Abstract
The discovery of new viruses is important for predicting their potential threats to the health of humans and other animals. A novel picornavirus was identified from oral, throat, and anal swab samples collected from belugas (Delphinapterus leucas), from Dalian Sun Asia Tourism Holding Co., China, between January and December 2018, using a metagenomics approach. The genome of this novel PicoV-HMU-1 strain was 8197 nucleotides (nt) in length, with a open reading frame (from 1091 to 8074 nt) that encoded a polyprotein precursor of 2328 amino acids. Moreover, the genomic length and GC content of PicoV-HMU-1 were within the ranges found in other picornaviruses, and the genome organization was also similar. Nevertheless, PicoV-HMU-1 had a lower amino acid identity and distinct host species compared with other members of the Picornaviridae family. Phylogenetic trees were constructed based on the P1 and 3D amino acid sequences of PicoV-HMU-1 along with representative members of the Picornaviridae family, which showed that PicoV-HMU-1 was related to unclassified bat picornaviruses groups. These findings suggest that the PicoV-HMU-1 strain represents a potentially novel genus of picornavirus. These data can enhance our understanding of the picornavirus genetic diversity and evolution.
Similar content being viewed by others
Introduction
Members of the family Picornaviridae are small, icosahedral, non-enveloped, positive-sense ( +) single-stranded RNA (ssRNA) viruses that can cause various symptoms, ranging from mild febrile illness to severe diseases of the respiratory, cardiac, hepatic, mucocutaneous, and central nervous systems1. According to the International Committee on Taxonomy of Viruses (https://talk.ictvonline.org/), the family Picornaviridae, order Picornavirales, currently consists of 158 species grouped into 68 genera, which include the genera Aphthovirus (foot-and-mouth disease virus), Cardiovirus (Cardiovirus A), Enterovirus (Enterovirus A and Rhinovirus A), Hepatovirus (Hepatovirus A), Limnipivirus (Limnipivirus A), and Parechovirus (Human parechovirus, Ljungan virus, and Sebokele virus).
Picornaviruses belong to the family Picornaviridae (https://www.ictv.global/report/picornaviridae) and have a genome that is 6.7–10.1 kilobases in size and contain a single open reading frame (ORF) that encodes a large polyprotein that is cleaved into structural and non-structural proteins by viral proteases. One exception is the genus Dicipivirus, which possesses two separated ORFs2. The characteristic organizational pattern of the picornaviral genome is VPg–5′-UTR–(L)–P1–P2–P3–3′-UTR–poly(A)2. The 5′-untranslated region (UTR) of picornaviruses is a structured functional region that contains an internal ribosomal entry site (IRES), which mediates the end-independent initiation of translation3. The single ORF encodes a polyprotein precursor that is divided into three regions (P1, P2, and P3), of which P1 encodes the capsid proteins, whereas P2 and P3 encode proteins involved in proteolytic processing and viral replication. The P1 region is derived from VP4 to VP1, the P2 region is derived from 2A to 2C, and the P3 region is derived from 3A to 3D1.
Advances in metagenomics have allowed the identification of new viral genomic sequences with clinical and epidemiological value. Metagenomic technology has facilitated virus discovery due to the enhanced capacity to detect viruses with an unknown genetic background. Recently, several novel picornaviruses were discovered within different hosts, including a novel picornavirus from Macaca mulatta in China identified by viral metagenomic analysis4. Furthermore, a novel bovine picornavirus was discovered in Japan5, and several picornaviruses, including three bat, one feline, and one canine picornaviruses were discovered in Hong Kong6,7,8. The discovery of novel picornavirus provides the foundation for virus traceability and evolutionary research. Overall, approximately 75% of emerging diseases are estimated to arise from zoonotic sources9. Zoological parks and aquariums provide a unique opportunity for emerging virus surveillance. The discovery of novel viruses may help predict their potential threat to the health of humans and other animals. In this report, we analyzed by high-throughput next-generation sequencing (NGS) a novel picornavirus identified in belugas (Delphinapterus leucas) from an aquarium.
Results
Metagenomic analysis
In total, 28 oral, throat, and anal swab samples from 12 captive belugas (Delphinapterus leucas) were acquired and were combined into two pools. We found new viruses for construction and NGS in one of the pools. A total of 1.91 Gb of nucleotide data (19,732,441 valid reads, 100 bp in length) were obtained. Reads that were classified as being from cellular organisms (such as bacteria, archaea, and eukaryotes) and those with no significant similarities to any amino acid (aa) sequences in the non-redundant (NR) nucleotide database were removed. The remaining 7517 reads were best-matched with viral proteins from the NR database, which included the families Herpesviridae, Podoviridae, Microviridae, Picornaviridae, Papillomaviridae, environmental samples viruses, ssRNA viruses, unclassified + ssRNA viruses, unclassified RNA viruses, and unclassified bacterial viruses (Fig. 1). There were 60 Picornaviridae-associated reads in the lane. The reads mapped to the picornaviral genome are shown in Supplementary Fig. S1 online.
Genome analysis
Based on the NGS-generated fragment sequences, primers were designed to obtain the intervening portions of the genome. The terminal sequences were then acquired using a 5' and 3' rapid amplification of cDNA ends (RACE) method. We obtained a picornaviral genome sequence, named the PicoV-HMU-1 strain, which was 8,197 bases in length and with a G + C content of 43.4% after exclusion of the polyadenylated tract. Both the 5'- (1090 bases) and 3'-ends (123 bases) of the genome contained UTRs. The genome contained a ORF of 6,984 bases that encoded a polyprotein precursor of 2328 aa. The sequenced PicoV-HMU-1 genome was predicted to encode the P1 (843 aa), P2 (694 aa), and P3 (791 aa) regions, which potentially encode the capsid proteins VP4, VP2, VP3, and VP1; the non-structural proteins 2A, 2B, and 2C, and 3A, 3B, 3C, and 3D, respectively (Fig. 2). The 5′-UTR of PicoV-HMU-1 was 1090 nt in length. According to the blast analysis, PicoV-HMU-1 shared high sequence identity with Bat picornavirus(HQ595344). We selected 362–1090 bp to analyse the secondary structure by comparison with the complete sequence of HQ595344, and hence we performed the secondary structure prediction shown in Fig. 3.
Phylogenetic analyses
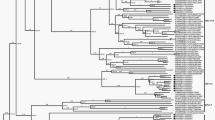
The phylogenetic trees were constructed based on the P1 and 3D aa sequences of PicoV-HMU-1 along with representative members of the Picornaviridae family and other unclassified viruses. In the P1 trees, PicoV-HMU-1 was close to unclassified bat picornavirus 3 and la_io_picornavirus_1 and formed a monophyletic branch (Fig. 4). In the 3D trees, PicoV-HMU-1 was also related to the unclassified bat picornaviruses 3 and formed an independent cluster (Fig. 5).
Phylogenetic analyses of the P1 regions of PicoV-HMU-1. Maximum likelihood mtREV with the Freqs (+ F) model, gamma distributed with invariant sites (G + I), and 1000 bootstrap replicates using MEGA 6 software. The virus identified in this study is indicated by the red circle. Bootstrap values are shown on the branches.
Phylogenetic analyses of the 3D regions of PicoV-HMU-1. Maximum likelihood mtREV with the Freqs (+ F) model, gamma distributed with invariant sites (G + I), and 1000 bootstrap replicates using MEGA 6 software. The virus identified in this study is indicated by the red circle. Bootstrap values are shown on the branches.
Prevalence of PicoV-HMU-1
All 28 swab samples from 12 captive belugas were tested for PicoV-HMU-1 by nested-polymerase chain reaction (PCR) of the VP1 proteins of PicoV-HMU-1, among which two throat and two anal swab samples were positive (14.29%) (Table 1).
Discussion
Previous viral outbreaks in marine mammals (such as morbillivirus infections in European seals and Eastern U.S. dolphins) have temporarily reduced their populations10. Morbillivirus infections have also been detected in marine mammals in the Canadian arctic, as evidenced by the presence of neutralizing antibodies in walruses11. In 2007, researchers in California identified the first sequence-confirmed picornavirus isolated from ringed seals, which was proposed to represented a member of a new picornavirus genus in the Picornaviridae family12. In the last year, Australian researchers demonstrated that the novel marine flaviviruses identified in crustaceans were more closely related to the terrestrial vector-borne flaviviruses than classical insect-specific flaviviruses13. Although their clinical relevance remains unclear, identification of novel viruses and their respective animal hosts is important for a better understanding of their genetic diversity, evolution, biology, and potential for cross-species transmission and emergence.
In this study, we discovered a new picornavirus within oral, throat, and anal swab samples from white whales using metagenomic technology. Genomic characterization revealed that the structural features of PicoV-HMU-1 are similar to those of other Picornaviridae family members, with its genomic length and GC content being within the ranges of those found in picornaviruses. Moreover, the genome organization was similar to that of other picornaviruses, with the characteristic gene order: 5′-VP4-VP2-VP3-VP1-2A-2B-2C-3A-3B-3Cpro-3Dpol-3′ (with 3Cpro and 3Dpol being 3C protease and 3D polymerase, respectively). Picornaviruses encode extensive RNA stem-and-loop structures in the 5′-UTR regions that are critical for viral replication.
The 5′-UTR of PicoV-HMU-1 was 1090 nt in length. According to the Bat picornavirus(HQ595344), We performed the secondary structure prediction with the secondary structure region (362–1090 bp). The secondary structure of the PicoV-HMU-1 sequence is different from that of all currently known IRESs (Fig. 3). At the nucleic acid level, the sequence was barely comparable with that of other known picornaviruses in the National Center for Biotechnology Information nucleotide database. Therefore, we suggest that this secondary structure is of a novel type of IRES. Pair-wise alignment of the aa sequences of PicoV-HMU-1 with those of other picornaviruses revealed that it shared high sequence identity with parabovirus A1 (P1: 48.1%; P2: 35.5%; P3: 57.5%), feline picornavirus (P1: 47.5%; P2: 40.7%; P3: 61.6%), and ovine picornavirus (P1: 45.8%; P2: 35.1%; P3: 53.1%) than with other representative viruses from the recognized genera within Picornaviridae. However, its highest sequence identity was with unclassified picornaviruses; bat picornavirus group 1–3 (P1: 450.0–46.9%; P2: 46.3–44.3;%; P3: 67.1–65.6%) (Table 2). Phylogenetic analysis confirmed that PicoV-HMU-1 was related to unclassified bat picornaviruses groups. Therefore, it is likely that these viruses may have shared a common ancestor, and started to evolve independently after terrestrial mammals entered the ocean. Moreover, these results suggest that the host of the novel picornavirus was exclusively the white whale.
According to our findings, PicoV-HMU-1 has a lower aa identity and a distinct host species compared with other members of the Picornaviridae family. Thus, we propose that the novel PicoV-HMU-1 is a new picornavirus genus. Nevertheless, the role of this picornavirus in the evolution, transmission, and biology of marine mammals warrants further investigation. Thus, it is important to document the existence of novel picornaviruses isolated from marine mammals. Moreover, future studies investigating the evolution, classification, and traceability of mammalian virology should pay increased attention to viruses carried by marine mammals.
Materials and methods
Sample collection
The sampling procedures were approved by the Ethics Committee of the Hainan Medical University (Approval number: HMUEC20180059). A total of 28 oral swabs, throat swabs, and swab samples of were collected from 12 captive belugas (Delphinapterus leucas) from Qingdao Polar Haichang Ocean Park and Dalian Sun Asia Tourism Holding Co., China, from January to December 2018. The samples were immersed in maintenance medium in virus-sampling tubes (Yocon, Beijing, China), transported to the laboratory within 24 h using cold chain transportation, and stored at − 80 ℃.
Viral nucleic acid library construction and next-generation sequencing
All 28 samples were combined into two pools based on sampling time and were passed through 0.45 µm filters. The collected filtrates were digested with DNase (Applied Biosystems, Waltham, MA, USA) to remove unprotected nucleic acids. Total RNA was extracted using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. cDNA was generated using Superscript III Reverse Transcriptase (Invitrogen, Waltham, MA, USA). Amplified viral nucleic acid libraries were analyzed on an HiSeq2500 sequencer (Illumina, San Diego, CA, USA) for a single read of 100 bp in length. The raw sequence reads were filtered using previously described criteria to obtain valid sequences14.
Taxonomic assignment
Sequence similarity-based taxonomic assignments were conducted as previously described14. Briefly, each read was evaluated for viral origin by conducting alignments with the non-redundant (NR) nucleotide and protein databases of the National Center for Biotechnology Information, using BLASTn and BLASTx, respectively (Expected value < 10−5, F: Filter query sequence, default = T). Taxonomies of aligned-reads with the best BLAST scores (E-value < 10−5) from all lanes were parsed and exported using the Metagenome Analyzer (MEGAN) 615.
Genome sequencing
Molecular clues provided by metagenomic analyses were used to classify sequence reads into a virus family or genus using MEGAN 6. Viral RNA was isolated using QIAamp Viral RNA Mini Kit (Qiagen), and cDNA was generated using random primer and Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Gene-specific primers were designed based on representative identified reads of the novel picornavirus while ensuring that they covered the partial genomes by nested-PCR amplification and Sanger sequencing. The remaining genomic sequences were obtained by amplification using a genome-walking kit (Takara, Kusatsu, Japan), and the full 5′- and 3′-end sequences were obtained by repeated amplification using a 5′-RACE system version 2.0 combo (Invitrogen) and 3′-Full RACE Core Set with PrimeScript RTase (Takara) according to the manufacturers’ instructions. All primer sequences were based on the newly obtained reads and newly amplified sequences. All primers used are shown in Supplementary Table S1 online.
Detection of picornavirus
Using the genomic sequences of the viruses obtained by amplification of the ends, specific primers for nested-PCR were designed to screen for picornavirus in the 28 swabs samples collected from 12 captive belugas. PCRs were performed using KOD One PCR Master Mix (Takara), and the first PCR round was primed with outer primers (F: 5′-AGCAGTTACCTTGCCCACG-3′ and R: 5′-TCCCTGCTCGCACCTTG-3′), and the second PCR round was primed with inner primers (F: 5′-CGCCTGAGACTGGTGT-3′ and R: 5′-TTGCCATTGGGTGTAA-3′). PCR product (1 µL) from the first round was used as template for the second round reaction. The thermal cycling conditions for the first round PCR were 98 ℃ for 5 min, followed by 35 cycles at 98 ℃ for 10 s, 50 ℃ for 5 s, 68 ℃ for 5 s, and a final elongation step at 68 ℃ for 1 min; and for the second round PCR were 98 ℃ for 5 min, followed by 35 cycles of 98 ℃ for 10 s, 58 ℃ for 5 s, 68 ℃ for 5 s, and a final elongation step at 68 ℃ for 1 min. Finally, the PCR products were analyzed by 1% agarose gel electrophoresis ultraviolet imaging. Positive samples were determined by the presence of 642 bp amplified products. All PCR products were further verified by Sanger sequencing.
Genome annotation and phylogenetic analysis
Picornaviruses containing IRES-like sequences in the 5′-UTRs were identified by BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) of viral sequences in the GenBank database. Secondary/tertiary structural elements in picornavirus 5′-UTRs were modeled using the aligned RNAalifold server of the ViennaRNA Web Services (http://rna.tbi.univie.ac.at/). MEGA 6.0 (http://www.megasoftware.net)15 was used to align the nt sequences and deduce the corresponding aa sequences using the MUSCLE package with default parameters. Phylogenetic trees showing the relationships among picornaviruses based on the amino acid sequences of P1 and 3D were generated using maximum likelihood mtREV with Freqs (+ F) model, gamma distributed with invariant sites (G + I), and 1000 bootstrap replicates. Routine sequence alignments were performed using Clustal Omega, EMBOSS Needle (both available at: http://www.ebi.ac.uk/Tools/), MegAlign and Lasergene (DNAstar, Madison, WI, USA), and T-coffee (http://www.tcoffee.org/) with manual curation. The nt sequences of the genomes and aa sequences of the ORF were deduced by comparison with those of other picornaviruses. The conserved protein families and domains were predicted using Pfam and InterProScan 5 (available at: http://www.ebi.ac.uk/services/proteins). Amino acid identities and genetic distances were calculated using the ML method and were performed using pairwise evolutionary distance calculation as the distance metric.
Animal rights statement
Animals were treated according to the guidelines of Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People’s Republic of China, 1988). The sampling procedure was approved by the Ethics Committee of the Hainan Medical University.
Data availability
The genome sequences of PicoV-HMU-1 are available at GenBank (https://www.ncbi.nlm.nih.gov/genbank/), with accession number: MW883077.
References
Jiang, P., Liu, Y., Ma, H. C., Paul, A. V. & Wimmer, E. Picornavirus morphogenesis. Microbiol. Mol. Biol. Rev. 78, 418–437. https://doi.org/10.1128/MMBR.00012-14 (2014).
Zell, R. et al. ICTV virus taxonomy profile: Picornaviridae. J. Gen. Virol. 98, 2421–2422. https://doi.org/10.1099/jgv.0.000911 (2017).
Jackson, R. J., Hellen, C. U. T. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. https://doi.org/10.1038/nrm2838 (2010).
Ao, Y., Zhou, Y., Li, D. & Duan, Z. A novel picornavirus identified in wild Macaca mulatta in China. Adv. Virol. 165, 495–504. https://doi.org/10.1007/s00705-019-04442-3 (2019).
Nagai, M. et al. Identification and complete genome analysis of a novel bovine picornavirus in Japan. Virus Res. 210, 205–212. https://doi.org/10.1016/j.virusres.2015.08.001 (2015).
Lau, S. K. et al. Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 85, 8819–8828. https://doi.org/10.1128/jvi.02364-10 (2011).
Lau, S. K. et al. Identification of a novel feline picornavirus from the domestic cat. J. Virol. 86, 395–405. https://doi.org/10.1128/JVI.06253-11 (2012).
Woo, P. C. Y. et al. Complete genome sequence of a novel picornavirus, canine picornavirus, discovered in dogs. J. Virol. 86, 3402–3403. https://doi.org/10.1128/jvi.07228-11 (2012).
Taylor, L. H., Woolhouse, M. E. J., Dye, C., Latham, S. M. & Woolhouse, M. E. J. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 356, 983–989. https://doi.org/10.1098/rstb.2001.0888 (2001).
Kennedy, S. Morbillivirus infections in aquatic mammals. J. Comp. Pathol. 119, 201–225. https://doi.org/10.1016/s0021-9975(98)80045-5 (1998).
Nielsen, O., Stewart, R. E. A., Measures, L., Duignan, P. & House, C. A morbillivirus antibody survey of Atlantic Walrus, Narwhal and Beluga in Canada. J. Wildl. Dis. 36, 508–517. https://doi.org/10.7589/0090-3558-36.3.508 (2000).
Kapoor, A. et al. A highly divergent Picornavirus in a marine mammal. J. Virol. 82, 311–320. https://doi.org/10.1128/jvi.01240-07 (2008).
Parry, R. & Asgari, S. Discovery of novel Crustacean and cephalopod flaviviruses: Insights into the evolution and circulation of flaviviruses between marine invertebrate and vertebrate hosts. J. Virol. https://doi.org/10.1128/JVI.00432-19 (2019).
Yang, J. et al. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J. Clin. Microbiol. 49, 3463–3469. https://doi.org/10.1128/JCM.00273-11 (2011).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. https://doi.org/10.1093/molbev/mst197 (2013).
Acknowledgements
This work was supported by the the Major Science and Technology Program of Hainan Province (grant No. ZDKJ202003), the Key Research and Development Plan of Hainan Province (grant No. ZDYF2020150), Hainan Provincial Natural Science Foundation of China (Grant No. 820QN267, 2019RC218, 820RC628), the National Natural Science Foundation of China (grant No. 32060015, 82060378), the Scientific Research Foundation of the Higher Education Institutions of Hainan Province, China (grant No. Hnky2021-40, Hnky2021-44), the Research project of Hainan academician innovation platform (grant No. YSPTZX202004), the Hainan talent development project (grant No. SRC200003), the National S&T Major Project “China Mega-Project for Infectious Disease” (Grant No. 2018ZX10711001), the Youth Innovation Promotion Association of Chinese Academy of Sciences (grant No. 2020363), and the Scientific and Technological Program of Strategic Priority in Chinese Academy of Sciences (grant No. XDB42030403).
Author information
Authors and Affiliations
Contributions
F.Y., .J.D., P.Z. designed the study. P.Z., G.W., W.Z., S.S., Y.H., R.P., X.H., Z.W., J.L., S.L., F.Y., X.C., F.D., L.N., Y.Z., C.T., G.L., S.L., S.B. collected the specimens and performed the experiments. J.D., F.Y., analyzed the data. J.D., F.Y., G.W. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, G., Huang, Y., Zhang, W. et al. Identification and genome analysis of a novel picornavirus from captive belugas (Delphinapterus leucas) in China. Sci Rep 11, 21018 (2021). https://doi.org/10.1038/s41598-021-00605-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-00605-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.