Abstract
An entomopathogenic fungus newly named Ophiocordyceps langbianensis was collected from Lang Biang Biosphere Reserve, located in Lam Dong Province, Vietnam. It is characterized as a species of Ophiocordyceps (Ophiocordycipitaceae, Hypocreales) having the unique characteristics of a cylindrical fertile part and several branched apical appendices. Each ascospore develops as two swollen, constricted part-spores. A phylogenetic analysis of multiple genes, including nrLSU, nrSSU, Rpb1, ITS and Tef, supported its systematic position in the genus of Ophiocordyceps; it is related to O. brunneipunctata. Based on morphological and phylogenetic analyses, O. langbianensis was confirmed as a new species from Vietnam.
Similar content being viewed by others
Introduction
The genus Ophiocordyceps, first established by Petch in 1931, belongs to the family Ophiocordycipitaceae, order Hypocreales, comprising approximately 250 species1,2. Originally, Ophiocordyceps was classified as a subgenus of Cordyceps by Kobayasi (1941, 1982) and Mains (1958)3,4,5. In 2007, Sung et al. established a new called family Ophiocordycipitaceae, comprising Ophiocordyceps, based on morphological and phylogenetic analyses6,7. The distinction of the genus Ophiocordyceps from Cordyceps was done due to the darkly pigmented stromata of Ophiocordyceps, which are pliant, wiry or fibrous and tough in texture, compared to the brightly pigmented stromata of Cordyceps7. Species of Ophiocordyceps are entomopathogenic on a wide range of insects. The hosts of species of Ophiocordyceps are the larvas of Coleoptera and Lepidoptera as well as the adults of Araneae, Diptera, Hemiptera, Hymenoptera, Odonata and Orthoptera3,4,5,6,7. Although Ophiocordyceps has worldwide distribution, the tropics and subtropics are where the highest numbers of the species are recorded. Moreover, it is considered that there is an underestimation of the number of Ophicordyceps species.
Vietnam is located in a tropical region with terrestrial ecosystems. The forests feature a rich biodiversity of both flora and fauna due to the tropical monsoon climate with high temperature and rainfall. This is a favorable environment for the development of entomopathogenic fungi. Lang Biang Biosphere Reserve is located in Lam Dong Province and comprises a vast primitive jungle with the Lang Bian Mountain at its core, one of Vietnam’s four biodiversity centers. During our expedition to discover the diversity of entomopathogenic fungi, we collected the sample DL0017. In this study, we introduce this specimen as a new species of Ophiocordyceps that parasitizes the larva of Coleoptera. We present a morphological description and phylogenetic analysis based on the phylogenetic construction of nuclear large ribosomal subunit (nrLSU), nuclear small ribosomal subunit (nrSSU) and RNA Polymerase II Subunit B1(rpb1) of species of Ophiocordyceps, including this new species.
Materials and methods
Fungal specimen collection
The specimen, DL0017, used for this study was collected from Lang Biang Biosphere Reserve (N 12°2′19.0″, E108°26′04.7″, elevation 1680 m) in 9th August, 2016. The specimen, including the host, was extracted carefully, noted, and photographed in the field using a digital camera. The specimen was immediately wrapped in wax paper, placed in a collection bag, and taken to the laboratory.
Cultivation techniques
According to the identification of conidia, phialides and colony coloration, the isolate cultures were grown on YMG media, composed of 4 g/l yeast extract (Sigma-Aldrich, Germany), 10 g/l malt extract (Sigma-Aldrich, Germany), 4 g/l glucose (Sigma-Aldrich, Germany), and incubated at 20 °C for a period of 20 days with PDA media (potato extract 4 g/l, dextrose 20 g/l, agar 15 g/l; Merck, Germany).
For fruit body induction, cultures were grown on millet substrate (millet/silkworm pupae powder = 20:1 (w/w)) and brown rice substrate (brown rice/silkworm pupae powder = 20:1 (w/w)) at 20 °C under 12 h light and 12 h darkness with relative humidity of over 90%.
Morphological study: macro- and micro-morphological analysis
Morphological observations were carried out and recorded according to the guidelines of Kobayasi and Sung et al.3,4,7. The macroscopic characteristics of the fresh fruit body were carefully observed, including the stipe, stroma, etc. Moreover, the color was noted according to Kornerup and Wanscher8. Additionally, the host insect was identified based on morphological characteristics, such as mandibulate mouthparts, antennae, shape of head and thorax. For the micro-morphological analysis, one or two perithecia were removed from the stroma and placed on a microscope slide in lactophenol-cotton blue to measure the sizes and shapes of the perithecia, asci and ascospores. Finally, the nomenclatural novelty and descriptions were deposited in MycoBank.
DNA extraction, PCR amplification, target gene sequencing
Genomic DNA was isolated by using the phenol/chloroform method (pH = 8)11. The fruiting body was incubated in a lysis buffer (2.0% SDS, Tris–HCl pH 8.0, 150 mM NaCl, 10 mM EDTA, 0.1 mg/ml Proteinase K) at 65 °C overnight. The supernatant was collected by centrifugation, and a volume of 700 μL of phenol/chloroform/isoamyl alcohol (25:24:1) was supplemented and centrifuged. The supernatant was collected and precipitated with absolute isopropanol. Finally, the isolated genomic DNA was stored in Tris–EDTA buffer at − 20 °C for further studies.
The primer pairs used to amplify nrLSU, nrSSU, rpb1, ITS and Tef regions are shown in Table 1. The final volume of PCR was done in a total of 15 μL with the thermal program: 1 cycle at 95 °C for 5 min, 40 cycles at 95 °C for 30 s, X °C for 30 s, 72 °C for 2 min, 1 cycle at 72 °C for 5 min (Note: X °C is the annealing temperatures for each target gene shown in Table 1); 5 μL aliquots of amplification product were electrophoresed on a 2.0% agarose gel and visualized in a UV transilluminator. The amplified product was sequenced at Nam Khoa (Vietnam) company.
Taxa and nrLSU, nrSSU, rpb1, ITS and tef sequences collection, DNA proofreading and phylogeny analysis
The data set of nrLSU, nrSSU, rpb1, ITS and tef sequences were established by sequences downloaded from Genbank (NCBI) and based on the previous data published by Sung et al.7. The nrLSU, nrSSU, rpb1, ITS and tef were noted with accession number, name of taxon and locality. The amplified DNA sequences were proofread to remove ambiguous signals at both ends by different software, including Seaview 4.2.12 and Chromas Lite 2.1.1. The phylogenetic tree was constructed based on neighbor-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML), using Molecular Evolutionary Genetics Analysis (MEGA) version 5. Additionally, the best evolution model was predicted using jModelTest.
Results
Taxonomy
Ophiocordyceps langbianensis T. D. Lao, T. A. H. Le & N. B. Truong, sp. nov.
Mycobank MB836716 Figs. 1, 2, 3.
Asexual states. (A) Ascospores germinating after 96 h; (B) Septate hyphae, branched, conidia in intercalary or terminal cells; (C) White colony after 45 days on YMG media; (D) Aerial hyphae with divergent phialides; (E) Elliptical conidia in chains after release from the phialide, (F, G) Stromata growing on cereal substrates.
Typification
VIETNAM. Lam Dong Province, Lang Bian Biosphere Reserve, Lang Bian mountain: N 12°02′19.0″, E108°26′04.7″; elevation 1680 m; humidity: over 85%; temperature: day 20 °C–22 °C, night: 14 °C–16 °C; collected between 9h00–15h00 of the day on 9 August, 2016, from the larva of a beetle of Coleoptera in moist soil surrounded by dried leaves. Truong B.N. DL0017 (Holotype DLU; Iso VNMN, DLU).
Distribution
Vietnam, only known from Lang Bian Mountain.
Etymology
“Langbianensis” refers to Lang Bian Mountain, Lam Dong province, Vietnam.
Host
On the larva of a beetle of Coleoptera. Larva: 28–32 mm long, hard-body, shiny, smooth, dark brownish yellow; body composed of 13 segments with black edges; larva with three pairs of jointed legs attached to thorax.
Habitat
Individuals of associated species appeared at the type locality, including pioneer species such as Acer laurinum (Aceraceae), Baccaurea harmandii (Euphorbiaceae), Castanopsis chinensis (Fagaceae), Eriobotrya poilanei (Rosaceae), Jasminum longisepalum (Oleaceae), Phoebe petelotii (Lauraceae) and Tetrastigma lanceolarium (Vitaceae).
Sexual morph
Stroma arising from the head of the host larva, solitary, rarely branched, 40–100 mm long; host covered with thin, tough layer of mycelium. Stipe filiform, cylindrical, 30–67 mm × 0.7–1.0 mm, pale yellow. Fertile portion, cylindrical, 7.0–14.0 mm × 1.5–2.0 mm, brownish yellow with dark brown ostiolar dots of perithecia. Apical appendices, pale yellow, 2–10 primary or secondary branches, 4.0–10.0 × 0.5 mm. Perithecia immersed, ovate or pyriform, 260–400 μm × 100–190 µm. Asci, cylindrical, 200–250 μm × 5.0–6.0 μm, with thickened cap. Ascospores filiform, multiseptate, articulated in long-chain after discharging, sometimes breaking into 1-celled part spores, cylindrical, swollen, two waist-like constrictions, 5–7.5 μm × 1.3–2 µm.
Asexual morph
Germination of ascospores after 48 h on PDA; white colony, slow growing on YMG and PDA media, 25.00 mm and 24.58 mm after 40 days (respectively); septate hyphae, branched, chlamydospores developing in intercalary or terminal cells. Aerial hyphae with divergent phialides; elliptical conidia in chains after release from phialide. Stromata without fertile part forming on cereal substrates. Minor differences in morphological characteristics of stromata developing from different substrates. Stromata, white, branched when developing on millet substrate; brownish yellow, solitary, rarely branched, when developing on brown rice substrate.
Amplification of nrLSU, nrSSU, rpb1, ITS and tef genes
Target genes, including nrLSU, nrSSU, rpb1, ITS and tef, were successfully amplified with corresponding primers (Table 1). The bands of 950-bp, 1102-bp, 803-bps, 700-bps, and 1030-bps corresponding to the amplified nrLSU, nrSSU, rpb1, ITS and tef were observed in the electrophoresis on 2.0% agarose gel. The PCR products were sequenced with the signal of the peaks in both strands of target genes; the sequence was significant, unique and good for reading.
The systematic concatenated nrLSU, nrSSU, rpb1, ITS and tef gene dataset
To construct a phylogeny of major lineages, representative taxa were chosen based on previous study7. The data set of nrLSU, nrSSU, rpb1, ITS and tef consisted of 50, 50, 46, 39 and 42 taxa representing the morphological and ecological diversity of genera in Ophiocordycipitaceae, Clavicipitaceae, and Cordycipitaceae, including the outgroup taxon Glomerella cingulata (Glomerellaceae, Glomerellales) (Table 2). A combined concatenated dataset consisting of 30 representative taxa was constructed based on the list of individual target genes.
Molecular phylogeny analysis
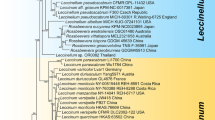
The sequences of nrLSU, nrSSU, rpb1, ITS and tef of DL0017 were similar to the representative sequence of Cordyceps brunneipunctata (similarity > 90%), with accession numbers of DQ518756, DQ522542, DQ522369, GU723777 and DQ522324. Sequences were aligned and edited using the MEGA 5.2. Gaps were excluded from the phylogenetic analysis. The dataset of representative taxa and DL0017 target gene sequence consisted of 451 bp for nrLSU, 674 bp for nrSSU, 392 bp for Rpb1, 158 bp for ITS and 790 bp for tef. The evolution model that was most fixed with nrLSU, nrSSU, Rpb1, ITS and tef were TN93 + G, K2 + G + I, T92 + G + I, K2 + G, and TN93 + G + I respectively. The phylogenetic trees were generated with Neighbor Joining (NJ), Maximum Parsimony (MP), and Maximum Likelihood (ML) methods with replication of 1000. Based on the NJ, MP, and ML phylogenetic trees, individual nrLSU, nrSSU, Rpb1, ITS, and tef of DL0017 clustered together with Ophiocordyceps brunneipunctata within separate branches with credible bootstrap (≥ 50%), suggesting that these species are related (Table 3).
Information from molecular phylogenetic analysis based on separate genes is not enough to reconstruct trees for higher classification compared to multigene analysis. Therefore, a combined data set, including 2,319 bp of five target genes, nrLSU-nrSSU-Rpb1-ITS-tef, was analyzed. The evolution model that was most fixed with the combined dataset was TN93 + G + I, as determined by MEGA 5.2. The phylogenetic trees, based on analysis of the combined data, could be broadly separated into three groups, which corresponded to the families of Clavicipitaceae, Ophiocordycipitaceae and Cordycipitaceae. In the phylogenetic tree, DL0017 clustered with Ophiocordyceps brunneipunctata with bootstraps of 100/100/100 (NJ/MP/ML phylogenetic tree) and formed a separate, monophyletic branch. Within this monophyletic branch, DL0017 and O. brunneipunctata clustered together closely, suggesting that these species were truly associated (Fig. 4). The molecular phylogenetic analysis confirmed that there were differences between DL0017 and other related species.
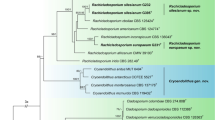
To confirm the authenticity of DL0017 as the most closely associated with Ophiocordyceps brunneipunctata, the reconstruction of Neighbor-Net network of DL0017 and its allies was performed. The Neighbor-Network analysis supported the results from the phylogenetic analysis (Fig. 5). The network presented three complex groups, corresponding to three families: Clavicipitaceae, Ophiocordycipitaceae and Cordycipitaceae. The DL0017 closely clustered with Ophiocordyceps complex. Additionally, speciation was observed between the cluster of DL0017 and O. brunneipunctata.
Comparison of Ophiocordyceps langbianensis with close species
In the phylogenetic analysis, the Ophiocordyceps langbianensis clustered with Ophiocordyceps brunneipunctata with high bootstrap support, suggesting a close relationship. To confirm the authenticity of DL0017 as a new species, we compared DL0017 and its close species, O. brunneipunctata. It differed from O. brunneipunctata by the morphological characteristics described in Table 4. Therefore, DL0017 was confirmed as a new species, namely O. langbianensis.
Discussion
Lang Biang Biosphere Reserve, located in Lam Dong Province, is classified as Vietnam’s biodiversity center and considered a hotspot of fungal biodiversity, including entomopathogenic fungi. During our expedition to validate the diversity of entomopathogenic fungi in Lang Biang Biosphere Reserve, the sample DL0017 was collected.
Morphological analysis indicated that DL0017, named Ophiocordyceps langbianensis, is a new taxon. Species belonging to the family Ophicordycipitaceae have stromata that are darkly pigmented or rarely brightly colored), tough, fibrous, pliant, and rarely fleshy. Additionally, asci are usually cylindrical with thickened ascus apex. Ascospores are usually cylindrical, multiseptate, and disarticulate into part-spores or non-disarticulating7. Our specimen shares these common characteristics.
Based on the phylogenetic analysis, the specimen DL0017 clustered with Ophiocordyceps brunneipunctata in Ophiocordycipitaceae12. However, the morphologies of these two species are different in many characteristics, including color, size of stroma, stipe, and dots in the fertile portion. The apical appendix of O. brunneipunctata lacks branching, while O. langbianensis has 2–10 branches. Additionally, the ascospores of O. brunneipunctata break into part-spores, while the ascospores of O. langbianensis stick together to form a multiseptate chain, separating into unicellular part-spores under a strong interaction force. Multiple gene sequences of the related Ophiocordyceps species were used in the phylogenetic analysis. A comparison was done among the species listed in Table 4 with respect to cylindrical fertile portion, embedded perithecia, and an apical appendix. Among them, only species of Cordyceps furcicaodata have a branch-forming apical appendix. In the comparison between Cordyceps furcicaodata and O. langbianensis, Cordyceps furcicaodata was found to be smaller than O. langbianensis. The stroma of Cordyceps furcicaodata arose from the middle of the host larva, while that of O. langbianensis arose from one end of the insect larva13. As mentioned above, ascospores of O. langbianensis stick together to form a multiseptate chain, which could only be ruptured into unicellular part-spores by a strong force, while ascospores of Cordyceps furcicaodata often break into unicellular part-spores.
The asexual morph of O. langbianensis consists of long and divergent phialides, elliptical conidia usually in chains considered paecilomyces-like or purpureocillium-like14,15. Conversely, O. bruneipunctata produced a mononematous hirsutella-like asexual morph from colonies after 3–4 weeks.
Conclusion
We successfully applied morphological characterization in combination with phylogenetic analysis of multiple genes, including nrLSU, nrSSU, rpb1, ITS, and Tef, to delimit sample DL0017, collected from Lang Biang Biosphere Reserve located in Lam Dong Province, Vietnam, as a new species named Ophiocordyceps langbianensis, belonging to the genus of Ophiocordyceps (Ophiocordycipitaceae, Hypocreales).
References
Petch, T. Notes on entomogenous fungi. Trans. Br. Mycol. Soc. 21, 34–67 (1973).
Spatafora, J. W. et al. New 1F1N species combinations in Ophiocordycipitaceae(Hypocreales). IMA Fungus. 6, 357–362. https://doi.org/10.5598/imafungus.2015.06.02.07 (2015).
Kobayasi, Y. The genus Cordyceps and its allies. Sci. Rep. Tokyo Bunrika Daigaku Sect. B 5, 53–260 (1941).
Kobayasi, Y. Keys to the taxa of the genera Cordyceps and Torrubiella. Trans. Mycol. Soc. Jpn. 23, 329–364 (1982).
Mains, E. B. North American entomogenous species of Cordyceps. Mycologia 50, 69–222 (1958).
Castlebury, L. A., Rossman, A. Y., Sung, G. H., Hyten, A. S. & Spatafora, J. W. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 108, 864–872. https://doi.org/10.1017/s0953756204000607 (2004).
Sung, G. H. et al. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. https://doi.org/10.3114/sim.2007.57.01 (2007).
Kornerup, A. & Wanscher, J. H. Methuen Handbook of Colour (Eyre Methuen, London, 1981).
Vilgalys, R. & Sun, B. L. Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc. Natl. Acad. Sci. USA 91, 4599–4603. https://doi.org/10.1073/pnas.91.10.4599 (1994).
White, T. J., Bruns, T., Lee, S. & Taylor, J. In PCR Protocols: A Guide to Methods and Applications 315–322 (Academic Press, London, 1990).
Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15, 532–537 (1993).
Liang, Z. Q., Liu, A. Y., Liu, M. H. & Kang, J. C. The genus Cordyceps and its allies from the Kuankuoshui Reserve in Guizhou III. Fungal Divers. 14, 95–101 (2003).
Hywel-Jones, N. L. Cordyceps brunneipunctata sp. nov. infecting larvae in Thailand. Mycol. Res. 99, 1195–1189 (1995).
Samson, R. A., Mahmood, T. The genus acrophialophora (fungi, moniliales). Acta. Bot. Neerl. 19(6), 804–808 (1970).
Luangsa-ard, J. et al. Purpureocillium, a new genus for the medically important Paecilomyces lilacinus. FEMS Microbiol. Lett. 321(2), 141–149 (2011).
Acknowledgements
We express our special thanks to National Foundation for Science and Technology Development (NAFOSTED) under the grant number 106-NN.06.2015.44, Vietnam; Ho Chi Minh City Open University for the genuine support throughout this research work under the grant number E2019.06.3. We also thank Dr. Hiep Minh Dinh, Son Kim Hoang, Hanh Van Trinh, Mai Hoang Nguyen, and Dr. Tien Van Tran for their assistance.
Author information
Authors and Affiliations
Contributions
N.B.T. collected the sample DL0017. T.D.L., T.A.H.L. conceived, planned and carried out the experiments and contributed to the interpretation of the results; T.D.L. took the lead in writing the manuscript. All authors provided critical feedback and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lao, T.D., Le, T.A.H. & Truong, N.B. Morphological and genetic characteristics of the novel entomopathogenic fungus Ophiocordyceps langbianensis (Ophiocordycipitaceae, Hypocreales) from Lang Biang Biosphere Reserve, Vietnam. Sci Rep 11, 1412 (2021). https://doi.org/10.1038/s41598-020-78265-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78265-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.