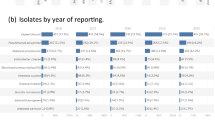
Abstract
Antimicrobial resistance (AMR) surveillance in bloodstream infections (BSIs) is challenging in low/middle-income countries (LMICs) given limited laboratory capacity. Other specimens are easier to collect and process and are more likely to be culture-positive. In 8102 E. coli BSIs, 322,087 E. coli urinary tract infections, 6952 S. aureus BSIs and 112,074 S. aureus non-sterile site cultures from Oxfordshire (1998–2018), and other (55,296 isolates) rarer commensal opportunistic pathogens, antibiotic resistance trends over time in blood were strongly associated with those in other specimens (maximum cross-correlation per drug 0.51–0.99). Resistance prevalence was congruent across drug-years for each species (276/312 (88%) species-drug-years with prevalence within ± 10% between blood/other isolates). Results were similar across multiple countries in high/middle/low income-settings in the independent ATLAS dataset (103,559 isolates, 2004–2017) and three further LMIC hospitals/programmes (6154 isolates, 2008–2019). AMR in commensal opportunistic pathogens cultured from BSIs is strongly associated with AMR in commensal opportunistic pathogens cultured from non-sterile sites over calendar time, suggesting the latter could be used as an effective proxy for AMR surveillance in BSIs.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) is among the top ten global health threats1, and is a particularly acute challenge in low- to middle-income countries (LMICs)2,3,4. Surveillance is a key tool to combat AMR, particularly in LMICs where lack of laboratory capacity prevents routine patient-level antimicrobial susceptibility testing5. The lack of surveillance data contributes to a pragmatic but broad-spectrum empiric treatment approach in hospitals with limited laboratory facilities2,6, generally resulting in overtreatment and acting as a selective pressure for AMR7. To date, surveillance capacity-building programmes have generally focused on implementing blood culture surveillance due to the high mortality of bloodstream infections (BSIs); however, blood cultures are a comparatively costly sample to process, require specialised laboratory equipment and trained staff due to the invasive nature of blood sampling, and can have slow turnaround times, reducing their utility for both clinical management and surveillance8. Additionally, the low positivity rate means that very high sample throughput is needed to confidently estimate AMR prevalence. Consequently, empirical treatment guidelines currently remain largely uninformed by AMR estimates from BSI surveillance in LMICs.
One solution to improve surveillance could be to assess AMR rates using data from non-invasive samples (e.g. urine, wound swabs), which are easier and faster to collect in large numbers, and easier to process in fledgling laboratories. Reflecting this, the 2020 WHO GLASS report includes AMR prevalence in urine cultures for key Gram-negative pathogens. However, associations between AMR rates in blood and other specimens have been studied in only a small number of longitudinal studies. Data from South Korea, the US and the UK have observed similar resistance rates in blood cultures and non-blood specimens (urine, faeces, skin/screening samples) for Escherichia coli, non-typhoidal Salmonella enterica and methicillin-resistant Staphylococcus aureus9,10,11,12. This raises the broader question as to whether, for multiple commensal opportunistic pathogen-drug combinations, the proportion of resistant BSIs could be estimated from the proportion of resistant isolates cultured from other body-sites consistently over calendar time (i.e. resistance trends), improving the power, feasibility and cost of culture-based AMR surveillance, and informing the development of locally empiric treatment guidelines13.
To evaluate this hypothesis, we first investigated the annual prevalence of and associations between AMR in commensal opportunistic pathogens isolated from BSIs and other body-sites for multiple pathogen-drug combinations in Oxfordshire [regional analysis, high-income country (HIC)]. We then extended the analysis, first to consider concurrent or previous isolates in the same patient, and second to consider the relevance of this approach in other world regions through publicly available datasets [Antimicrobial Testing Leadership and Surveillance (ATLAS)14] and collaborating hospitals/programmes in LMICs.
Results
AMR prevalence across antimicrobial classes in E. coli UTIs is strongly associated with AMR prevalence in E. coli BSIs in Oxfordshire, UK
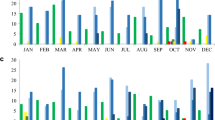
We focused initially on E. coli as the most common bacteria causing BSIs and UTIs2,15,16. We included 8,102 E. coli BSIs and 322,087 E. coli UTIs (40-fold higher) from Oxfordshire (Supplementary Fig. S1); four antibiotics (amoxicillin, co-amoxiclav, ciprofloxacin, and trimethoprim) were consistently tested in both between 1998 and 2018. Whilst absolute resistance prevalence was generally slightly higher in bloodstream isolates, trends in the proportion of resistant BSIs and UTIs were very similar, notably tracking substantial fluctuations over time in trimethoprim resistance (Fig. 1, Supplementary Fig. S2, Supplementary Table S1), with resistance rates in the same year most strongly correlated (i.e. maximum cross-correlation at lag 0; Supplementary Table S1). For these four antibiotics, there was no evidence of strong variation in the relationship between resistance rates over time.
Resistance prevalence in E. coli and S. aureus in blood versus non-blood cultures in Oxfordshire, UK, 1998–2018 maximum cross-correlation at time lag 0 in 3/4 drugs (ciprofloxacin, co-amoxiclav, trimethoprim) for E. coli and 4/6 drugs for S. aureus (ciprofloxacin, erythromycin, gentamicin, oxacillin); cross-correlation 0.77 at lag 1 (0.75 at lag 0), 0.95 at lag 0, 0.96 at lag 0, 0.80 at lag 0 for E. coli and 0.95 at lag 0, 0.93 at lag 0, 0.54 at lag 0, 0.75 at lag 0, 0.69 at lag − 4, 0.60 at lag 3 for S. aureus from top to bottom respectively (full results in Supplementary Table S1).
For a further eight antibiotics (aztreonam, ceftazidime, ceftriaxone, co-trimoxazole, ertapenem, gentamicin, meropenem and piperacillin-tazobactam) consistently tested between 2013 and 2018, again, resistance rates were slightly higher in BSIs and showed reasonable concordance with resistance in UTIs (Supplementary Fig. S3, Supplementary Tables S1, S2), with prevalences within 10% of each other in 109/132 (89%) drug-years (Fig. 2, left-hand panel, Table 1). Overall for E. coli, resistance prevalence in BSIs was highly correlated with resistance prevalence in UTI (CCC = 0.93 (95% CI 0.91–0.95) (Table 1). Agreement was also relatively high across our four pre-defined resistance prevalence categories which might affect empirical prescribing practice, with 83/132 (63%) drug-years in the same prevalence category and 45/132 (34%) in adjacent prevalence categories (Fig. 2, right-hand panel).
Association between resistance prevalence in blood versus non-blood cultures for (a) E. coli, (b) Klebsiella spp., (c) S. aureus, (d) S. pneumoniae, (e) E. faecalis and E. faecium, Oxfordshire 1998–2018 Left: Scatterplot of the proportion of resistant non-blood cultures versus the proportion of resistant blood cultures per year for all antibiotics. Each dot reflects resistance prevalence for one antibiotic in one specific year, coloured by antibiotic type. Line of identity shown in blue, with associated ± 5% and ± 10% agreement discordance intervals. Additional lines are drawn to denote 0.05 (5%), 0.1 (10%) and 0.2 (20%) resistance prevalence thresholds for blood (horizontal lines) versus non-blood (vertical lines) cultures for ease of visualisation of agreement between clinically meaningful resistance categories in these two types of samples. Right: Stacked bars representing the data in the left-hand panel grouped as categorical resistance prevalence ranges, classifying resistance prevalence in blood cultures by the proportion in urine/non-sterile site (i.e. non-blood) cultures.
Considering matched pairs of isolates from individual patients, there was no evidence that the modestly different resistance rates seen in E. coli BSIs and UTIs overall were due to differential sampling of intrinsically different underlying populations, given high but similarly imperfect agreement in susceptibility (86–100% across 11 antibiotics for concurrent infections, 80–94% for previous infections, Table S3). In particular, considering the closest prior E. coli UTI 3–90 days before a BSI, > 80% UTIs resistant to amoxicillin, co-amoxiclav, ciprofloxacin, and trimethoprim shared the same resistance phenotype in the subsequent BSI.
AMR prevalence across antimicrobial classes in S. aureus from sterile and non-sterile sites is strongly associated with AMR prevalence in S. aureus BSIs in Oxfordshire, UK
Next we considered similar analyses for S. aureus, as one of the commonest causes of Gram-positive BSI and skin/soft tissue infections. Between 1998 and 2018, there were 6952 S. aureus BSIs and 166,179 S. aureus non-blood cultures in Oxfordshire: 54,105 (33%) from sterile sites and 112,074 (67%) from non-sterile sites (8- and 16-fold higher respectively, Supplementary Fig. S1). Resistance trends for ciprofloxacin, erythromycin and oxacillin were similar for the different sample types, and varied substantially over time (Fig. 1, Supplementary Table S1, Supplementary Fig. S4), again supporting the possibility of using resistance prevalence in non-blood cultures as a proxy for resistance prevalence in blood cultures. Agreement in resistance prevalence between blood/non-blood samples was poorer for gentamicin and tetracycline resistance, although resistance rates were low (< 15%). As might be expected, given the propensity of S. aureus to cause purulent metastatic infection in association with BSI, agreement was slightly better between resistance prevalence in blood and other sterile site cultures than blood and non-sterile site cultures (Supplementary Fig. S5).
As non-sterile site cultures are easier to collect and would be more likely to be taken in settings with limited healthcare capacity, and agreement with resistance prevalence in BSIs was reasonable, we focussed on this group of isolates. Overall, resistance prevalence was more similar in bloodstream and other non-sterile infection sites in S. aureus than E. coli (Supplementary Tables S1, S2), but there were also slightly stronger effects of calendar time for S. aureus, reflecting generally higher rates of resistance in blood compared with non-sterile site cultures during the MRSA epidemic in the mid-2000s, which subsequently became more similar (Supplementary Fig. S3).
For S. aureus, including nine other antibiotics (amoxicillin, clindamycin, co-trimoxazole, daptomycin, fusidic acid, linezolid, moxifloxacin, mupirocin, teicoplanin) tested between 2013 and 2018 only, resistance prevalences in blood and non-sterile site cultures were within 10% of each other in 167/180 (93%) drug-years, with 132 (73%) drug-years in the same resistance category and 45 (25%) in adjacent categories (Fig. 2, Table 1). Overall resistance prevalence in blood was highly correlated with resistance prevalence in non-sterile isolates [CCC = 0.97 (0.95–0.97)] (Table 1).
Within individual patients, overall agreement between resistance in concurrent blood and non-sterile cultures was extremely high (96–100%) for S. aureus, with ciprofloxacin, erythromycin and oxacillin having < 5% of resistant blood cultures with susceptible non-sterile cultures and < 4% susceptible blood cultures with resistant non-sterile sites cultures (Supplementary Table S3).
AMR prevalence across antimicrobial classes in Klebsiella spp., Streptococcus pneumoniae and Enterococcus spp. from blood and non-blood specimens is also strongly associated in Oxfordshire, UK
Although numbers were smaller than for E. coli and S. aureus, broadly similar associations in resistance prevalence between blood/non-blood isolates were seen for Klebsiella spp. (2536 BSIs and 24,192 non-blood cultures) (Fig. 2, Table 1, Supplementary Figs. S6, S7, Supplementary Tables S1, S2), and several additional common Gram-positive pathogens, including S. pneumoniae (1703 BSIs and 7369 non-blood cultures), E. faecalis (1218 BSIs and 15,317 non-blood cultures) and E. faecium (934 BSIs and 2027 non-blood cultures) (Fig. 2, Table 1, Supplementary Fig. S8, Supplementary Tables S1, S2). For Klebsiella spp. resistance prevalences in blood and non-blood isolates were within 10% of each other in 79/80 (99%) drug-years, with 51 (64%) drug-years in the same resistance category and 26 (32%) in adjacent categories (Fig. 2, Table 1); for S. pneumoniae 79/81 (98%), 63 (78%), 15 (19%), respectively; for E. faecalis 35/36 (97%), 34 (94%), 2 (6%), respectively; for E. faecium 9/12 (75%) and with all 12 drug-years in the same resistance category.
AMR prevalence in blood and non-blood sites across antimicrobial classes and common pathogens is also strongly associated within 17 countries in the ATLAS dataset
In the ATLAS dataset, there were 401 and 155 country-drug-years with > 100 E. coli and S. aureus isolates from HICs, respectively, from both bloodstream and urinary/non-sterile infections (Supplementary Table S4). For E. coli, time trends could be estimated over 7 years or more in four countries for 11 antibiotics, and were broadly similar (Fig. 3; S. aureus in Supplementary Fig. S9). Across all drugs, prevalence of resistance in E. coli BSIs and UTIs were within 10% of each other in 376/401 (94%) country-drug-years (Fig. 4, left-hand panel, Table 1); 326 (81%) country-drug-years were in the same resistance category and 74 (18%) in adjacent categories (Fig. 4, right column). For S. aureus, respective figures were 147/155 (95%), 154 (99%) and 1 (1%). Agreement was similarly high in middle-income countries where smaller numbers of isolates were available (Fig. 4, Table 1, Supplementary Figs. S10, S11), for E. coli 213/268 (80%), 234 (87%), 31 (12%), respectively, for S. aureus 308/344 (90%), 289 (84%), 47 (14%), respectively; and also in other pathogens (Table 1, Supplementary Figs. S12–S16), for K. pneumoniae 146/228 (64%), 170 (75%), 53 (23%), respectively, for P. aeruginosa 31/36 (86%), 32 (89%), 4 (11%), respectively, and for E. cloacae 45/61 (74%), 49 (80%), 10 (16%) respectively,.
Association between resistance prevalence in blood versus non-blood cultures for: (a) E. coli, (b) S. aureus, in the ATLAS dataset, split into high-income countries and middle-income countries. Left: Scatterplot of the proportion of resistant non-blood cultures versus the proportion of resistant blood cultures per year for all antibiotics. Each dot reflects resistance prevalence for one antibiotic in one specific year, coloured by antibiotic type. Line of identity shown in blue, with associated ± 5% and ± 10% agreement discordance intervals. Additional lines are drawn to denote 0.05 (5%), 0.1 (10%) and 0.2 (20%) resistance prevalence thresholds for blood (horizontal lines) versus non-blood (vertical lines) cultures for ease of visualisation of agreement between clinically meaningful resistance categories in these two types of samples. Right: Stacked bars representing the data in the left-hand panel grouped as categorical resistance prevalence ranges, classifying resistance prevalence in blood cultures by the proportion in urine/non-sterile site (i.e. non-blood) cultures.
AMR prevalence in blood and non-blood sites across antimicrobial classes and common commensal opportunistic pathogens is also associated within three LMICs
Considering hospital/programme-level LMIC datasets, numbers were smaller with 6154 isolates in total only (Supplementary Table S5 contains the minimum and maximum number of blood and non-blood cultures per year per pathogen for each dataset, minimum 5, maximum 327). Estimates were therefore more variable, but both time trends (Fig. 5, Supplementary Fig. S17) and comparisons of individual country-drug-years (Fig. 6, Table 1, Supplementary Fig. S18) supported overall findings that resistance profiles in non-blood culture isolates correlate well with those in blood culture isolates over time. For the datasets from Cambodia, Laos and Thailand, E. coli resistance prevalences in blood and non-blood isolates were within 10% of each other in 27/70 (39%), 10/20 (50%) and 77/120 (64%) drug-years respectively, with 63/70 (90%), 20/20 (100%), 77/120 (64%) drug-years respectively in the same resistance category and 4/70 (6%), 0/20 (0%), 34/120 (28%) in adjacent categories (Fig. 6, Table 1). For S. aureus, for Cambodia respective figures were 39/48 (81%), 30 (62%), 15 (31%), for Laos 8/10 (80%), 7 (70%), 3 (30%), and for Thailand 72/84 (86%), 55 (65%) and 21 (25%) respectively.
Association between resistance prevalence in E. coli blood versus urine cultures in three hospitals/programmes in LMICs. Left: Scatterplot of the proportion of resistant non-blood cultures versus the proportion of resistant blood cultures per year for all antibiotics. Each dot reflects resistance prevalence for one antibiotic in one specific year, coloured by antibiotic type, with 21 dots per antibiotic (one for each year 1998–2018). Line of identity shown in blue, with associated ± 5% and ± 10% agreement discordance intervals. Additional lines are drawn to denote 0.05 (5%), 0.1 (10%) and 0.2 (20%) resistance prevalence thresholds for blood (horizontal lines) versus non-blood (vertical lines) cultures for ease of visualisation of agreement between clinically meaningful resistance categories in these two types of samples. Right: Stacked bars representing the data in the left-hand panel grouped as categorical resistance prevalence ranges, classifying resistance prevalence in blood cultures by the proportion in urine/non-sterile site (i.e. non-blood) cultures.
Discussion
Here, we show that across countries and World Bank income status (i.e. high, middle, low income status), AMR prevalence in commensal opportunistic pathogens cultured from non-blood samples could potentially be used as a proxy for AMR prevalence in BSIs within a given setting. Focusing on two of the most common bacteria causing serious infections, E. coli and S. aureus, we found that, whilst not perfect, agreement between AMR rates in blood cultures and in other clinical cultures was good for most antibiotics, and, importantly, consistently associated over time. Similar results were observed for Klebsiella spp., S. pneumoniae, E. faecalis, E. faecium, P. aeruginosa and E. cloacae. This could be of particular relevance to reducing the costs and logistics of AMR surveillance in LMICs, and improving the speed with which AMR prevalence data can be generated, given that urine and skin swab samples are easy to collect and more straightforward to culture than blood, with higher positivity rates. Reliable local AMR prevalence data could then be used to better inform the appropriateness of different empirical antimicrobial guidelines.
Whilst many studies estimate resistance prevalence in various infection sites, to our knowledge, very few studies have directly compared resistance in blood versus non-blood sites, and these essentially focus only specific drug-pathogen combinations9,11,12. In 2011, Health Protection Scotland and National Services Scotland published a protocol for the surveillance of AMR in UTIs, arguing that it would be more relevant and feasible to monitor emerging multidrug resistant strains and changes in proportions of resistance in UTIs than in BSIs17. However, the hypothesised relationship was not analysed using the data collected18. The WHO piloted a similar approach but did not compare with resistance rates in BSIs19. The use of metagenomics on pooled colonisation samples as a proxy measure for AMR prevalence in populations has also been proposed as a strategy for rapidly generating reliable AMR prevalence estimates to inform prescribing guidelines20.
Current efforts on AMR surveillance in LMICs are directed towards building laboratories and training staff to carry out antimicrobial susceptibility testing on blood cultures, arguing that if this can be achieved, other sample cultures will become possible too. However, setting up blood culture processing is very expensive, both in terms of infrastructural and running costs, and because the microbiological yield from blood cultures is low, it will be many years until sufficient data on AMR prevalence can be accumulated for surveillance purposes. Incorporating blood culture testing in public hospitals may also be significantly biased by differences in the ability of patients to pay2. Longitudinal surveillance data on AMR prevalence in BSIs is available at a limited number of research sites, but these programmes cover a limited geographic area and may not be representative of other locations in a country.
Collecting specimens from urine/skin surface/genital sites is cheaper and less invasive than sampling blood, and therefore substantially easier to do on large numbers from different communities, as well as having higher positivity rates, increasing the utility of microbiological sampling using these sample types for AMR surveillance. This is true even in HICs, as seen in this study, with much narrower confidence intervals around AMR prevalence estimates derived from much larger numbers of non-blood Oxfordshire isolates. Even allowing for the greater manual processing needed for non-blood cultures, yield in terms of resistance antibiograms is almost certainly far greater from the same number of non-sterile specimens compared to blood specimens. Whilst the specific positivity rates and proportion of pathogens from each species will vary across settings, as an example based on Oxfordshire data 1998–2016, obtaining 100 E. coli isolates would only require the testing of 361 urine samples (assuming 37% are culture-positive and 75% of these are E. coli), but 7693 blood cultures (assuming 13% are positive and 10% of these are E. coli.
A major strength of our study is the different datasets used and the different commensal opportunistic pathogens studied (with varying AMR prevalence), and the robustness of findings across these, including detailed, continuous surveillance of one large region (Oxfordshire), a global analysis across 17 countries (ATLAS), and specific evaluations in three LMICs—although the datasets in these three countries were relatively small. A crucial underpinning assumption is that pathogens causing non-bloodstream infections are representative of pathogens causing BSI in terms of antibiogram, either because the source of infection is commonly from commensal colonising organisms that become pathogenic opportunistically or because both types of infection arise from a similar reservoir; this assumption seems reasonable. If the former is more common, then one might hypothesise that this would occur more often from resistant colonising organisms as these are more likely to be unsuccessfully treated with antibiotics in the community, potentially explaining the generally slightly higher resistance in bloodstream compared to non-bloodstream specimens. One limitation of our study is that we only had enough numbers to analyse the relationship for eight commensal opportunistic pathogens. Our results may therefore not generalise to all other pathogens, although there was no indication that this would be the case from the ones that we were able to include, and, by definition, the included pathogens are the most common causing infections. An important limitation of our approach could be that different population sub-groups were sampled for blood and other specimens; however, our analysis of blood/non-blood samples from the same patients suggests this is unlikely to cause major bias. We have also not considered AMR surveillance encompassing phenotypic profiles (i.e. patterns of resistance within one isolate across different antibiotics), as opposed to discrete pathogen-drug combinations. We also pooled all samples together: future work could consider whether relationships are generalizable between different population sub-groups (e.g. by age, nosocomial/community infections). Finally, the resistance prevalence categories we considered were slightly arbitrary, which is why we considered two different approaches (varying margins of error and varying categorical thresholds).
Although different absolute resistance rates were observed in blood versus non-blood samples for many pathogen-drug combinations, cross-correlations were generally high, as was agreement within 10% and across AMR prevalence categories that might affect empiric antimicrobial prescribing practice. Several biological reasons for the lack of perfect agreement are plausible: for example, a large proportion of E. coli BSIs have a urinary focus, but appropriate empiric treatment generally limits progression to BSI, in contrast to resistant, and perhaps sub-optimally treated UTIs. Hence one might expect the proportion of drug-resistant BSIs to be higher, but underlying time trends to track each other, as we observed. Performance did vary slightly between different pathogen-drug combinations as well as between different locations. However, the totality of evidence across all the datasets supports the value of using non-sterile site specimens for antimicrobial surveillance, regardless of the income of the country.
In HICs, susceptibility testing still typically takes at least 48 h from sampling to result, during which empiric antimicrobials are administered, potentially leading to poorer outcomes if the infection is caused by a resistant organism4. However, increasing use of electronic health records means it is becoming much simpler to check for previous UTIs and use this to guide empiric treatment at the patient-level. Our paired analysis illustrates the potential of this approach based on an arbitrarily chosen interval of 3–90 days prior to the BSI; future work could investigate how the strength of this association varies further back in time, or using the most resistant rather than most recent urinary isolate. Similarly, Yelin et al.21 found that incorporating demographic information with susceptibility results of previous UTIs and antibiotic purchases in a machine learning model improved guidance of empirical treatment for new community-acquired UTIs.
AMR surveillance plays a key role in optimising the use of antibiotics, developing empiric treatment guidelines, and improving antimicrobial stewardship. Lack of microbiology facilities and unwillingness of patients to have samples taken (often due to cost) have been identified as key factors influencing antibiotic use22,23. LMICs face numerous challenges in setting up surveillance systems similar to those in HICs, including lack of coherent governance, budget, technical expertise, information technology systems and co-ordination. Although capacity-building and optimising diagnostic infrastructures in LMICs is clearly important, our study highlights that a bridging strategy using body-sites which are easier to sample, cheaper, faster and easier to culture compared to blood could provide a more rapidly scalable approach to AMR surveillance, providing evidence for empiric treatment recommendations. This sampling strategy could also be amenable to intermittent population survey-type approaches as opposed to continuous surveillance. Using population-level surveillance as a stepping-stone complementary to developing adequate infrastructure to support individual patient-level testing could be considered a pragmatic first step to informing AMR prevalence estimates and empiric treatment guidelines in resource-limited settings.
Materials and methods
Experimental design
Oxfordshire, UK, dataset (regional analysis, high-income country)
We obtained antibiograms (i.e. phenotypic profiles) for E. coli, Klebsiella spp., Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus faecalis and Enterococcus faecium from the Infections in Oxfordshire Research Database (IORD)24, from 1 January 1998 to 31 December 2018. IORD includes all microbiology tests performed in Oxfordshire, UK, with a population size of ~ 680,000 individuals, and has Research Ethics Committee and Confidentiality Advisory Group approval (19/SC/0403, 19/CAG/0144) as a de-identified electronic research database. We compared E. coli and Klebsiella cultured from BSIs versus urinary tract infections (UTI); and isolates from BSIs versus all other body-sites for other pathogens (i.e. non-BSIs; excluding surveillance swabs), further sub-divided into sterile versus non-sterile samples for S. aureus (Supplementary Methods); total 558,616 isolates. We estimated yearly resistance prevalence for all antimicrobials with susceptibility data for > 65% samples in that year (mostly > 80%). In Oxfordshire, testing used manual disk diffusion before 2013, and thereafter automated testing with the BD Phoenix™ Automated Microbiology System (Becton Dickinson, NJ, USA). Binary susceptibility (i.e. susceptible/resistant) phenotypes were defined following laboratory standard operating procedures in place at the time of any given test, and using relevant European Committee on Antimicrobial Susceptibility testing (EUCAST) breakpoints for all specimen types25, dropping a very small number of intermediate isolates. We did not de-duplicate isolates by patient, in order to reflect the data that would be easily available from routine laboratory systems.
Statistical analysis
For each pathogen-drug combination within the Oxfordshire dataset, we first estimated the proportion resistant in blood versus other samples in each calendar year (with 95% confidence intervals (CI)) using Lin’s concordance correlation coefficient (CCC) for comparisons26. We used time-series cross-correlation functions to identify the time interval at which the correlation between resistance prevalence was strongest. As perfect agreement is unrealistic, we also considered agreement within what might be considered clinically acceptable error by infectious disease specialists (± 5% and ± 10% difference in prevalence). In addition, for each pathogen-drug combination, we considered four arbitrary categories of AMR prevalence which might affect empirical prescribing practice, specifically: < 5% resistance prevalence (most prescribers would readily prescribe the antibiotic empirically), 5–10% resistance prevalence (most would prescribe for mild infections), 10–20% resistance prevalence (prescribers might consider alternative antibiotics as first-line empiric choices) and > 20% resistance prevalence (many prescribers would not prescribe if alternative options were available). These thresholds are arbitrary and so we also present exact estimates in Figures to demonstrate robustness to their choice. We used logistic random-effects meta-analysis to estimate the overall difference between resistance prevalence in the two sample types (i.e. blood versus non-blood), treating every calendar year as an independent study, assessing heterogeneity across years using the I2 statistic27. We used meta-regression to estimate the effect of calendar year on the proportion of resistant bloodstream isolates (with its standard error), assuming the proportion of resistant isolates from other infection sites was known, given their greater numbers. We also used meta-regression to directly estimate the association between log odds of resistance in bloodstream infections (outcome, with its standard error) and the log odds of resistance in other infection sites (explanatory variable, fixed). (See Supplementary Material for full details).
Resistance rates could differ between blood and other infection sites because of differences in the underlying populations being sampled. A secondary analysis therefore considered matched pairs of isolates from the same patient, selecting the closest culture from a different site up to 3 days before or 2 days after the blood culture11, assessing concordance using McNemar’s test. We also considered whether susceptibility of a previous E. coli UTI could predict susceptibility of a subsequent E. coli BSI, by selecting the temporally closest urine culture taken between 3 and 90 days before the blood culture11,28 and considering major/very major error rates of predictions29. While the time intervals are arbitrary, they have been used in the literature before and have been guided by the standard definition of a recurrent UTI episode11,28.
Antimicrobial Testing Leadership and Surveillance (ATLAS) dataset (country-level analyses, low/middle/high-income countries)
We also analysed pathogen-drug combinations at the country level (pooling multiple hospitals/regions) using the ATLAS dataset, comprising 103,559 isolates from 17 countries between 2004 and 201714. Intermediate isolates were considered non-susceptible. We considered all years where at least 30 isolates were tested for a given drug in LMICs (95% CI width around prevalence always < 37%); and 100 isolates for HIC (width < 20%) given greater numbers from HIC and strong inverse association between numbers tested and resistance prevalence in HIC, suggesting preferential testing of resistant isolates. Resistance proportions, time-series cross-correlations, extent of agreement, and meta-analyses/regression were performed as for the Oxfordshire dataset above.
Cambodia, Laos, Thailand datasets (local level analyses, low-middle income countries)
Finally, we also analysed microbiology data through collaborative programmes at Angkor Hospital for Children (Siem Reap, Cambodia), Mahosot Hospital, a large national referral centre (Vientiane, Laos), and the Shoklo Malaria Research Unit, serving migrant and refugee populations on the Thailand-Myanmar border (Mae Sot, Thailand), including antibiotics tested for > 40% of the isolates across all study years, and comprising 6154 isolates, 2008–2019. Whilst these are not entirely representative of the countries, they have information on resistance in both bloodstream and other specimens, information generally not available elsewhere. Here, antimicrobial susceptibility testing was conducted using disk diffusion and results interpreted using Clinical and Laboratory Standards Institute (CLSI) 2019 breakpoints. Intermediate isolates were considered non-susceptible. Extended spectrum beta-lactamase (ESBL) status was confirmed using the double disk method (ceftazidime ± clavulanate and cefotaxime ± clavulanate) as a ≥ 5 mm increase in a zone diameter for either agent with clavulanic acid. Resistance proportions, time-series cross-correlations, extent of agreement, and meta-analyses/regression were performed as for the Oxfordshire dataset above.
We conducted all analyses using R 3.5.1, using the metafor package for meta-analyses30.
Data availability
ATLAS data is publicly available14. Data from Oxfordshire are available from the Infections in Oxfordshire Research Database, subject to an application meeting the ethical and governance requirements of the database (contact email iord@ndm.ox.ac.uk). Requests for data from individual hospitals in Laos, Cambodia and Thailand should be made to Vilada Chansamouth, Paul Turner, Clare L Ling respectively. All statistical analyses were performed using standard functions in the following R packages: ggplot2 (version 3.1.0), metafor (version 2.1-0). Code used for data analysis is available upon request from the corresponding author.
References
World Health Organisation. Ten Threats To Global Health In 2019 (2019). https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019. (Accessed on Nov 25, 2021).
Vernet, G. et al. Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg. Infect. Dis. 20, 434–441 (2014).
Dk, B. A view on antimicrobial resistance in developing countries and responsible risk factors. Int. J. Antimicrob. Agents 24, 105–110 (2004).
Laxminarayan, R., Bhutta, Z., Duse, A., Brien, T., Okeke, I., Pablo-Mendez, A., et al. 2003 Drug Resistance P1031–51. In Disease Control Priorities in Developing Countries 2nd ed (eds Jamison, D., Breman, J., Measham, A., Alleyne, G., Claeson, M., Evans, D., et al.) (The International Bank For Reconstruction And Development/The World Bank, 2003).
Ja, A., Ntemgwa, M. & An, A. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 6, 47 (2017).
Vlieghe, E., Bal, A., Gould, I. Surveillance of Antibiotic Resistance in Developing Countries: Needs, Constraints and Realities 463–475 (Antimicrobial Resistance in Developing Countries, 2010).
Ca, H. & Kariuki, S. Antimicrobial resistance in developing countries. BMJ 317, 647–650 (1998).
Ac, S., Nc, G., Islam, J., Sj, P. & Jag, S. AMR surveillance in low and middle-income settings—A roadmap for participation in the global antimicrobial surveillance system (Glass). Wellcome Open Res. 2, 92 (2017).
Lee, H. et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 To April 2017: First one-year report from Kor-Glass. Euro Surveill. 23, 1800047 (2018).
Shakeri, H. et al. Establishing statistical equivalence of data from different sampling approaches for assessment of bacterial phenotypic antimicrobial resistance. Appl. Environ. Microbiol. 84, e02724-17 (2018).
Vihta, K. et al. Trends over time in Escherichia coli bloodstream infections, urinary tract infections, and antibiotic susceptibilities in Oxfordshire, UK, 1998–2016: A study of electronic health records. Lancet Infect. Dis. 18, 1138–1149 (2018).
Walker, S., Te, P., O’connor, L., Dw, C. & Wyllie, D. Are there better methods of monitoring MRSA control than bacteraemia surveillance? An observational database study. PLoS One 3, E2378 (2008).
Ac, S. et al. Supporting surveillance capacity for antimicrobial resistance: Laboratory capacity strengthening for drug resistant infections in low and middle income countries. Wellcome Open Res. 2, 91 (2017).
Wellcome Trust. Wellcome Data Re-Use Prize—Antimicrobial Testing Leadership And Surveillance. (2019). https://www.Synapse.Org/#!Synapse:Syn17009517/Wiki/585653 (accessed 29 May 2021).
Edelsberg, J. et al. Prevalence of antibiotic resistance in us hospitals. Diagn. Microbiol. Infect. Dis. 78, 255–262 (2014).
Poolman, J. & Anderson, A. Escherichia coli and Staphylococcus aureus: Leading bacterial pathogens of healthcare associated infections and bacteremia in older-age populations. Expert. Rev. Vaccines 17, 607–618 (2018).
Health Protection Scotland, National Services Scotland. Protocol for Surveillance of Antimicrobial Resistance in Urinary Isolates in Scotland. (2011). https://hpspubsrepo.blob.core.windows.net/hps-website/nss/2821/documents/1_protocol-forsurveillance-of-amr-in-urinary-isolates-in-scotland-v1.0.pdf. (Accessed on Nov 25, 2021).
Health Protection Scotland, Information Services Division. Scottish Antimicrobial Use and Resistance in Humans in 2015 (2016). https://www.isdscotland.org/Health-Topics/Prescribing-and-Medicines/Publications/2016-08-30/2016-08-30-SAPG-2015-Report.pdf. (Accessed on Nov 25, 2021).
World Health Organization. Community-based surveillance of antimicrobial use and resistance in resource-constrained settings, Report On Five Pilot Projects (2009). https://Apps.Who.Int/Iris/Handle/10665/70036. (Accessed on Nov 25, 2021).
Auguet, O. et al. Metrics for public health perspective surveillance of bacterial antibiotic resistance in low- and middle-income countries. Biorxiv https://doi.org/10.1101/2020.02.10.941930v1 (2020).
Yelin, I. et al. Personal clinical history predicts antibiotic resistance of urinary tract infections. Nat. Med. 25, 1143–1152 (2019).
Kotwani, A., Wattal, C., Katewa, S., Pc, J. & Holloway, K. Factors influencing primary care physicians to prescribe antibiotics in Delhi India. Fam. Pract. 27, 684–690 (2010).
Ganguly, N. et al. Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J. Med. Res. 134, 281–294 (2011).
Finney, J., Walker, A., Peto, T. & Wyllie, D. An efficient record linkage scheme using graphical analysis for identifier error detection. BMC Med. Inform. Decis. Mak. 11, 7 (2011).
European Committee on Antimicrobial Susceptibility Testing. 2019 Clinical Breakpoints for Bacteria V 9.0. http://www.Eucast.Org/Clinical_Breakpoints/. (Accessed on Nov 25, 2021).
Li, L. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45, 255–268 (1989).
Higgins, J., Thompson, S., Deeks, J. & Altman, D. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Cc, B. et al. Containing antibiotic resistance: decreased antibiotic-resistant coliform urinary tract infections with reduction in antibiotic prescribing by general practices. Br. J. Gen. Pract. 57, 785–792 (2007).
U.S. Department Of Health and Human Services Food and Drug Administration Center for Devices and Radiological Health. Guidance for Industry and Fda: Class Ii Special Controls Guidance Document: Antimicrobial Susceptibility Test (Ast) Systems. https://www.fda.gov/medical-devices/guidance-documents-medical-devices-and-radiation-emitting-products/antimicrobial-susceptibility-test-ast-systems-class-ii-special-controls-guidance-industry-and-fda. (Accessed on Nov 25, 2021).
Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48 (2010).
Acknowledgements
We thank all the people of Oxfordshire who contribute to the Infections in Oxfordshire Research Database. Research Database Team: R Alstead, C Bunch, DCW Crook, J Davies, J Finney, J Gearing (community), L O’Connor, TEA Peto (PI), TP Quan, J Robinson (community), B Shine, AS Walker, D Waller, D Wyllie. Patient and Public Panel: G Blower, C Mancey, P McLoughlin, B Nichols. We thank Liam Shaw for discussion about the ATLAS dataset and feedback on the manuscript. We thank the diagnostic microbiology laboratory staff at Angkor Hospital for Children, Mahosot Hospital Microbiology Laboratory, and the Shoklo Malaria Research Unit clinic and laboratory staff.
Funding
This study was funded by National Institute for Health Research (NIHR). Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial. Resistance (NIHR200915) at the University of Oxford in partnership with Public. Health England (PHE) and was supported by the Oxford NIHR Biomedical Research Centre. ASW is an NIHR Senior Investigator. The report presents independent research. funded by NIHR. The views expressed in this publication are those of the authors and not necessarily those of the NHS, NIHR, the Department of Health or Public Health England.
Author information
Authors and Affiliations
Contributions
K.-D.V., N.J.W., T.E.A.P., and A.S.W. designed the study. T.P.Q. prepared extracts from the Infections in Oxfordshire Research Database. K.-D.V. obtained data from the Antimicrobial Testing Leadership and Surveillance (ATLAS). Data from the Angkor Hospital for Children was provided by P.T., from the Mahosot Hospital Microbiology Laboratory by M.V., E.A.A., V.C., and from the Shoklo Malaria Research Unit by C.L. K.-D.V., T.E.A.P. and A.S.W. analysed the data. K.-D.V., T.E.A.P., and A.S.W. prepared the figures. K.-D.V., D.E., T.E.A.P., and A.S.W. prepared the first draft of the manuscript. All authors commented on the data and its interpretation, revised the content critically, and approved the final version.
Corresponding author
Ethics declarations
Competing interests
DWE reports personal fees from Gilead, outside the submitted work. All other authors report no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vihta, KD., Gordon, N.C., Stoesser, N. et al. Antimicrobial resistance in commensal opportunistic pathogens isolated from non-sterile sites can be an effective proxy for surveillance in bloodstream infections. Sci Rep 11, 23359 (2021). https://doi.org/10.1038/s41598-021-02755-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-02755-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.