Abstract
Gastric cancer (GC) is a common cancer and the leading cause of cancer-related death worldwide. To improve the diagnosis and treatment of GC, it is necessary to identify new biomarkers by investigating the cellular and molecular mechanisms. In this study, miR-30c-5p expression was significantly down-regulated in GC tissues by comprehensive analysis using multiple databases. The target genes of miR-30c-5p with up-regulated expression level in GC were identified, including ADAM12 (a disintegrin and metalloproteinase12), EDNRA (the Endothelin receptor type A), STC1 (stanniocalcin 1), and CPNE8 (the calcium-dependent protein, copine 8). The expression level of ADAM12 was significantly related to depth of invasion (p = 0.036) in GC patients. The expression level of EDNRA was significantly related to grade (P = 0.003), depth of invasion (P = 0.019), and lymphatic metastasis (P = 0.001). The expression level of CPNE8 was significantly related to grade (P = 0.043) and TNM stage (P = 0.027).Gene set enrichment analysis showed that they might participate in GC progression through cancer-related pathways. CIBERSORT algorithm analysis showed that their expressions were related to a variety of tumor-infiltrating immune cells. The higher expression of those target genes might be the independent risk factor for poor survival of GC patients, and they might be potential prognostic markers in GC patients.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is one kind of common cancer and the second most common cause of death compared with other cancers worldwide1. The occurrence and development of gastric cancer are affected by genetic and environmental factors, including family history, individual genotype, dietary, obesity, helicobacter pylori infection, and so on2. Gastric cancer can occur in any part of the stomach, more than half of which occur in the gastric antrum. The vast majority of gastric cancer is adenocarcinoma, without obvious symptoms at the early stage. Gastric cancer is often similar to the symptoms of chronic gastric diseases such as gastritis and gastric ulcer, or causes nonspecific symptoms such as epigastric discomfort and belching, so it is easy to be ignored in the early phase. With the growth of the tumor, more obvious symptoms appear when it affects gastric function. With the progress of the disease, epigastric pain worsens, and appetite decreases. Patients with advanced gastric cancer often have anemia, weight loss, malnutrition, and even cachexia3. The overall 5-year survival rate of patients with GC is poor, because many GC patients have entered the advanced stage of cancer at the time of diagnosis4. At present, the maim approaches for the treatment of GC include surgery, chemotherapy, immunotherapy, and targeted molecular therapy5. For example, CXCR5 + CD8 + T was suggested to be used as a biomarker and a therapeutic target in GC, because of the association between CXCR5 + CD8 + T and better clinical outcomes in GC patients6. To improve the diagnosis and treatment of GC, it is necessary to identify and validate new biomarkers for GC by investigating the cellular and molecular mechanisms of gastric cancer7.
MicroRNA (miRNA), a type of small endogenous non-coding RNA consisting of 19–25 nucleotides, can regulate gene expression by degrading or inhibiting the translation of its target transcripts8. In recent years, increasing studies showed that abnormal expressions of multiple miRNAs cloud play roles in the proliferation, migration, apoptosis, and invasion of gastric cancer. For example, miR-301a-5p had vital functions by targeting Scinderin (SCIN) mRNA in the development and progression of gastric cancer9, and miR-486-5p could modulate gastric cancer cell proliferation, migration and tumor progression by targeting follistatin-like 3 (FSTL3)10.
The miR-30 family consists of 5 highly conserved members: miR-30a, miR-30b, miR-30c, miR-30d, and miR-30e. The miRNAs of miR-30 family have a common sequence near the 5 'end and different compensation sequences near the 3' end, and they might affect the proliferation, metastasis, invasion, and drug resistance of GC cells by targeting the corresponding genes11. The previous studies showed that the microRNA of miR-30c-5p, one mature form of the miR-30 family members, might act as a tumor suppressor in the physiological activities and development processes of a variety of cancers (arrest of cell cycle, inhibition of invasion and migration, and induction of apoptosis). The published data on GC12, breast cancer13, colon cancer14, prostate cancer15, and pancreatic cancer16 indicated that the expression levels of miR-30c-5p were lower in cancer tissues than that in adjacent tissues. Moreover, the expression level of miR-30c-5p was also related to the prognosis of cancer. It was identified that decreased miR-30c-5p expression was related to prolonged survival of malignant mesothelioma patients17. Although the published reports suggested miR-30c-5p might play a role in gastric cancer, the target genes of miR-30c-5p in gastric cancer should be further investigated.
In this study, an attempt was made to investigate potential diagnostic and prognostic values of miR-30c-5p target genes in gastric cancer by using a filtration step analysis and integrated bioinformatics tools. Firstly, according to the data from Gene Expression Omnibus (GEO) database and The Cancer Genome Atlas (TCGA) database, an association between miR-30c-5p expression and gastric cancer risk was evaluated. Secondly, the multiple databases were combined to predict and identify the target genes of miR-30c-5p. Thirdly, the expression levels of miR-30c-5p target genes were validated by using data from TCGA. Fourthly, the overall survival (OS) of the differentially expressed target gene and univariate Cox regression analysis were carried out to explore the potential prognostic values of miR-30c-5p target genes in GC patients. Finally, tumor immune cells in TCGA gastric cancer samples with the expression of miR-30c-5p target genes were analyzed by the CIBERSORT algorithm. The results of this study would be helpful to reveal the potential target genes of miR-30c-5p, which might be used as diagnostic and prognostic markers in gastric cancer.
Results
Association between miR-30c-5p expression level and gastric cancer risk
According to the inclusion and exclusion criteria, 11 datasets from GEO database (GSE2373918, GSE2659519, GSE2870020, GSE3374321, GSE7877522, GSE94882, GSE9941523, GSE9341524, GSE6312125, GSE5439726, and GSE3007027) and TCGA data were identified to determine the association between miR-30c-5p expression level and the risk of GC. The flowchart of data acquisition was shown in Supplementary Fig. S1. The detail characteristics of the GEO datasets and TCGA data were shown in Supplementary Table S1, including 781 GC patients and 248 normal individuals. To analyze the expression of miR-30c-5p, the standardized mean difference (SMD) was calculated using a random-effect model. The negative SMD [SMD = − 0.42; 95% confidence interval (CI): (− 0.81, − 0.04); P < 0.05)] indicated that the expression of miR-30c-5p was downregulated in GC tissues (Fig. 1). In addition, Begg's test showed that no significant publication bias was observed in this study (p > 0.05).
Four miR-30c-5p target genes identified with up-regulated expression levels in GC
To predict miR-30c-5p target genes, the online tools and databases of TargetScan28, miRDB29, miRWalk30, and DIANA31 were used. Somewhat unique target genes were predicted by each online prediction tool. A total of 5444 target genes were identified from TargetScan, 1312 genes from miRDB, 1573 genes from miRWalk, and 1761 genes from DIANA. Among them, 357 common target genes were obtained (Supplementary Fig. S2A). The list of target genes was shown in Supplementary Spreadsheets 1. In this study, GSE54129 and GSE118916, which were global gene expression analyses of gastric cancer by microarrays, were used to screen for up-regulated differentially expressed genes in GC. According to the previous section result of that miR-30c-5p was down-regulated expressed in GC cases, the miR-30c-5p target genes should theoretically be up-regulated expressed in GC patients. According to the criteria of adjusted P < 0.05 and logFC ≥ 1.5, 763 up-regulated expressed genes in GC samples were identified from GSE54129 (Supplementary Spreadsheets 2), and 266 up-regulated genes were identified from GSE118916 (Supplementary Spreadsheets 3). The Venn diagram result showed that four miR-30c-5p target genes were highly expressed in gastric cancer, including ADAM12 (a disintegrin and metalloproteinase12), EDNRA (the Endothelin receptor type A), STC1 (stanniocalcin 1), and CPNE8 (the calcium-dependent protein, copine 8) (Supplementary Fig. S2B).
Validation of expression levels of the four mRNA targets of miR-30c-5p using TCGA data
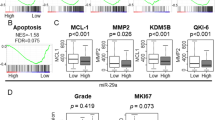
According to GSE54129 and GSE118916 data, the expression levels of four mRNA targets of miR-30c-5p were significantly up-regulated expressed in GC tissues. In this study, the transcriptional data from the TCGA database were adopted to validate the expression levels of miR-30c-5p target genes (ADAM12, EDNRA, STC1, and CPNE8) in GC tissues. The results showed that miR-30c-5p target genes were all highly expressed in GC tissues compared with normal gastric tissues (Fig. 2).
Relationship between expression levels of miR-30c-5p target genes and clinicopathological features
The correlation of expressions of miR-30c-5p target genes with clinicopathological staging characteristics were evaluated by using information of GC patients from the TCGA database. All GC samples were split into two groups (high expression group vs low expression group) based on median target gene expression levels. Relationships between expression levels and clinicopathological parameters in GC patients were listed in Table 1. The results showed that the high expression level of ADAM12 was significantly associated with depth of invasion (p = 0.036) in GC patients. The high expression level of EDNRA was essentially identified with tumor grade (P = 0.003), depth of invasion (P = 0.019), and lymphatic metastasis (P = 0.001) in GC patients. The high expression level of CPNE8 was significantly associated with grade (P = 0.043) and TNM stage (P = 0.027) in GC patients. However, the expression of STC1 was not associated with any clinicopathological features.
Association of miR-30c-5p target genes with overall survival and prognosis of gastric cancer and assessment of diagnostic value
The prognostic value of miR-30c-5p target gene expression levels in GC was analyzed using the GEPIA or Kaplan–Meier plotter online analysis website. As shown in Supplementary Fig. S3 and Supplementary Fig. S4, the miR-30c-5p target gene high expression level group of miR-30c-5p target genes were predict poor prognosis by using the GEPIA or Kaplan–Meier plotter online tools. In addition, univariate Cox regression analysis revealed the role of miR-30c-5p target genes in the prognosis of gastric cancer. The results showed that age (P = 0.0072), TNM stage (P = 0.0002), lymph node metastasis (P = 0.0012), ADAM12 expression (P = 0.0052), EDNRA expression (P = 0.0125), STC1 expression (P = 0.0086), and CPNE8 expression (P = 0.00081) were significantly related with the prognostic of GC patients (Table 2). Overall, the miR-30c-5p target gene was an unfavorable prognostic factor marker. As shown in Supplementary Fig. S5, the AUC values of ADAM12, EDNRA, STC1, and CPNE8 were 0.963 (95% CI = 0.973–0.990), 0.717 (95% CI = 0.634–0.800),0.782 (95% CI = 0.705–0.859), and 0.585 (95% CI = 0.490–0.680), respectively. It suggested that ADAM12, EDNRA, and STC1 had high value in the diagnosis of gastric cancer.
Gene set enrichment analysis of miR-30c-5p target genes
GSEA_4.1.0 enrichment software32 was used for analysis the potential molecular mechanism of the miR-30c-5p target genes in gastric carcinogenesis, and the enrichment pathway was set to be significant for nominal (NOM) p < 0.05 and false discovery rate (FDR) q < 0.05. In the high expression group of miR-30c-5p target genes , the enriched terms involved in the significant up-regulation of cell adhesion and tumorigenesis were "ECM receptor interaction", "focal adhesion", "JAK signaling pathway", and "MAPK signaling pathway". The detail information was listed in Supplementary Spreadsheets 4 and Supplementary Fig. S6.
Association between the miR-30c-5p target genes and tumor infiltrating immune cells in gastric cancer
In order to explore the correlation between the expressions of miR-30c-5p target genes and tumor-infiltrating immune cells (TIC) abundance, CIBERSORT algorithm33 was used to screen the proportion of immune cell subsets. TA total of 375 GC patients with adjust P < 0.05 were selected for the following analysis (Supplementary Fig. S7A), the correlation of 22 tumor-infiltrating immune cells analysis demonstrated the relationship between the GXYLT2 expression and immune cells infiltration (Supplementary Fig. S7B). The difference and correlation between ADAM12 expression and tumor-infiltrating immune cells showed that 12 tumor-infiltrating immune cells were significantly correlated with ADAM12 expression (Fig. 3). Among them, macrophases M0, macrophases M2, mast cells activated, neutrophils and NK cells resting were positively correlated with ADAM12 expression. B cells memory, B cells naive, mast cells resting, monocytes, plasma cells, T cells CD4 memory resting and T cells regulatory (Tregs) were negatively correlated with ADAM12 expression. In this study, 9 tumor-infiltrating immune cells were significantly correlated with the expression of CPNE8, among which B cells naive, mast cells resting, monocytes, T cells CD4 memory resting, and T cells regulatory (Tregs) were positively correlated with the expression of CPNE8. CPNE8 expression was negatively correlated with NK cell resting, T cells CD4 memory resting, T cells CD8, and T cell follicular helper (Fig. 4). And 7 tumor-infiltrating immune cells were significantly correlated with EDNRA expression, among which macrophases M2 and mast cells rest were positively correlated with EDNRA expression. The expression of EDNRA was negatively correlated with B cells memory, plasma cells, T cells CD4 memory activated, T cells CD8, and T cells follicular helper (Fig. 5). The association analysis identified that macrophases M0 was positively correlated with STC1 expression, and B cells memory was negatively correlated with STC1 expression (Fig. 6).
The association between the proportion of tumor immune cells and ADAM12 expression. (A) Violin plots present the ratio differentiation of 22 immune cells between gastric cancer tumor samples with low or high ADAM12 expression relative to the median ADAM12 expression level, and Wilcoxon rank sum was used for significance testing. (B) Venn diagram showing immune cells with significant differences and correlations with ADAM12 expression. (C) Scatter plots show that the proportion of immune cells in 12 tumors was correlated with ADAM12 expression (P < 0.05).
The association between the proportion of tumor immune cells and CPNE8 expression. (A) Violin plots present the ratio differentiation of 22 immune cells between gastric cancer tumor samples with CPNE8 expression. (B) Venn diagram showing immune cells with significant differences and correlations with CPNE8 expression. (C) Scatter plots show that the proportion of immune cells was correlated with CPNE8 expression (P < 0.05).
The association between the proportion of tumor immune cells and EDNRA expression. (A) Violin plots present the ratio differentiation of 22 immune cells between gastric cancer tumor samples with EDNRA expression. (B) Venn diagram showing immune cells with significant differences and correlations with EDNRA expression. (C) Scatter plots show that the proportion of immune cells was correlated with EDNRA expression (P < 0.05).
The association between the proportion of tumor immune cells and STC1 expression. (A) Violin plots present the ratio differentiation of 22 immune cells between gastric cancer tumor samples with STC1 expression. (B) Venn diagram showing immune cells with significant differences and correlations with STC1 expression. (C) Scatter plots show that the proportion of immune cells was correlated with STC1 expression (P < 0.05).
Discussion
GC is a common cancer, and maintains high morbidity and mortality in all cancers34. Most patients missed the opportunity for radical surgery and received chemotherapy, radiotherapy, or targeted therapy, so the prognosis of patients with GC was poor. Abnormal gene expression may be related to the prognosis of patients35. It is necessary to discover new molecular markers for detection, surveillance, and prognostication of GC. Numerous studies have shown that miRNA can regulate gene expression by playing a key role in carcinogenesis or tumor suppression36. For example, miR-296-5p and miR-28-3p showed elevated and decreased expression in the blood of GC patients, respectively, and both high miR-296-5p expression and low miR-28-3p expression predicted poor GC patient survival37. To our knowledge, the expression levels of miRNA-30c-5p were decreased in the different types of cancers, and the miR-30c-5p might be as a tumor suppressor gene by inhibiting proliferation, migration, invasion, and metastasis of cancer cells. In pancreatic cancer progression, miR-30c-5p might play a role by inhibiting twinfilin 1 (TWF1) gene 16. MiRNA-30c-5p could suppress breast cancer survival by targeting KRAS Proto-Oncogene38, or histone deacetylase 9 (HDAC9) gene39. A recent study reported that miR-30c-5p was significantly decreased in the blood of HCC patients and could effectively discriminate HCC patients from healthy individuals40. There were also several studies on miR-30c-5p in GC. The previous reports showed that the expression level of miR-30c-5p was decreased in GC cell lines, and it could degrade metastasis-associated protein 1 (MTA1) to inhibit epithelial mesenchymal transition (EMT) process41. Moreover, the overexpression of miR-30c-5p could inhibit proliferation and induced apoptosis in GC cells12. Taken together, miR-30c-5p might act as a tumor suppressor to inhibit GC development.
In this study, the result showed that miR-30c-5p expression was remarkably lower in GC tissues (SMD = -0.42). Based on the target gene prediction tools, four miR-30c-5p target genes (ADAM12, EDNRA, STCI, and CPNE8) were identified to relate to the prognosis of GC. And then, the relationship between the expression levels of these four target genes and the diagnostic value and prognosis of gastric cancer were validated by using TCGA data. A significant association was also observed between expression levels of miR-30c-5p target genes and patient survival. Moreover, ADAM12 expression was significantly related with depth of invasion, EDNRA expression was significant related with the grade, depth of invasion, and lymphatic metastasis, and CPNE8 expression was related with grade and TNM stage in GC patients. GC patients with high expression of miR-30c-5p target genes had poor overall survival. Results of the ROC curve showed that ADAM12 and EDNRA, and CPNE8 had high value in the diagnosis of gastric cancer. These results showed that the miR-30c-5p target gene might act as an oncogene to promote the occurrence of GC.
ADAM12, one member of the ADAM (metalloproteinase) protein family, has extracellular metalloproteinase and intracellular signaling properties42. In this study, ADAM12 was significantly increased in GC and was related to prognosis. Moreover, it had a high value in the diagnosis of GC (AUC = 0.963). Another study reported that ADAM12 expression was higher in pancreatic cancer than in normal pancreatic tissue, and increased expression of ADAM12 had poor survival43. ADAM12 was also highly expressed in breast cancer and could promote cell invasion, migration, and epithelial-mesenchymal transition44.
EDNRA is a G-protein-coupled receptor for endothelin. It has been demonstrated to be overexpressed in multiple tumors, including epithelial ovarian cancer, bladder cancer cells, and prostate cancer45,46,47. EDNRA was significantly increased in bladder cancer and is related to metastasis and poor prognosis in cystic cancer patients45. It has been confirmed that EDNRA was highly expressed in GC cells, and miR-200c could regulate the proliferation and invasion of GC cells by inhibiting the expression of EDNRA48. It is consistent with our bioinformatics analysis results. Here, we found that the EDNRA mRNA was significantly higher in GC tissues compared with normal gastric tissues. What’s more, the high expression of EDNRA was associated with the survival rate and prognosis of GC and had a certain value for the diagnosis of GC (AUC = 0.717).
Stanniocalcin-1 (STC1) is a secreted glycoprotein hormone49. Several studies have reported that STC1 was closely related to the development of multiple cancers. In glioblastoma, in vitro experiments showed that STC1 could regulate glioblastoma invasion and metastasis through the TGF-β/Smad 4 signaling pathway50. In triple negative breast cancer, overexpression of STC-1 could activate the JNK/c-Jun signaling pathway to increase the invasiveness of triple-negative breast cancer cells51. In GC, high expression of STC1 in GC has been confirmed, and STC1 may play a carcinogenic role in hypoxia GC through B cell lymphoma-2 (Bcl-2) imbalance52. However, another study showed that STC1 overexpression might inhibit malignant biological behavior through PI3K/Akt signal transduction and NF-κB phosphor-P65 Ser53653. STC1 may play a regulatory role in different tumors as a tumor suppressor. According to the results of this bioinformatics analysis, STC1 was highly expressed in GC and was related to prognosis. It might also be helpful for the diagnosis of GC (AUC = 0.782).
CPNE8, a member of the Copine family, is related to a variety of tumor development. Studies have shown that CPNE8 knockout dramatically inhibits the growth of ovarian clear cell carcinoma54. In early cervical cancer (CC), CPNE8 expression was significantly increased in CC cells with knockdown of long non-coding RNA RP11-396F22.1, suggesting that RP11-396F22.1 might enhance tumor aggressiveness by negatively regulating CPNE855. So far, there is no report on the molecular mechanism of cpne8 in gastric cancer. In this study, CPNE8 was highly expressed in GC, which was related to prognosis.
Abundant pathways were found in samples with high expression of miR-30c-5p target genes, and some up-regulated pathways were identified to be associated with cell adhesion and tumors. Significant up-regulated terms and pathways included “focal adhesion”, “ECM-receptor interaction”, “Jak-STAT signaling pathway”, “TGF-beta signaling pathway”, “MAPK signaling pathway”, and so on. Among them, the interaction of ECM could directly or indirectly affect cell activity, such as metastasis and invasion of malignant tumor56. MAPK pathway was considered to be an important mechanism of gastric cancer. DNA damage inducible transcript 4 (DDIT4) could promote gastric cancer proliferation and tumorigenesis through the MAPK pathways57. These results suggested that miR-30c-5p target genes might contribute to the pathways involved in cancer, and they might be involved in the occurrence of GC, with a poor survival rate of GC patients.
Tumor microenvironment (TME) plays an increasingly important role in tumor development, in which immune cells play an important role. As in the activated state, CTLs kill target cells through granule exocytosis and Fas ligand (FasL)—mediated apoptosis induction, and secrete interferon-α (IFN-α) and tumor necrosis factor-γ (TNF-γ) to induce cancer cell cytotoxicity58. In the tumor microenvironment, NK cells mainly release perforin and granzyme to mediate tumor killing reaction and induce apoptosis of target cells59. In the context of immunotherapy, synergistic effects of PD-1/PD-L1 checkpoint inhibitors and vascular endothelial growth factor (VEGF)/vascular endothelial growth factor receptor (VEGFR) have been demonstrated, and combination therapy improves tumor response, as well as improves overall survival of gastric cancer patients60. In the present study, the correlation between the expression of miR-30c-5p target genes and tumor-infiltrating immune cells was explored in gastric cancer. The analysis indicated that macrophages M2 was positively correlated with ADAM12 and EDNRA expression. It had been reported that apolipoprotein E (ApoE) was a highly specific and effective protein in M2 macrophage derived exosomes, and contributes to tumor cell migration61, and M2 macrophages could be considered as an independent poor prognostic factor in all samples62. In this study, the expression of CPNE8 was positively correlated with T cells regulatory (Tregs). The previous report showed that Treg from GC could decompose ATP into adenosine, then induced apoptosis and inhibited the proliferation of CD8 + T cells through A2AR pathway, which further leads to immune escape of GC63.To sum up, combined with the results of this study, the crosstalk between miR-30c-5p target gene expression and some immune cells could promote the development of gastric cancer.
There are several limitations to this study. Firstly, a significant limitation is the limited number of sample sizes. The specimens from the patients and controls were not very large. Secondly, the accuracy differences between the GEO database and TCGA database might affect the interpretation of the results. Thirdly, the potential interaction between GC risk factors and miR-30c-5p target genes was not determined. To identify the mechanisms of the miR-30c-5p target genes in the GC occurrence and progression, more studies with larger samples are urgently needed.
In this study, we confirmed that the expression level of miR-30c-5p was lower in gastric cancer than in normal gastric tissues. The results demonstrated for the first time that GXYLT2 might play a role during the process of the pathogenesis in gastric cancer via several signaling pathways, and the expression level of GXYLT2 was correlated with infiltrating immune cells. The univariate and multivariate cox regression analysis suggested that GXYLT2 expression might be the independent risk factor for poor survival of GC patients, and GXYLT2 might be a potential prognostic marker in GC patients.
In this study, we confirmed that the expression level of miR-30c-5p was lower in gastric cancer than in normal gastric tissues. Moreover, four miR-30c-5p target genes were identified with up-regulated expression levels in GC and the poor prognosis of GC patients. The miR-30c-5p target gene might play roles during the process of the pathogenesis in gastric cancer via several signaling pathways, and the expression levels were correlated with infiltrating-immune cells. The higher expression might be the independent risk factor for poor survival of GC patients, and they might be potential prognostic markers in GC patients.
Methods
Analysis of miR-30c-5p expression in gastric cancer
The miR-30c-5p expression level was obtained in GC from GEO and TCGA databases. The keywords (gastric or stomach), (cancer or adenocarcinoma or tumor), and (miR or miRNA or microRNA) were used to identify the relative GEO data set. The studies included should meet the following criteria: (1) study on gastric cancer; (2) a cohort or case–control design; (3) the miR-30c-5p expression level detected from gastric cancer tissues or normal control tissues. The exclusion criteria were: (1) sample size in each group less than 5; (2) without original data of miR-30c-5p expression; (3) cell or animal experiments only; (4) diseases not about tumors or cancers. RNA-seq data of GC in the TCGA database were also used in this study. Heterogeneity between samples was analyzed by the I2 test and Cochrane's Q test. The combined effect amount was the standardized mean difference (SMD), and the overall effect size was evaluated by Cohen's classification. Begg’s regression was used to evaluate the publication bias.
Prediction of miR-30c-5p target gene
The miR-30c-5p target gene was predicted by using four online tools, which were miRDB29 (http://www.mirdb.org/), TargetScan28 (http://www.targetscan.org/vert_71/), miRWalk30 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html), and DIANA31 (http://www.microrna.gr/microT-CDS). The predicted common target genes from the four online websites were obtained by Venn diagram. Venn diagram was drawn by venndiagram package of R software.
Analysis of differentially expressed mRNAs in gastric cancer
The microarray data of GSE54129 and GSE118916 on gastric cancer were obtained from the GEO dataset. The GEO2R tool was used to identify differentially expressed genes between GC and normal tissues. The highly expressed genes in GC samples were chosen according to logFC ≥ 1.5 and adjusted P < 0.05. The common portion of highly expressed mRNAs and predicted miR-30c-5p target genes were screened out by using Venn diagrams.
Analysis of the correlation between miR-30c-5p target gene expression levels and clinicopathological features
The mRNA expression profiles of 375 gastric cancer patients and 32 healthy people were acquired from the TCGA database. "Limma package" and "beeswarm package" were used to detect the expression levels of miR-30c-5p target genes. The clinicopathological data of GC patients included the patient's gender, age, tumor grade, TNM stage, depth of invasion, lymph node metastasis, and distant metastasis. According to the median expression of the target gene of miR-30c-5p, patients with GC were further divided into two groups (high and low) to analyze the correlation between miR-30c-5p target gene expression levels and clinicopathological features.
Analysis and evaluation of prognostic value and clinical diagnostic value of miR-30c-5p target gene in GC patients
The GEPIA browser and Kaplan Meier plotter were adopted to analyze the prognostic value of target gene expression in GC patients. The Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) is an online analysis website based on the TCGA database64. The Kaplan–Meier plotter (http://kmplot.com/analysis/) is an online survival analysis tool that can estimate the effect of individual genes on GC survival based on the GEO database65. Based on the median expression levels of miR-30c-5p target genes in all samples, GC patients were divided into two groups (high and low) for survival prognosis analysis. And log-rank P values and hazard ratios (HRs) were calculated for 95% confidence intervals. In this study, univariate Cox regression analysis was carried out to identify prognostic factors by using the "survival and survminer package" of R software. The receiver operating characteristic curve (ROC) was drawn by R software "pROC package" to evaluate the diagnostic value of ROC in gastric cancer.
Gene set enrichment analysis for miR-30c-5p target gene
According to the median expression of miR-30c-5p target gene, 375 GC patients from TCGA data were divided into two groups (high expression and low expression). GSEA detected the top-ranked gene enrichment pathways in the two groups. The number of permutations of genes was set to 1000 in the GSEA analysis. The normalized enrichment score (NES), false detection rate (FDR), and nominal (NOM) P-value were used to adjudicate each enriched pathway.
Correlation analysis between miR-30c-5p target gene and tumor-infiltrating immune cells in gastric cancer
According to the CIBERSORT algorithm, the proportion of immune cells and the number of infiltrations in all gastric cancer samples in the TCGA database were calculated by using “e1071 package” and “preprocesscore package” of R software. and then samples were screened according to P < 0.05. The correlation between immune cells and miR-30c-5p target genes was calculated according to “corrplot package” of the R software, and the statistical significance was tested by Pearson coefficient. The immune cells with significant expression of miR-30c-5p target gene were obtained by Venn diagram.
Statistical analysis
Statistical analysis was performed using R version 4.0.2 software. The mRNA differential expression of miR-30c-5p target gene was analyzed by non-paired t test. A ROC curve was established to evaluate the diagnostic value of miR-30c-5p target gene mRNA expression level for GC. Pearson chi-square test was adopted to analyze the relationship between miR-30c-5p target genes and clinical characteristic variables. The Wilcoxon rank-sum test was applied for the analysis of the difference between target gene expression and tumor-infiltrating immune cells in gastric cancer, and the Pearson coefficient was used for the correlation analysis. The differences were statistically significant when P < 0.05.
Ethics approval
The data used in this study were obtained from the publicly available datasets, such as GEO database (https://www.ncbi.nlm.nih.gov/geo), and The Cancer Genome Atlas (https://portal.gdc.cancer.gov).
Data availability
Publicly available datasets were analyzed in this study, these can be found in GEO database (https://www.ncbi.nlm.nih.gov/geo), and The Cancer Genome Atlas (https://portal.gdc.cancer.gov). The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary materials.
References
Claassen, Y. H. M. et al. Impact of upfront randomization for postoperative treatment on quality of surgery in the CRITICS gastric cancer trial. Gastric Cancer 22, 369–376. https://doi.org/10.1007/s10120-018-0875-1 (2019).
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. & Lordick, F. Gastric cancer. Lancet 396, 635–648. https://doi.org/10.1016/S0140-6736(20)31288-5 (2020).
Shah, S. C., Piazuelo, M. B., Kuipers, E. J. & Li, D. AGA clinical practice update on the diagnosis and management of atrophic gastritis: Expert review. Gastroenterology https://doi.org/10.1053/j.gastro.2021.06.078 (2021).
Durães, C., Almeida, G. M., Seruca, R., Oliveira, C. & Carneiro, F. Biomarkers for gastric cancer: Prognostic, predictive or targets of therapy?. Virchows Arch. 464, 367–378. https://doi.org/10.1007/s00428-013-1533-y (2014).
Catanese, S. & Lordick, F. Targeted and immunotherapy in the era of personalised gastric cancer treatment. Best Pract. Res. Clin. Gastroenterol. 50–51, 101738. https://doi.org/10.1016/j.bpg.2021.101738 (2021).
Wang, J. et al. Intratumoral CXCR5(+)CD8(+)T associates with favorable clinical outcomes and immunogenic contexture in gastric cancer. Nat. Commun. 12, 3080. https://doi.org/10.1038/s41467-021-23356-w (2021).
Wang, S., Wu, M. & Ma, S. Integrative analysis of cancer omics data for prognosis modeling. Genes https://doi.org/10.3390/genes10080604 (2019).
Pritchard, C. C. et al. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 5, 492–497. https://doi.org/10.1158/1940-6207.Capr-11-0370 (2012).
Huang, Y. et al. MiR-301a-5p/SCIN promotes gastric cancer progression via regulating STAT3 and NF-kappaB signaling. J. Cancer 12, 5394–5403. https://doi.org/10.7150/jca.59747 (2021).
Dai, Z. T. et al. Regulation of follistatin-like 3 expression by miR-486–5p modulates gastric cancer cell proliferation, migration and tumor progression. Aging https://doi.org/10.18632/aging.203412 (2021).
Croset, M. et al. miRNA-30 family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis-associated genes. Cancer Res. 78, 5259–5273. https://doi.org/10.1158/0008-5472.CAN-17-3058 (2018).
Han, W., Mu, Y., Zhang, Z. & Su, X. Expression of miR-30c and BCL-9 in gastric carcinoma tissues and their function in the development of gastric cancer. Oncol. Lett. 16, 2416–2426. https://doi.org/10.3892/ol.2018.8934 (2018).
Lai, Y. H. et al. Collagen triple helix repeat containing-1 negatively regulated by microRNA-30c promotes cell proliferation and metastasis and indicates poor prognosis in breast cancer. J. Exp. Clin. Cancer Res. 36, 92. https://doi.org/10.1186/s13046-017-0564-7 (2017).
Zhang, Q. et al. Role of microRNA-30c targeting ADAM19 in colorectal cancer. PLoS ONE 10, e0120698. https://doi.org/10.1371/journal.pone.0120698 (2015).
Ling, X. H. et al. MicroRNA-30c serves as an independent biochemical recurrence predictor and potential tumor suppressor for prostate cancer. Mol. Biol. Rep. 41, 2779–2788. https://doi.org/10.1007/s11033-014-3132-7 (2014).
Sun, L. L., Cheng, M. & Xu, X. D. MicroRNA-30c inhibits pancreatic cancer cell proliferation by targeting twinfilin 1 and indicates a poor prognosis. World J. Gastroenterol. 25, 6311–6321. https://doi.org/10.3748/wjg.v25.i42.6311 (2019).
Busacca, S. et al. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am. J. Respir. Cell Mol. Biol. 42, 312–319. https://doi.org/10.1165/rcmb.2009-0060OC (2010).
Oh, H. K. et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin. Cancer Res. 17, 2657–2667. https://doi.org/10.1158/1078-0432.Ccr-10-3152 (2011).
Lim, J. Y. et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J. Gastroenterol. 19, 7078–7088. https://doi.org/10.3748/wjg.v19.i41.7078 (2013).
Tseng, C. W., Lin, C. C., Chen, C. N., Huang, H. C. & Juan, H. F. Integrative network analysis reveals active microRNAs and their functions in gastric cancer. BMC Syst. Biol. 5, 99. https://doi.org/10.1186/1752-0509-5-99 (2011).
Carvalho, J. et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J. Pathol. 228, 31–44. https://doi.org/10.1002/path.4032 (2012).
Yu, B. et al. MiR-148a functions as a tumor suppressor by targeting CCK-BR via inactivating STAT3 and Akt in human gastric cancer. PLoS ONE 11, e0158961. https://doi.org/10.1371/journal.pone.0158961 (2016).
Song, Y. X. et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat. Commun. 8, 289. https://doi.org/10.1038/s41467-017-00304-1 (2017).
Sierzega, M. et al. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of miR-21 and miR-331. Br. J. Cancer 117, 266–273. https://doi.org/10.1038/bjc.2017.190 (2017).
Zhang, X. et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS ONE 10, e0116979. https://doi.org/10.1371/journal.pone.0116979 (2015).
Chang, H. et al. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver 9, 188–196. https://doi.org/10.5009/gnl13371 (2015).
Kim, C. H. et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med. Genom. 4, 79. https://doi.org/10.1186/1755-8794-4-79 (2011).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife https://doi.org/10.7554/eLife.05005 (2015).
Chen, Y. & Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127–D131. https://doi.org/10.1093/nar/gkz757 (2020).
Shin, C. et al. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol. Cell 38, 789–802. https://doi.org/10.1016/j.molcel.2010.06.005 (2010).
Paraskevopoulou, M. D. et al. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 41, W169–W173. https://doi.org/10.1093/nar/gkt393 (2013).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102, 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782. https://doi.org/10.1038/s41587-019-0114-2 (2019).
Ferlay, J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359-386. https://doi.org/10.1002/ijc.29210 (2015).
Simonian, M., Mosallayi, M. & Mirzaei, H. Circulating miR-21 as novel biomarker in gastric cancer: Diagnostic and prognostic biomarker. J. Cancer Res. Ther. 14, 475. https://doi.org/10.4103/0973-1482.175428 (2018).
Chen, Y. et al. Oncogenic and tumor suppressive roles of microRNAs in apoptosis and autophagy. Apoptosis 19, 1177–1189. https://doi.org/10.1007/s10495-014-0999-7 (2014).
Guo, Y. et al. Diagnostic and prognostic value of serum miR-296-5p and miR-28-3p in human gastric cancer. Cancer Biother. Radiopharm. https://doi.org/10.1089/cbr.2020.4144 (2020).
Tanic, M. et al. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS ONE 7, e38847. https://doi.org/10.1371/journal.pone.0038847 (2012).
Liang, Z., Feng, A. & Shim, H. MicroRNA-30c-regulated HDAC9 mediates chemoresistance of breast cancer. Cancer Chemother. Pharmacol. 85, 413–423. https://doi.org/10.1007/s00280-019-04024-9 (2020).
Wong, V. C. et al. Hallmark microRNA signature in liquid biopsy identifies hepatocellular carcinoma and differentiates it from liver metastasis. J. Cancer 12, 4585–4594. https://doi.org/10.7150/jca.59933 (2021).
Cao, J. M., Li, G. Z., Han, M., Xu, H. L. & Huang, K. M. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed. Pharmacother. 93, 554–560. https://doi.org/10.1016/j.biopha.2017.06.084 (2017).
Kveiborg, M., Albrechtsen, R., Couchman, J. R. & Wewer, U. M. Cellular roles of ADAM12 in health and disease. Int. J. Biochem. Cell Biol. 40, 1685–1702. https://doi.org/10.1016/j.biocel.2008.01.025 (2008).
Veenstra, V. L. et al. ADAM12 is a circulating marker for stromal activation in pancreatic cancer and predicts response to chemotherapy. Oncogenesis 7, 87. https://doi.org/10.1038/s41389-018-0096-9 (2018).
Huang, X. et al. Adam12 and lnc015192 act as ceRNAs in breast cancer by regulating miR-34a. Oncogene 37, 6316–6326. https://doi.org/10.1038/s41388-018-0410-1 (2018).
Laurberg, J. R. et al. High expression of GEM and EDNRA is associated with metastasis and poor outcome in patients with advanced bladder cancer. BMC Cancer 14, 638. https://doi.org/10.1186/1471-2407-14-638 (2014).
Pflug, B. R. et al. Endothelin-1 promotes cell survival in renal cell carcinoma through the ET(A) receptor. Cancer Lett. 246, 139–148. https://doi.org/10.1016/j.canlet.2006.02.007 (2007).
Kar, S. P. et al. Genome-wide meta-analyses of breast, ovarian, and prostate cancer association studies identify multiple new susceptibility loci shared by at least two cancer types. Cancer Discov. 6, 1052–1067. https://doi.org/10.1158/2159-8290.Cd-15-1227 (2016).
Wei, W. et al. miR-200c regulates the proliferation, apoptosis and invasion of gastric carcinoma cells through the downregulation of EDNRA expression. Int. J. Mol. Med. 41, 1619–1626. https://doi.org/10.3892/ijmm.2017.3317 (2018).
Jellinek, D. A. et al. Stanniocalcin 1 and 2 are secreted as phosphoproteins from human fibrosarcoma cells. Biochem. J. 350(Pt 2), 453–461 (2000).
Xiong, Y. & Wang, Q. STC1 regulates glioblastoma migration and invasion via the TGF-β/SMAD4 signaling pathway. Mol. Med. Rep. 20, 3055–3064. https://doi.org/10.3892/mmr.2019.10579 (2019).
Han, J., Jeon, M., Shin, I. & Kim, S. Elevated STC-1 augments the invasiveness of triple-negative breast cancer cells through activation of the JNK/c-Jun signaling pathway. Oncol. Rep. 36, 1764–1771. https://doi.org/10.3892/or.2016.4977 (2016).
Wang, Y. et al. Stanniocalcin-1 promotes cell proliferation, chemoresistance and metastasis in hypoxic gastric cancer cells via Bcl-2. Oncol. Rep. 41, 1998–2008. https://doi.org/10.3892/or.2019.6980 (2019).
Pan, X. et al. STC1 promotes cell apoptosis via NF-κB phospho-P65 Ser536 in cervical cancer cells. Oncotarget 8, 46249–46261. https://doi.org/10.18632/oncotarget.17641 (2017).
Nagasawa, S. et al. Systematic identification of characteristic genes of ovarian clear cell carcinoma compared with high-grade serous carcinoma based on RNA-sequencing. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20184330 (2019).
Zhao, Y. et al. Overexpression of long non-coding RNA RP11–396F22.1 correlates poor prognosis of patients with early-stage cervical cancer. Am. J. Transl. Res. 10, 684–695 (2018).
Cambier, S. et al. A role for the integrin alphavbeta8 in the negative regulation of epithelial cell growth. Cancer Res. 60, 7084–7093 (2000).
Du, F. et al. DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. Cancer Commun. 38, 45. https://doi.org/10.1186/s40880-018-0315-y (2018).
Farhood, B., Najafi, M. & Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 234, 8509–8521. https://doi.org/10.1002/jcp.27782 (2019).
Habif, G., Crinier, A., André, P., Vivier, E. & Narni-Mancinelli, E. Targeting natural killer cells in solid tumors. Cell. Mol. Immunol. 16, 415–422. https://doi.org/10.1038/s41423-019-0224-2 (2019).
Voron, T. et al. Control of the immune response by pro-angiogenic factors. Front. Oncol. 4, 70. https://doi.org/10.3389/fonc.2014.00070 (2014).
Zheng, P. et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 9, 434. https://doi.org/10.1038/s41419-018-0465-5 (2018).
Liu, X. et al. Regulatory T cells and M2 macrophages present diverse prognostic value in gastric cancer patients with different clinicopathologic characteristics and chemotherapy strategies. J. Transl. Med. 17, 192. https://doi.org/10.1186/s12967-019-1929-9 (2019).
Shi, L. et al. Adenosine generated by regulatory T cells induces CD8(+) T cell exhaustion in gastric cancer through A2aR pathway. Biomed. Res. Int. 2019, 4093214. https://doi.org/10.1155/2019/4093214 (2019).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102. https://doi.org/10.1093/nar/gkx247 (2017).
Szász, A. M. et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7, 49322–49333. https://doi.org/10.18632/oncotarget.10337 (2016).
Funding
This research was funded by Graduate Innovation Project of Bengbu Medical College (Byycxz20002), and Key Projects of University Excellent Young Talents Support Plan (gxyqZD2018061). The funders had no involvement in the design, conduct, or interpretation of this study.
Author information
Authors and Affiliations
Contributions
Conception and design: Y.G.; Data acquisition: S.H., H.L., J.Z. and S.L.; Statistical analysis and interpretation of data: S.H., H.Z. and Y.G.; Drafting the manuscript: S.H. and Y.G.; Figures and tables: S.H., H.L. and Y.G. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hu, S., Liu, H., Zhang, J. et al. Effects and prognostic values of miR-30c-5p target genes in gastric cancer via a comprehensive analysis using bioinformatics. Sci Rep 11, 20584 (2021). https://doi.org/10.1038/s41598-021-00043-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-00043-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.