a loss of p200, with a residual non-specific band migrating at a slightly higher molecular weight (Fig. 2c). The expression level of the p75 band was unchanged, as expected based on our targeting strategy (Fig. 2c).
CRISPR/Cas9 genomic editing precludes the existence of a p75 CUX1 isoform. (a) CRISPR/Cas9 editing approach to selectively delete p200 in the KG-1 cell line, which only has one copy of CUX1. (b) Immunoblot for CUX1 in KG-1 single cell clones edited with a gRNA targeting exon 4 of CUX1 using the ABE217 antibody. (c) Immunoblot for CUX1 in KG-1 single cell clones edited with a gRNA targeting exon 4 of CUX1 using the B-10-HRP antibody. (d) CRISPR/Cas9 editing approach targeting exon 23 of CUX1 to delete all CUX1 isoforms in the KG-1 cell line. (e) Immunoblot for CUX1 in a KG-1 single cell clone edited with a gRNA targeting exon 23 of CUX1 using the ABE217 antibody (n = 3). (f) Immunoblot for CUX1 in a KG-1 single cell clone edited with a gRNA targeting exon 23 of CUX1 or HPRT using the B-10 antibody (n = 3).
CRISPR/Cas9 genomic editing of the predicted intronic p75 transcriptional start site does not eliminate the p75 isoform band. (a) CRISPR/Cas9 editing approach using two gRNAs flanking the predicted intronic p75 TSS. (b) PCR screening strategy for identifying single cell clones with deletion of the p75 putative TSS. PCR products from primer pairs spanning the first cut site or the second cut site in intron 20 indicate the lack of a successful deletion. A PCR product from primers spanning the deletion site indicates a successful deletion. (c) Immunoblot for CUX1 using the B-10 antibody in a KG-1 single cell clone with successful deletion of the p75 TSS (n = 3).
As a control, we designed a gRNA targeting exon 23 of CUX1 (gEx23.1) to introduce frameshift mutations in and edit all CUX1 transcripts (Fig. 2d). As exon 23 is shared by all isoforms, including p75, it would be expected to disrupt all bona fide CUX1 proteins. Transfection with gEx23.1 followed by single cell cloning identified a clone with a single base pair insertion generating a frameshift mutation. We blotted the gEx23.1-edited KG-1 clone with ABE217 and observed that the band for p200 CUX1 shifted downward, consistent with a predicted C-terminal truncation of ~ 28 kDa (Fig. 2e). The B-10 antibody binds an epitope after the exon 23 gRNA cut site, thus all gEx23-edited isoforms will be undetectable with B-10 (Fig. 1b). Indeed, probing with B-10 demonstrates abolished expression of the p200 CUX1 band, yet persistent p75 (Fig. 2f). The fact that the 75 kDa band remained indicates that it is not encoded by the CUX1 locus.
We similarly targeted CUX1 using two different exon 23 gRNAs in primary human CD34+ HSPCs, and saw no change in expression of any bands other than p200 CUX1 (Fig. S5). The residual p200 protein in the gEx23 edited lanes is consistent with ~ 75–80% editing in these bulk populations. Overall, these data indicate that hematopoietic cells express p200 CUX1 and not p75.
We considered the possibility that p75 does not contain exon 23, perhaps due to alternative splicing. As a different approach, we designed a pair of gRNAs flanking the predicted p75 intronic TSS reported to be ~ 2.5 kb upstream of exon 21 (Fig. 3a)12. We reasoned that eliminating the putative intronic TSS would eliminate transcription of the p75 isoform while leaving p200 unperturbed. We deleted approximately 2.56 kb of intronic DNA, leaving 79 base pairs intact proximal to exon 21. Using a PCR strategy to screen the expected deletion, we generated a successfully deleted single-cell clone (KG-1 Δp75 #21, Fig. 3b). Immunoblotting, however, indicated no change in p75 (Fig. 3c). This suggests that this 2.56 kb segment of DNA does not harbor the putative p75 TSS, and is incongruent with the prior report12. In summary, these experiments show that the presumptive p75 isoform contains neither exon 23 nor a TSS within 2.56 kb upstream of exon 21 as originally described, further implicating the p75 band as a western blotting artefact.
Proteomics approaches do not support the existence of p75 CUX1
We considered that the p75 artefact results from the denaturing conditions of western blotting. To test this, we performed a CUX1 immunoprecipitation which is performed with proteins in their native state. After immunoprecipitation of CUX1 in KG-1 cells with the B-10 antibody, we probed the resulting blots again with B-10. While the p75 band is present in the input control, we do not observe it in the immunoprecipitate (Fig. 4a). This result is consistent with p75 being an artefact of western blotting.
Proteomic approaches do not support the existence of p75 CUX1. (a) B-10 immunoblot after B-10 immunoprecipitation of CUX1 in the KG-1 cell line. KG-1 cells were also immunoprecipitated with a mouse IgG antibody as a negative control. Input control is also included (n = 3). Blot is cropped on the edge to remove a redundant lane. (b) p200 and p75 regions (red rectangles) were excised from Imperial stained SDS/PAGE of two whole cell extract replicates in the KG-1 cell line, and two B-10 immunoprecipitated CUX1 replicates from the KG-1 cell line. (c) Schematic showing the number and distribution of peptides mapping to CUX1 and CASP protein sequences from the immunoprecipitated CUX1 samples (n = 2). Heat map at the bottom depicts the number of each peptide detected across replicates. Peptides with ambiguous assignments are shown in both potential candidate isoforms. (d) Schematic showing the number and distribution of peptides mapping to CUX1 and CASP protein sequences from the whole cell lysate samples. Heat map at the bottom depicts the number of each peptide detected across replicates. Peptides with ambiguous assignments are shown in both potential candidate isoforms.
We turned to unbiased proteomics to search for any CUX1 peptides within the 75 kDa region. We subjected KG-1 whole cell lysates and CUX1 immunoprecipitates to SDS-PAGE. We excised regions corresponding to 200 kDa and 75 kDa and performed LC–MS/MS (Fig. 4b). In the 200 kDa region after CUX1 immunoprecipitation, we observed 51 peptides (21 unique, shown as blue rectangles, Fig. 4c; Table S1) summed across replicates that mapped to CUX1/CASP protein sequences. Because p200, p75 and CASP partially share exons, some peptides are ambiguous and thus map to both p200 and CASP or both p200 and p75. However, as CASP is a 77 kDa protein and p75 is 75 kDa, we can infer that all ambiguous peptides in the 200 kDa region are in fact p200. In the 75 kDa region of the immunoprecipitated samples, we observed one peptide that ambiguously mapped to the N-terminal region of CUX1/CASP, but observed no peptides mapping to the p75 region (Fig. 4c; Table S2). We think this might be a degradation product from the p200 band for this sample, as it was a low-scoring peptide of low intensity, compared to the corresponding peptide in the p200 sample. These data confirm that p75 CUX1 does not immunoprecipitate with anti-CUX1, consistent with Fig. 4a.
We assessed the proteomic analysis of whole cell lysates, which are agnostic to antibody selection (Fig. 4b, first two lanes). The p200 band contained 8 peptides (5 unique) which, based on a priori knowledge of relative protein migrations, we ascribed to p200 (Fig. 4d; Table S3). Within the p75 band isolated from lysates, we observed 7 peptides (3 unique) that all unambiguously map to CASP. We did not detect any peptides from the p75 region in the lysates that mapped to p75 CUX1 (Fig. 4d; Table S4). Taken together, we conclude that there is no proteomic evidence of the p75 isoform in a representative AML cell line that possesses the p75 protein band.
No p75 CUX1 is detected at the RNA level in human AML and breast cancer cell lines
It remained possible that the p75 protein is below the level of detection of mass spectrometry. As a more sensitive test, we looked for the presence of p75 CUX1 at the mRNA level by RT-PCR, using two primer sets (pairs 2 and 4 and pair 3 and 4) spanning intron 20 and exon 22 (Fig. 5a, b). Primer pair 3 and 4 was previously reported to detect the p75 transcript12. We assessed cDNA from three AML cell lines (K562, Kasumi-1, and KG-1) and three human breast cancer cell lines described to express p75 (T47D, MDA-MB-231, and MCF-7). Primers for GAPDH and all CUX1 isoforms (primer pair 1 and 4 and pair ex23 and ex24) served as positive controls. We did not detect any bands with the previously reported p75 primers (3 and 4) or with our primers (2 and 4, Fig. 5c). We confirmed that primer 4 was functional, as it successfully amplified p200 (primer pair 1 and 4), indicating that the lack of a p75 transcript was not due to primer design issues. Overall, we do not detect a p75 transcript in either human AML cell lines or the breast cancer cell lines previously reported to express p7512.
No p75 CUX1 is detected at the RNA level in human AML and breast cancer cell lines. (a) Schematic of primers used to detect p75. (b) Table of anticipated PCR products and whether they are unique to the p75 isoform. (c) Reverse-transcriptase PCR products after 30 cycles of PCR spanning the intron 20 region of CUX1 using cDNA reverse-transcribed from mRNA in 6 different cell lines (K562, Kasumi-1, KG-1, T47D, MDA-MB-231 and MCF-7) (n = 3). cDNA from the KG-1 Δp75 #21 cell line was used as a negative control for p75 mRNA expression. GAPDH, p200-specific primers (1 and 4) and CUX1 primers spanning all isoforms (ex23 and 24) were used as positive controls. Gel is cropped for clarity.
Functional genomics consortia datasets lack epigenetic or transcriptional evidence for a CUX1 p75 intronic transcriptional start site
Heretofore, our analysis has encompassed thirteen cell types. We sought to extend our analysis to comprehensively assess additional normal and malignant tissue types. As such, we leveraged consortia-generated functional genomics datasets for evidence of p75 CUX1 across a variety of cell types. We first assessed epigenetic marks that canonically decorate promoters. Promoters and enhancers both have H3K27ac, H3K4me3 and H3K4me1 deposition, although enhancers have higher levels of H3K4me128. All three of these marks are present at the p200 TSS across seven Tier 1 ENCODE cell lines (Fig. 6a). However, we did not observe any H3K4me3 peaks in the intron 20 region of CUX1, including in MCF7, previously reported to express p75 (Fig. 6a). We observed some H3K27ac and H3K4me1 peaks in intron 20, but these peaks were not at the predicted TSS and not in MCF7 cells (Fig. 6a). Additionally, we also looked at the ChromHMM track in MCF7 cells. ChromHMM is a computational approach that integrates experimental ChIP-seq datasets for different histone marks into a hidden Markov model to assign chromatin states genome-wide29. The MCF7 ChromHMM track shows an active TSS (red region) at exon 1a and 1b but only weak transcription (dark green) and no promoter regions in the intron 20 region (MCF7 ChromHMM track, Fig. 6a). Thus, epigenetic features of promoters are not present at the putative p75 TSS, even in a cell type documented to express p75.
Functional genomics consortia datasets lack epigenetic or transcriptional evidence for a CUX1 p75 intronic transcriptional start site. (a) CUX1 Refseq gene model is aligned with tracks of indicated transcriptional and epigenetic marks (including H3K4me1, H3K4me3 and H3K27ac ChIP-seq, DNase hypersensitivity tracks, CAGE-seq data in 5 ENCODE cell lines, POL2RA ChIP-seq tracks, and transcription factor ChIP-seq tracks). Tracks highlighted in red are derived from a cell type previously reported to express p75 CUX1. The ENCODE RNA-seq data in 9 cell lines was overlaid using the Transparent setting in the same track. (b) CAGE-seq data from the FANTOM browser at exon 1 and intron 20 of CUX1 in 975 human primary cells and cancer cell lines. (c) RNA-seq peaks for CUX1 expression in human breast tissue, MCF7 and T-47D cell lines from Burge lab sequencing data37 visualized on the UCSC Genome Browser. No intronic reads contiguous with exon 21 are observed.
Promoters are also characterized by accessible chromatin, transcription factor binding, RNA-polymerase II (POL2RA) occupancy, and CpG islands30,31,32,33. In line with this, the p200 TSS has pronounced DNase hypersensitivity, transcription factor occupancy, POLR2A peaks, and a CpG island (Fig. 6a). However, there is a paucity of these signals in intron 20 and they are not enriched in the previously mapped p75 TSS. There is one DNase accessibility site in MCF7 near the predicted p75 TSS, but it does not have any of the other features of a promoter. These data are all inconsistent with an intron 20 alternative TSS.
Efforts have been made to comprehensively annotate promoters by synthesizing functional genomic datasets and curated databases. One such catalog is the Eukaryotic Promoter Database (EPD), which correctly annotates both exon 1a and 1b but not any promoters within intron 20 (Fig. 6a “EPDnew Promoters” track)34.
We confirmed this observation with next-generation sequencing-based cap-analysis gene expression (CAGE-seq), which captures 5′ capped mRNA transcripts to map TSSs35. CAGE-seq identifies 5′ capped mRNA in 5 different ENCODE cell lines (GM12878, H1-hESC, K562, HepG2 and MCF7) at exons 1a and 1b of CUX1 yet none within intron 20, including in MCF7 (Fig. 6a). Similar results were observed from CAGE-seq data from an atlas of 975 human primary cells and cancer cell lines (Fig. 6b)36. In aggregate, these data do not support an alternative intronic TSS.
The presumptive p75 transcript expresses intronic sequence proximal to exon 2112. We mined RNA-seq datasets to identify reads spanning this region in MCF7 and T-47D human breast cancer cell lines reported to express p756,12,37. All exon 21 sequencing reads abruptly end at the intron 20/exon 21 border, inconsistent with the p75 transcript (Fig. 6c). In the MCF7 cell line, we observed some sequencing reads within the intron 20 region, but these were not conserved across four different replicates and were not contiguous with exon 21 reads37 (Fig. 6c). These data do not support the existence of a p75 mRNA containing intronic sequence.
Finally, we note that no gene assembly or gene prediction database for coding or non-coding RNA annotate a p75 isoform. NCBI Refseq, CCDS, GENCODE, Ensembl gene predictions, AUGUSTUS, or ORFeome all lack a p75 transcript (Fig. S6)38,39,40,41,42. Collectively, these extensive datasets spanning thousands of samples provide no evidence of a p75 CUX1 transcript.
Discussion
Next-generation sequencing and CRISPR/Cas9 genome editing has revolutionized biomedical research in many ways. Perhaps less appreciated, however, is the role of these technologies in establishing the legitimacy of research findings. CRISPR/Cas9 editing, for instance, has invalidated cancer dependencies, drug targets, and viral receptors, as some illustrative examples43,44,45. In this report, we employ the power of functional genomics and CRISPR editing to demonstrate that the CUX1 gene does not encode a p75 isoform as described12. This conclusion is buttressed via multiple orthogonal approaches including biochemical studies with several antibodies, proteomics and extensive mining of functional genomic datasets across a plethora of cell types. Taken together, our data is inconsistent with a p75 CUX1 isoform arising from an intronic TSS and suggests that prior reports were based on a western blotting artefact12.
Other studies support our conclusion. p75 was first identified in HeLa cells, HEK293 cells, breast cancer cell lines and mouse thymus12. The original manuscript identifying human CUX1 generated antiserum against the entire CUX1 protein, yet western blotting of HeLa cells identified only p20046. Probing HEK293 cells with an antibody against the C-terminus region of CUX1 shows no p75 in another study19. In a report that p75 CUX1 causes polycystic kidney disease, the endogenous p75 expression was never documented at the protein level; all subsequent experiments were performed by over-expressing p75 cDNA47. Probing a western blot of entire Drosophila embryos for the highly conserved CUX1 ortholog, Cut, only reveals the full-length protein48. While p200 CUX1 protein increases after TGF-β treatment in normal lung fibroblasts, p75 does not49. Finally, in a study that reported that androgen-resistant prostate cancer cell lines upregulate p200, p75 was unchanged50. Collectively, these studies either fail to document an endogenous p75 protein, or uncouple the biology of p200 from p75.
It is unclear what the p75 cDNA product previously reported represents12. Perhaps it is a result of recursive splicing, where long introns are spliced in a sequential manner in tissue-specific contexts leading to a lag in splicing of intron 20 material51. Alternatively, studies of nascent transcription indicate that splicing does not always occur in the order of transcription, and introns that are spliced later temporally tend to be longer and have higher RNA-binding protein occupancy52. In keeping with this notion, intron 20 is the sixth longest intron in CUX1, and has an elevated RNA-binding protein occupancy compared to most other introns in the gene and thus may be spliced later than other introns in CUX152. It is conceivable that the p75 cDNA product previously observed represents an intermediate, incompletely spliced p200 cDNA.
In our analysis of hematopoietic cells, we only document the expression of the p200 CUX1 isoform. We cannot comment on the validity of p110 or other short isoforms (p80, p90 and p150). As these isoforms are generated post-translationally, functional genomics datasets are ineffective in determining their legitimacy. Future studies of these putative isoforms in other tissue types should employ stringent techniques such as CRISPR/Cas9 mutagenesis to ensure against being misled by western blotting artefacts.
Our finding calls into question studies that ascribed oncogenic functions to p75 CUX1. Many of these publications did not study endogenous p75, but instead employed overexpression models, which can confound results12,18,19,53,54. We speculate that overexpression of CUX1 has dominant negative effects. For instance, in mice, both overexpression of p75 and knockout or knockdown of CUX1 leads to myeloproliferative disease19,24,55. One interpretation of these seemingly incongruent findings is that artificial overexpression of p75 either interferes with the stoichiometry of endogenous CUX1 protein complexes or blocks full-length CUX1 from binding to its target genes. Indeed, p75 CUX1 has increased DNA-binding affinity compared to endogenous p200 CUX156. The net effect of CUX1 overexpression may be the disruption of endogenous CUX1 tumor suppressor activity. In support of this model, the p150 CUX1 isoform was found to exert a dominant negative phenotype upon p200 CUX114. In this light, the use of overexpression systems to characterize p110 may also misattribute this isoform with oncogenic properties16,18,53,57,58.
There are relatively fewer reports that p200 CUX1 is oncogenic. p200 CUX1 has been shown to promote cell line migration, invasion, and evasion of apoptosis17,59. p200 transgenic mice develop organ hyperplasia60. 7q copy number gains and CUX1 overexpression has been documented in primary cancers6,61,62. However, these later findings should be interpreted with caution. Chromosome 7 also encodes oncogenes, including EGFR (on 7p), and BRAF, CDK6, and EZH2 (on 7q). Thus, in cancers with chromosome 7 copy number gains, the driver may be a true oncogene, while CUX1 is a passenger. Indeed, rigorous pan-cancer gene-level analysis of copy number alterations and mutation patterns in primary patient samples reveal CUX1 genetic changes are significantly characteristic of a tumor suppressor gene20,21,22. There is now a growing body of work that CUX1 is tumor suppressive21,23,24,25,27,63.
Given the importance of CUX1 in development and disease across a wide variety of tissue types, it is critical to carefully dissect and understand the genomic structure of the CUX1 locus and encoded protein. The complexity of the gene has led to confusion in the field resulting in serious inaccuracies, most recently by Xu, et. al.64. We expect that our current study will help rectify these obstacles going forward.
Methods
Human cell culture
KG-1, Mono7, ML-2, HL-60, Kasumi-1, and UoCM1 were grown at 37 °C in Gibco RPMI 1640 media supplemented with 20% fetal bovine serum (FBS) and 1X antibiotic–antimycotic (Gibco). K562, U937 and T-47D cell lines were grown in Gibco RPMI 1640 media supplemented with 10% FBS and 1X antibiotic–antimycotic. NIH-3T3 fibroblasts, MCF7 and MDA-MB-231 breast cancer cell lines were grown in Gibco DMEM media supplemented with 10% FBS and 1X antibiotic–antimycotic. All cell lines were verified by STR analysis at the NUSeq Core Facility at Northwestern University.
Human mobilized peripheral blood CD34+ HSPCs from multiple healthy donors were purchased from the Fred Hutchinson Co-operative Center for Excellence in Hematology (Seattle, WA, USA). CD34+ HSPCs were expanded in StemSpan SFEMII base media supplemented with CC110 culture supplement for 1–3 days prior to electroporation.
Generation of CUX1-GFP tagged KG-1 cell line
pCUX1.1.0-gDNA (Addgene plasmid #112434; RRID: Addgene_112434) and pCUX1-donor plasmids (Addgene plasmid #112338; RRID: Addgene_112338) were a gift from Kevin White. The CUX1 homology arms were chemically synthesized by Gibson assembly and comprised 0.6 kb upstream and 1 kb downstream CUX1 sequence flanking the stop codon. The GFP tag is a LAP tag that contains a TEV protease cleavage site, an S peptide, the EGFP coding sequence, followed by an IRES and a kanamycin resistance element65. The LAP-GFP tag was PCR amplified from a separate plasmid, and assembled with the CUX1 donor sequence using Gibson assembly. KG-1 cells were transfected with 0.5 μg of each plasmid using the Neon® Transfection System (Invitrogen by Life Technologies, Waltham, MA, USA). The electroporation settings used for transfection was 1650 V, 20 ms pulse width, 1 pulse, and electroporation was performed according to manufacturer instructions. Cells were cultured in 0.5 mg/ml G418 after 7 days for 3 weeks to select for a transfected population. Primers used to confirm correct integration of the GFP tag are listed below:
-
EGFP 5′ primer: 5′-CATGAAGCAGCACGACTTCT-3′.
-
EGFP 3′ primer: 5′-CTGCTTGTCGGCCATGATATAG-3′.
-
5′ primer spanning homology arm and EGFP: 5′-GGAACCTATCGAATGGGAGTTC-3′.
-
3′ primer spanning homology arm and EGFP: 5′-AAGTCGTGCTGCTTCATGT-3′.
Immunoblots
Cells were lysed in RIPA buffer (50 mM Tris–HCl, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM EDTA and 1% NP-40 substitute). 1% Halt™ Protease Inhibitor Cocktail (Thermo Fisher) was added to the lysis buffer before use. The lysates were passed through a 25-gauge needle, incubated on ice for 20 min, with frequent vortexing, and clarified by centrifugation (5000 g, 10 min at 4 °C). Total protein of the resulting supernatant was quantified using the Bradford assay at 595 nm wavelength, with BSA used to generate the standard curve. 10–15 µg of protein was subjected to SDS-PAGE and probed with anti-CUX1 antibody conjugated to HRP (B-10-HRP, mouse mAb derived against aa 1308–1332, Santa Cruz, 1:1000 in 5% milk/TBST) and visualized using ECL substrate. β-Actin was detected with anti-β-actin-HRP (C4, Santa Cruz, 1:3000 in 5% milk/TBST). Other antibodies used to probe for CUX1 expression include ABE217 (rabbit polyclonal antibody derived against aa 861, 1:1000 in 5% milk/TBST), and PUC (rabbit polyclonal antibody generated in-house that recognizes aa 1223–1242, 1:1000 in 5% milk/TBST). GFP (D5.1) rabbit mAb #2956 (Cell Signaling Technology, Product #2956S, 1:1000 in 5% FBS/TBST) was used to probe for GFP-tagged CUX1. Nitrocellulose membrane (Thermo Scientific, Catalog number: 88018) was used for protein transfer and ECL substrate (Thermo Scientific, Catalog number: 34579) was used for visualizing the proteins of interest.
gRNA design
All gRNAs were designed using the Broad Institute’s sgRNA designer tool, and generated gRNA sequences were verified using Synthego’s Verify Guide Design tool. gRNA sequences used in this study were purchased from Synthego, and the sequences are listed below:
-
CUX1 exon 4 gRNA: 5′-UGCACUGAGUAAAAGAAGCA-3′.
-
CUX1 intron 20 5′ gRNA 1: 5′-GUAUUUCACGAUUCAGCCAA-3′.
-
CUX1 intron 20 3′ gRNA 1: 5′-CUUUGGGUCAUACAUUGGCA-3′.
-
CUX1 intron 20 5′ gRNA 2: 5′-AUGGCACAAAUCCACGCCAC-3′.
-
CUX1 intron 20 3′ gRNA 2: 5′-AUACUAAUUAAACGCUCUGU-3′.
-
CUX1 exon 23.1 (NLS) gRNA: 5′-GCUGUGCCGCCGCUUCAUGU-3′.
-
CUX1 exon 23.2 (HD) gRNA: 5′-CCAGCUGAAGAAACCCCGG-3′.
-
HPRT gRNA: 5′-GCAUUUCUCAGUCCUAACA-3′.
gRNA transfections
gRNA:Cas9 ribonucleoprotein (RNP) complexes were formed by mixing 1.8 μL of each gRNA at 100 μM and 1.5 μL Cas9 nuclease at 20 μM in 15 μL of electroporation buffer R. RNP complexes were incubated at room temperature for 10 min. 2 × 105 KG-1 cells were pelleted and washed twice with PBS. The cells were then suspended in the RNP complex, and electroporation was performed using the Neon® Transfection System (Invitrogen by Life Technologies, Waltham, MA, USA). The optimized electroporation settings used for transfecting the KG-1 cell line was 1700 V, 20 ms pulse width, 1 pulse, and electroporation was performed according to manufacturer instructions. For CD34+ HSPCs, 0.71 uL Cas9 nuclease at 20uM was mixed with 2.39 uL gRNA at 30 uM and 0.9 uL of electroporation buffer T. RNP complexes were formed by incubating for at least 15 min at room temperature. 200,000 CD34+ cells in 8 uL of Buffer T were then added to the RNPs, and 10ul of the mixture electroporated and immediately cultured in SFEMII + CC110. AAVS1 gRNA was used as a negative control. Electroporation settings used for CD34 + HSPCs was 1600 V, 10 ms pulse length, 3 pulses.
To examine the clonality of transfected cell lines, a serial dilution approach was adapted from Corning Life Sciences. Edited single cell clones were also established by sorting single cells into a 96-well plate on the AriaIIIu cell sorter.
PCR confirmation of gRNA editing
The following primers were used to amplify the gRNA cut site from genomic DNA to verify editing efficiency:
-
CUX1 intron 20 5′ primer for first cut site: 5′-AGCGCCCCTGTTTAGTTCTC-3′.
-
CUX1 intron 20 3′ primer for first cut site: 5′-GAGCCACCACGAAACTCAGA-3′.
-
CUX1 intron 20 5′ primer for second cut site: 5′-CCCTCATGTTAAGCCTTCCGA-3′.
-
CUX1 intron 20 3′ primer for second cut site: 5′-CACTGGAACTCATCGGGGAC-3′.
-
CUX1 exon 4 gRNA 5′ primer: 5′-CCCTCCTAGACCCTGAGCTT-3′.
-
CUX1 exon 4 gRNA 3′ primer: 5′-TTCATGTGTCCTGCACTCCC-3′.
-
CUX1 exon 23 gRNA 5′ primer: 5′-GGTGAGGACCTGACATTCCG-3′.
-
CUX1 exon 23 gRNA 3′ primer: 5′-GGACCAAGGAACGGACCAAT-3′.
Reverse-transcriptase PCR
RNA was extracted from 500,000 cells of the K562, Kasumi-1, KG-1, T47D, MDA-MB-231 and MCF-7 cell lines using Trizol, precipitated using chloroform and 70% ethanol, and purified using the RNeasy Mini kit (QIAGEN cat no 74104). 500 ng of RNA from each cell line was used to synthesize cDNA using the Thermo Scientific Maxima™ H Minus cDNA Synthesis Master Mix Kit (Thermo Scientific cat no M1661). 1 µL of this synthesized cDNA was then used in the qPCR reaction. Primers were used specific to the p200 CUX1 isoform, the p75 CUX1 isoform, and GAPDH primers as a housekeeping control. The primers for the p75 transcript were obtained from the paper that originally described the existence of this isoform12. In addition, we also designed our own primers spanning various regions of intron 20 of CUX1. Controls tested include a no template water control, and a no RT enzyme control. 30 cycles of PCR were performed. Primer sequences are listed below:
-
GAPDH forward primer: 5′-ACCACAGTCCATGCCATCAC-3′.
-
GAPDH reverse primer: 5′-TCCACCACCCTGTTGCTGTA-3′.
-
p200 + p75 CUX1 forward primer: 5′-CCGGAGGAGAAGGAGGCGCT-3′.
-
p200 + p75 CUX1 reverse primer: 5′-AGCTGTCGCCCTCCGAGCTG-3′.
-
CUX1 forward primer (primer 1): 5′-CCACTCCGTGACATCGCTC-3′.
-
p75 CUX1 forward primer (primer 2): 5′-CCCTCATGTTAAGCCTTCCGA-3′.
-
p75 CUX1 forward primer (primer 3): 5′-GCTATTTTCAGGCACGGTTTCTC-3′.
-
p75 CUX1 reverse primer (primer 4): 5′-TCCACATTGTTGGGGTCGTTC-3′.
Immunoprecipitation
CUX1 was immunoprecipitated from the KG-1 cell line, and then blotted for CUX1 again to query whether short CUX1 isoforms were able to be pulled down by the anti-CUX1 antibody. 100 × 106 cells were spun down for a CUX1 pulldown and a control IgG pulldown each. Cells were lysed in hypotonic buffer (5 mM EDTA, 5 mM EGTA, 5 mM Tris–Cl) with protease inhibitor added (Roche complete mini-EDTA free). Pellets were passed through a 20-gauge needle and incubated on ice, then spun down. The supernatant was removed, and the pellet was resuspended in RIPA buffer with protease inhibitor added (Roche Complete). Pellets were again passed through a 27-gauge needle, incubated on ice and subsequently spun down. The supernatant was collected, and then incubated overnight at 4 °C on a rocker with either 12 µg of the anti-CUX1 antibody (B-10, Santa Cruz) or a rabbit IgG antibody. Protein A/G beads (Santa Cruz) were then added the following day to the supernatant, and incubated at 4 °C on a rocker for 1 h. The immunoprecipitated protein was then spun down, washed in cold PBS, resuspended in loading buffer, and subjected to SDS-PAGE.
Sample preparation for LC–MS/MS
For the whole cell lysate samples, 20 μg of whole cell extract from KG-1 cells (determined by Bradford assay, Thermo #1856209 using λ595 nm) was loaded onto a 4–12% MOPS buffered 1D SDS-PAGE gel (Invitrogen NP0336BOX) and run at ~ 200 V for ~ 45 min. For the IP samples, 30% of the IP eluate (30 μL/100 μL) was loaded. The gel was stained with Imperial Stain (Thermo #24615) for 1 h at room temperature. For the whole cell lysate samples, the “p200” sections were excised from the gel by sterile razorblade (MW range 150–225 kDa) and the “p75” sections were excised in the MW range of ~ 60–75 kDa and in-gel trypsin digested as described below. For the IP samples, gel sections were excised as follows: “p200” sections MW range (~ 150–250 kDa) and “p75” sections MW range (~ 65–90 kDa) and in-gel trypsin digested as described below.
Trypsin digestion
Trypsin digestion was adapted from methods previously published65,66. In brief, gel sections were washed in dH2O and destained overnight using 100 mM NH4HCO3 (Sigma #285099) pH 7.5 in 50% acetonitrile (Fisher A998SK-4). A reduction step was performed by addition of 100 μL 50 mM NH4HCO3 pH 7.5 and 10 μL of 200 mM tris(2-carboxyethyl) phosphine HCl (Sigma #C4706-2G) at 37 °C for 30 min. The proteins were alkylated by addition of 100 μL of 50 mM iodoacetamide (Sigma #RPN6320V) prepared fresh in 50 mM NH4HCO3 pH 7.5 buffer and allowed to react in the dark at 20 °C for 30 min. Gel sections were washed in Millipore water, then acetonitrile, and vacuum dried. Trypsin digestion was carried out overnight at 37 °C with 1:50–1:100 enzyme–protein ratio of sequencing grade-modified trypsin (Promega #V5111) in 50 mM NH4HCO3 pH 7.5, and 20 mM CaCl2 (Sigma #C-1016). Peptides were extracted with 5% formic acid (Sigma #F0507-1L) in aqueous and 75% organic (ACN) combined and vacuum dried. Peptides were cleaned up using C18 spin columns (Thermo #89870) and sent to the Mayo Clinic Medical Genome Facility Proteomics Core for HPLC and LC–MS/MS data acquisition via Q-Exactive Orbitrap (Thermo).
LC–MS/MS via MaxQuant
LC–MS/MS was performed using adapted methods previously published67. In brief, peptide samples were re-suspended in Burdick & Jackson HPLC-grade water containing 0.2% formic acid (Fluka #60-006-17), 0.1% TFA (Pierce #28903), and 0.002% Zwittergent 3–16 (Millipore Sigma #693023), a sulfobetaine detergent that contributes the following distinct peaks at the end of chromatograms: MH+ at 392, and in-source dimer [2 M+ H+] at 783, and some minor impurities of Zwittergent 3–12 seen as MH+ at 336. The peptide samples were loaded onto a 100 μm × 40 cm PicoFrit column self-packed with 2.7 μm Agilent Poroshell 120, EC-C18, washed, then switched in-line with a 0.33 uL Optimize EXP2 Stem Traps, packed spray tip nano column packed with Halo 2.7 μm Pep ES-C18, for a 2-step gradient. Mobile phase A was water/acetonitrile/formic acid (98/2/0.2) and mobile phase B was acetonitrile/isopropanol/water/formic acid (80/10/10/0.2). Using a flow rate of 350 nL/min, a 90 min, 2-step LC gradient was run from 5% B to 50% B in 60 min, followed by 50–95% B over the next 10 min, hold 10 min at 95% B, back to starting conditions and re-equilibrated.
Electrospray tandem mass spectrometry (LC–MS/MS) was performed at the Mayo Clinic Proteomics Core on a Thermo Q-Exactive Orbitrap mass spectrometer, using a 70,000 RP (70 K Resolving Power at 400 Da) survey scan in profile mode, m/z 340–1800 Da, with lockmasses, followed by 20 MSMS HCD fragmentation scans at 17,500 resolution on doubly and triply charged precursors. Single charged ions were excluded, and ions selected for MS/MS were placed on an exclusion list for 60 s. An inclusion list (generated with in-house software) consisting of expected Cux1 sequences was used during the LC–MS/MS runs.
Database searching
Tandem mass spectra MS/MS samples were analyzed using MaxQuant (Max Planck Institute of Biochemistry, Martinsried, Germany; version 1.6.17.0). MaxQuant was set up to search the 210308_SPROT_Human_UP5640.fasta database assuming the digestion enzyme strict trypsin. MaxQuant was searched with a fragment ion mass tolerance of 20 PPM and a parent ion tolerance of 20 PPM. Maxquant 1FDR results files were processed in Perseus (version 1.6.14.0) for the proteingroups.txt in addition to being imported into Scaffold version 5.0.1.
Criteria for protein identification
Scaffold (version Scaffold_5.0.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted at 1% FDR by the Peptide Prophet algorithm (Keller et al. Anal. Chem. 2002;74(20):5383–5392) with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at 1% FDR and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm. Proteins sharing significant peptide evidence were grouped into clusters.
Data availability
The mass spectrometry proteomic data sets (MK1) and (MK3) were uploaded to the ProteomeXchange consortium via the PRIDE partner repository with the dataset identifier PXD027527.
References
Stastna, M. & Van Eyk, J. E. Analysis of protein isoforms: Can we do it better?. Proteomics 12(19–20), 2937–2948 (2012).
Lievens, P. M. J., Tufarelli, C., Donady, J. J., Stagg, A. & Neufeld, E. J. CASP, a novel, highly conserved alternative-splicing product of the CDP/cut/cux gene, lacks cut-repeat and homeo DNA-binding domains, and interacts with full-length CDP in vitro. Gene 197(1–2), 73–81 (1997).
Bürglin, T. R. & Cassata, G. Loss and gain of domains during evolution of cut superclass homeobox genes. Int. J. Dev. Biol. 46(1), 115–123 (2002).
Malsam, J., Satoh, A., Pelletier, L. & Warren, G. Golgin tethers define subpopulations of COPI vesicles. Science 307(5712), 1095–1098 (2005).
The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407), 330–337 (2012).
Ramdzan, Z. M. & Nepveu, A. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat. Rev. Cancer 14(10), 673–682 (2014).
Gillingham, A. K., Pfeifer, A. C. & Munro, S. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol. Biol. Cell. 13(11), 3761–3774 (2002).
Lowe, M. The physiological functions of the Golgin vesicle tethering proteins. Front. Cell Dev. Biol. 7, 94 (2019).
Sansregret, L., & Nepveu, A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene 412(1–2), 84–94 (2008).
Goulet, B. et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/CUX transcription factor. Mol. Cell 14(2), 207–219 (2004).
Moon, N. S. et al. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 21(18), 6332–6345 (2001).
Goulet, B. et al. Characterization of a tissue-specific CDP/CUX isoform, p75, activated in breast tumor cells. Can. Res. 62, 6625–6633 (2002).
Truscott, M. et al. Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J. Biol. Chem. 282(41), 30216–30226 (2007).
Maitra, U., Seo, J., Lozano, M. M. & Dudley, J. P. Differentiation-induced cleavage of Cutl1/CDP generates a novel dominant-negative isoform that regulates mammary gene expression. Mol. Cell. Biol. 26(20), 7466–7478 (2006).
Goulet, B., Truscott, M. & Nepveu, A. A novel proteolytically processed CDP/CUX isoform of 90 kDa is generated by cathepsin L. Biol. Chem. 387(9), 1285–1293 (2006).
Kedinger, V. et al. p110 CUX1 homeodomain protein stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E-cadherin and occludin. J. Biol. Chem. 284(40), 27701–27711 (2009).
Michl, P. et al. CUTL1 is a target of TGFβ signaling that enhances cancer cell motility and invasiveness. Cancer Cell 7(6), 521–532 (2005).
Cadieux, C., Yao, L., Vadnais, C., Drossos, M. & Nepveu, A. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Can. Res. 18, 7188–7198 (2009).
Cadieux, C. et al. Transgenic mice expressing the p75 CCAAT-displacement protein/cut homeobox isoform develop a myeloproliferative disease-like myeloid leukemia. Can. Res. 66(19), 9492–9501 (2006).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155(4), 948–962 (2013).
Wong, C. C. et al. Inactivating CUX1 mutations promote tumorigenesis. Nat. Genet. 46(1), 33–38 (2014).
Gao J., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling, 6(269), (2013).
McNerney, M. E. et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood 121(6), 975–983 (2013).
An, N. et al. Gene dosage effect of CUX1 in a murine model disrupts HSC homeostasis and controls the severity and mortality of MDS. Blood 131(24), 2682–2697 (2018).
Supper, E. et al. Cut-like homeobox 1 (CUX1) tumor suppressor gene haploinsufficiency induces apoptosis evasion to sustain myeloid leukemia. Nat. Commun. 12, 2482 (2021).
Goulet, B. et al. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol. Cancer Res. 5, 899–908 (2007).
Imgruet, M. K. et al. Loss of a 7q gene, CUX1, disrupts epigenetic-driven DNA repair and drives therapy-related myeloid neoplasms. Blood 138, 790–805 (2021).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473(7345), 43–49 (2011).
Ernst, J. & Kellis, M. ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods. 9, 215–216 (2012).
Weintraub, H. & Groudine, M. Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976).
Karabacak Calviello, A., Hirsekorn, A., Wurmus, R., Yusuf, D., & Ohler, U. Reproducible inference of transcription factor footprints in ATAC-seq and DNase-seq datasets via protocol-specific bias modeling. Preprint at bioRxiv, 284364 (2018).
Gardiner-Garden, M. & Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 196(2), 261–282 (1987).
Ioshikhes, I. P. & Zhang, M. Q. Large-scale human promoter mapping using CpG islands. Nat. Genet. 26(1), 61–63 (2000).
Dreos, R., Ambrosini, G., Perier, R. C. & Bucher, P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 41(D1), D157–D164 (2013).
Shiraki, T. et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U.S.A. 100(26), 15776–15781 (2003).
The FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456(7221), 470–476 (2008).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, 501–504 (2005).
Pruitt, K. D. et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 19(8), 1506 (2009).
Harrow, J. et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22(9), 1760–1774 (2012).
Hubbard, T. et al. The Ensembl genome database project. Nucleic Acids Res. 30(1), 38–41 (2002).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5), 637–644 (2008).
Lin, A., Giuliano, C. J., Sayles, N. M., & Sheltzer, J. M. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. eLife, 6, 1–17 (2017).
Lin, A., et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 11(509) (2019).
Chen, J. et al. Ephrin receptor A2 is a functional entry receptor for Epstein–Barr virus. Nat. Microbiol. 3(2), 172–180 (2018).
Neufeld, E. J., Skalnik, D. G., Lievens, P. M. & Orkin, S. H. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat. Genet. 1, 50–55 (1992).
Cadieux, C. et al. Polycystic kidneys caused by sustained expression of Cux1 isoform p75. J. Biol. Chem. 283(20), 13817–13824 (2008).
Blochlinger, K., Bodmer, R., Jan, L. Y. & Jan, Y. N. Patterns of expression of Cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4(8), 1322–1331 (1990).
Ikeda, T. et al. Transforming growth factor-β-induced CUX1 isoforms are associated with fibrosis in systemic sclerosis lung fibroblasts. Biochem. Biophys. Rep. 7, 246–252 (2016).
Dorris, E. R. et al. The transcription factor CUX1 negatively regulates invasion in castrate resistant prostate cancer. Oncotarget 11(9), 846–857 (2020).
Sibley, C. R. et al. Recursive splicing in long vertebrate genes. Nature 521(7552), 371–375 (2015).
Drexler, H. L., Choquet, K. & Churchman, L. S. Splicing kinetics and coordination revealed by direct nascent RNA sequencing through nanopores. Mol. Cell 77(5), 985-998.e8 (2020).
Vadnais, C. et al. Autocrine activation of the Wnt/β-Catenin pathway by CUX1 and GLIS1 in breast cancers. Biol. Open 3(10), 937–946 (2014).
Siam, R. et al. Transcriptional activation of the LatsI tumor suppressor gene in tumors of CUX1 transgenic mice. Mol. Cancer 8, 60 (2009).
Sinclair, A. M. et al. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 98(13), 3658–3667 (2001).
Moon, N. S., Bérubé, G. & Nepveu, A. CCAAT displacement activity involves CUT repeats 1 and 2, not the CUT homeodomain. J. Biol. Chem. 275(40), 31325–31334 (2000).
Sansregret, L. et al. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol. Cell. Biol. 26(6), 2441–2455 (2006).
Harada, R. et al. Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. 36(1), 189–202 (2008).
Ripka, S. et al. CUX1: Target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut 59(8), 1101–1110 (2010).
Ledford, A. W. et al. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27kip1 expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev. Biol. 245(1), 157–171 (2002).
Franch-Expósito, S., et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. eLife, 9, 1–22 (2020).
Wilson, M. A. et al. Copy number changes are associated with response to treatment with carboplatin, paclitaxel, and sorafenib in melanoma. Clin. Cancer Res. 22(2), 374–382 (2016).
Aly, M. et al. Distinct clinical and biological implications of CUX1 in myeloid neoplasms. Blood Adv. 3(14), 2164–2178 (2019).
Xu, A. et al. Overexpressed P75CUX1 promotes EMT in glioma infiltration by activating β-catenin. Cell Death Dis. 12(2), 1–15 (2021).
Poser, I. et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 (2008).
Truman, A. W. et al. The quantitative changes in the yeast Hsp70 and Hsp90 interactomes upon DNA damage. Data Brief 2, 12–15 (2014).
Wolfgeher D, et al. The dynamic interactome of human Aha1 upon Y223 phosphorylation. Data Brief 5, 752–755 (2015).
Lyon, S. M. et al. A method for whole protein isolation from human cranial bone. Anal. Biochem. 515, 33–39 (2016).
Acknowledgements
The authors thank Dr. Angela Stoddart for critical reading of the manuscript. The authors are grateful for the services and assistance provided by the following University of Chicago core facilities supported by the Cancer Center Support Grant (P30 CA014599): Cytometry and Antibody Technology (CAT) Facility (RRID: SCR_017760) and the Genomics Facility. We would like to thank Dr. Jennifer Moran from the Institute for Genomics and Systems Biology for providing the pCUX1-donor plasmid. We would also like to acknowledge the Mayo Clinic Medical Genome Facility Proteomics Core for generating the proteomics data and the work of mass spectrometrist Ken Johnson for assisting with instrument method setup and data generation. This work was supported in part by an American Cancer Society Research Scholar Grant (M.E.M), an American Society of Hematology Junior Faculty Scholar Award (M.E.M), the National Institutes of Health (HL142782 and CA231880, M.E.M.), a Department of Defense Congressionally Directed Medical Research Program PRCRP Impact Award (W81XWH-20-1-0556, M.E.M, S.J.K.), the Brinson Foundation (M.E.M.), and The University of Chicago Cancer Research Foundation Women’s Board (M.K., M.E.M.).
Funding
Funding was provided by DOD Peer Reviewed Cancer Research Program, American Cancer Society, American Society of Hematology and National Institutes of Health and Brinson Foundation.
Author information
Authors and Affiliations
Contributions
M.K. and M.E.M. conceived of the study. M.K., M.D.S., J.T.B., and D.J.W. performed experiments. S.K. contributed to experiments. M.K. analyzed data. D.J.W. analyzed the proteomics data. S.J.K. and M.E.M. supervised the study. M.K. and M.E.M. wrote the manuscript, and M.D.S., J.T.B., D.J.W. and S.J.K. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.J.K. is a founder of OncoSenescence, Transnostics and Cell IDx. The other authors declare no conflicting interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krishnan, M., Senagolage, M.D., Baeten, J.T. et al. Genomic studies controvert the existence of the CUX1 p75 isoform. Sci Rep 12, 151 (2022). https://doi.org/10.1038/s41598-021-03930-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-03930-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.