Abstract
Aberdeen Angus calves were sacrificed from immediately post-birth up to 96 days of age (DOA) and ileal samples were collected for microbial, histological and immunological analyses. Firmicutes bacteria were established immediately in the ileum of calves after birth and remained the dominant phyla at all time points from birth until 96 DOA. Temporal shifts in phyla reflected significantly increased Bacteroidetes at birth followed by temporal increases in Actinobacteria abundance over time. At a cellular level, a significant increase in cell density was detected in the ileal villi over time. The innate cell compartment at birth was composed primarily of eosinophils and macrophages with a low proportion of adaptive T lymphocytes; whereas an increase in the relative abundance of T cells (including those in the intra-epithelial layer) was observed over time. The ileal intestinal cells were immunologically competent as assessed by expression levels of genes encoding the inflammasome sensor NLRP3, and inflammatory cytokines IL1A, IL1B and IL33—all of which significantly increased from birth. In contrast, a temporal reduction in genes encoding anti-inflammatory cytokine IL10 was detected from birth. This study provides an integrated baseline of microbiological, histological and immunological data on the immune adaptation of the neonatal ileum to microbial colonisation in calves.
Similar content being viewed by others
Introduction
Recent studies focussing on the detection and characterisation of the microbiome that confronts every neonate has led to an expansion in our understanding of the prokaryotic species which live in and on eukaryotes, including humans1. In livestock species, characterisation has been more limited but a predominant focus on the gastro-intestinal (GI) tract of ruminants has led to comprehensive characterisation of the initial aerobic and facultative anaerobic bacteria such as Staphylococcus, Streptococcus, Enterococcus and Enterobacteria which utilise available oxygen and permit the subsequent establishment of anaerobes2,3,4. In the days and weeks following birth the establishment of methanogens, fungi and protozoa occurs5. It is now known that the neonatal ruminant GI tract represents a series of unique ecological niches harbouring distinct microbiomes that will play key roles in digestion, health and animal production of the adult animal6,7,8.
In the hours and days following birth, the neonatal calf receives only a liquid diet of colostrum followed by milk which provides early microbes which colonise the intestine9. At this stage, a specialised structural adaptation of the ruminal wall—known as the oesophageal groove, routes milk directly to the abomasum and ruminal development does not occur until solid food is introduced. Along with passive transfer of maternal antibodies, colostrum and milk also deliver growth factors (insulin-like growth factor-IGF-1 and hormones) to the neonate which influence gut development by increasing epithelial cell proliferation and villi and crypt growth in the small intestine10. Despite the detailed knowledge now available on the prokaryotic complexity within the neonate, how the host recognises, responds and adapts to it over time, remains poorly understood. Furthermore, this initial colonisation has important consequences for multiple complex physiological processes, not least of which is the appropriate development of the immune system, and all of which must be tightly regulated to prevent inflammation-associated tissue pathology and death.
While anatomically formed pre-partum, functional development of the ruminant immune system occurs post-partum. Prior to adaptive immune system development, the neonate principally relies on the innate immune system for protection from infection. It is now recognised that these early host-microbe interactions also contribute to the programming of the immune system5. Although the rapid colonisation of the mucosal surfaces including the host intestine by microbial communities might be expected to cause pathological inflammation, the mechanisms preventing uncontrolled inflammation and maintaining homeostasis are not fully understood. Some regulatory strategies have been identified including a mechanism by which microorganisms produce signals to promote maturation of regulatory immune cells and prevent hyper-activation of the immune response11,12. The term ‘inflammatory anergy’ is used to describe the temporary silencing of inflammatory responses during the establishment of a protective microbiome. However, the empirical evidence for these mechanisms are lacking in non-model organisms.
In cattle, the richest site of immune cell deposition along the intestine is in the ileum. The ileum is part of the small intestine which in its submucosa contains a high concentration of lymphoid nodules called Peyer's patches. These nodules form the gut-associated lymphoid tissue that is referred to as the “immune sensor” of the intestine13. These immune centres lined by follicle-associated epithelium (FAE) are critical for the distinction between pathogenic and commensal bacteria. Part of the epithelial layer contains specialised M cells which function as transporter cells (luminal proteins as well as antigens) from the intestinal lumen to the immune cells, and thereby serve as a gateway to tolerance or immune activation. These interactions are mediated via cellular receptors on resident epithelial cells as well as immune cells, referred to as Toll-like receptors (TLRs). Epithelial cells lining the GIT of germ-free animals had reduced expression of TLRs, suggesting commensal gut microbes promote the expression of these receptors14. Lotz et al. reported that down-regulation of the TLR4 signalling pathway which detects bacterial LPS prevents an inflammatory response15. One of the few studies conducted in calves describes down-regulated mRNA expression levels for several TLRs and antimicrobial peptides in the small intestine in the weeks after birth compared to calves at 6 months16. Additionally, IL1 and IL6 gene expression in the calf jejunum has been recently shown to be affected by method of birth17. In cattle, FAE of Peyer's patches have a critical role in trafficking not only colostral leukocytes into neonatal circulation18 but also infectious disease antigens including mycobacteria and thereby play a critical role in the immune response to these agents19. A number of problematic enteric infections in cattle are known to target the M cells in the intestine20,21,22, therefore suggesting that host–pathogen interactions in this region are of critical importance. However, there is limited knowledge on the pattern of tissue and immune system maturation in early life and how colonising microorganisms interact with the host to promote homeostasis.
The aim of this study was to comprehensively characterise the combined changes in microbial diversity and the development of the immune response occurring in conjunction with morphological development of the ileum in calves, from birth until 96 days of age. Understanding the integration, orchestration and ontogeny of host-microbe interactions in early life is key to the design of new strategies to protect calves against opportunistic bacterial and viral infections.
Results
Effect of age on ileal bacterial community dynamics of new-born calves
Bacterial 16S amplicon sequencing generated 29,169,392 total sequences and after initial processing, an average of 788,362 high-quality sequences were obtained per ileal sample, with read numbers ranging from 207,609 to 3,071,093 (median = 583,507). The vast majority of reads were successfully classified with only 0.39% 3.4% at 20.87% remaining unclassified at phylum, family and genus levels, respectively.
Twenty bacterial phyla were identified in the ileum of calves throughout early life (Supplementary Table 1). However, only 5 of those were present at abundances > 5% at any one sampling time point and are shown in Fig. 1A. Firmicutes dominated the ileum of new-born calves and these remained the most dominant phyla across the 96 day sampling window, accounting for 83% of total bacterial sequences detected at birth and ranging from 53–86% across time points up to 96 DOA. Proteobacteria (7.86%), Verrucomicrobia (5.61%) Bacteroidetes (1.96%), Actinobacteria (0.81%), Chlamydiae (0.39%) and Synergistes (0.28%) phyla combined to make up the remaining fractions of bacterial sequences (combined total 17%) at birth. At family level, Streptococcaceae were strongly dominant with 76.21% of all bacterial sequences assigned to this family. Other families present at high relative abundance included Verrucomicrobiaceae (5.6%), Pasteurellaceae (3.37% ± 0.33%), Enterobacteriaceae (2.12%), Campylobacteriaceae (1.59%), Lachnospiraceae (1.28%), Veillonellaceae (0.53%) and Synergistaceae (0.28%). Together these 8 families comprised approximately 91% of all bacterial sequences detected. At birth, the majority of all sequences (71.21%) were assigned to the genus Streptococcus, with Akkermansia (5.6%), Lactococcus (4.92%), Gallibacterium (2.9%), Escherichia (2.11%) and Campylobacter (1.59%) the next most relatively abundant genera. All other genera were present in lower relative abundances, each comprising less than 1% of total bacterial sequences obtained from the ileum samples.
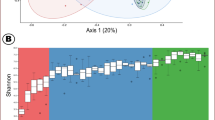
(A) Mean relative abundance (%) of sequences assigned to dominant bacterial phyla present in the ileum of calves at 6 sampling time points (birth—0, 7, 14, 21, 28 and 96 days of age, DOA). N = 3, mean ± SEM. Statistical significance is denoted as *, **, *** and **** for P values < 0.05, < 0.01, < 0.001 and < 0.0001, respectively. For clarity of interpretation, only significant changes from birth (0 DOA) are shown. (B) Non-metric multidimensional scaling (nMDS) plot showing clustering of bacterial community structure, at genus level, in the ileum of calves based on age.
Non-dimensional multiscaling (nMDS) and PERMANOVA were used to determine if significant differences existed between bacterial community structures in the ileum of calves across different age groups up to 96 DOA. Age-related clustering of bacterial community profiles was observed at phylum, family and genus level, but only clustering at genus level is shown in the nMDS plot displayed in Fig. 1B. Bacterial community structures in calves at 96 DOA can be observed to cluster quite a distance away from communities at all other time points. Similarly, bacterial communities in new-born calves (0 DOA) and calves at 7 DOA were very distinct from all other time points, with the exception of a couple of outliers. PERMANOVA analysis confirmed that bacterial community structures present in the ileum of calves differed at all 6 time points as observed on the nMDS plot (P = 0.0001).
Bacterial phyla contributing to differences in community structures at the different time points were identified by similarity percentage (SIMPER) analysis (Supplementary Table 2). Although Firmicutes was the dominant phylum at all times sampled, changes in the relative abundance of this phylum were not the main driver of dissimilarity in community structures. The main contributor to differences in community structures at most time points was another dominant phylum, Bacteriodetes. The relative abundance of this phylum increased from birth to 7 DOA and this explained 19.7% of the dissimilarity between community structures of new-born and calves at 7 DOA. Decreases in the relative abundance of Verrucomicrobia, Chlamydiae, Synergistetes and Fibrobacteres also contributed to the dissimilarity in bacterial community structures at the different time points. The relative abundance of Proteobacteria declined from birth until 28 DOA and while this phylum didn’t contribute to any differences observed at birth, it did contribute after that point. In contrast, the relative abundance of Actinobacteria increased from birth to 28 DOA but had declined by 96 DOA when it explained ~ 11% of the dissimilarity observed between bacterial communities in 28-day and 96-day old calves.
At genus level, relative abundance of Streptococcus, Gallibacterium, Akkermansia and Lactococcus in the ileum was lower at 7 DOA compared to birth, while relative abundances of Bacteroides, Prevotella, Subdoligranulum and Faecalbacterium were all increased. These bacteria explained roughly 15% of the dissimilarity between bacterial community structure in the ileum of new-born calves and week-old calves. Sequences assigned to the genera Subdoligranulum and Bacteroides had decreased by 14 DOA relative to other bacteria. Bifidobacterium, Sharpea, Olsenella, Veillonella, Prevotella and Gallibacterium all increased in relative abundance at 14 DOA and together explained over 15% of the dissimilarity between bacterial community profiles at 7 DOA and at 14 DOA.
At 21 DOA, the relative abundance of Streptococcus, Lactococcus, Veillonella and Clostridium sensu stricto had all decreased whereas Succinivibrio, Allobaculum, Faecalibacterium, Sutterella and unclassified genera from Alphaproteobacteria and Bacteroides had all increased. When combined, these genera accounted for 15% of the dissimilarity in ileum bacterial structure between time points. At 28 DOA, the relative abundance of Succinivibrio, Allobaculum, Faecalibacterium, Prevotella and unclassified Alphaproteobacteria and Bacteroides again decreased while Romboutsia and Sharpea increased. Finally at 96 DOA, there were decreases in the relative abundances of Bifidobacterium, Sharpea and Streptococcus while relative abundances of Saccharofermentans, Lysinibacillus, Turicibacter, Escherichia and unclassified genera from Ruminococcaceae, Lachnospiraceae and Candidatus Saccharibacteria all increased and accounted for almost 14% of the dissimilarity calculated in bacterial community structure in the ileum of calves 28 days after birth (pre-weaning) and the ileum of calves 96 days after birth (post-weaning).
At phylum level, bacterial richness and diversity (based on the Shannon–Wiener index) decreased between birth and 7 DOA (P = 0.0005 and P = 0.003 respectively). Bacterial richness returned to levels similar to those in new born calves on 14 DOA (P = 0.004) (Supplementary Table 3). At the genus level, bacterial richness, evenness and diversity also dropped between birth and 7 DOA (P = 0.0005) and fluctuated thereafter. Interestingly, despite these fluctuations no significant differences (P > 0.05) were observed in bacterial richness, evenness or diversity at phylum or genus level between the ileum of new-born calves and 96-DOA calves.
Morphological changes in the ileum of new-born calves from birth to 96 DOA
The length of villi in the ileum of calves significantly decreased between birth and 7 DOA (P = 0.008, Fig. 2A) but there was no significant difference in villus length at 96 DOA compared to new-born calves (P = 0.8, Fig. 2B). The width of the villi was similar across all age groups.
(A) Villi length and (B) width (mm) in the ileum of calves at 6 sampling time points (birth—0, 7, 14, 21, 28 and 96 days of age, DOA). (C) Average immune cell counts (macrophages, eosinophils, T lymphocytes), including the epithelial layer (intra-epithelial lymphocytes, IEL) per villus. N = 3 calves per group. Statistical significance is denoted as *, **, *** and **** for P values < 0.05, < 0.01, < 0.001 and < 0.0001, respectively. For clarity of interpretation, only significant changes from birth (0 DOA) are shown.
Overall, the average number of immune cells per ileal villi markedly increased from 14 DOA with significant increases at 14 (P < 0.05), 21 and 28 DOA (P < 0.0001) (Supplementary Table 4). By 96 DOA, total cell numbers almost doubled in number showing significant sustained cellular infiltration into the villi (P < 0.0001, Fig. 2C). Neutrophils were observed in low numbers in the lamina propria mucosa with occasional exocytosis in two animals each at D7, D14 and D96 and one animal each at D21 and D28 (data not shown). Eosinophils and macrophages were found distributed evenly throughout the lamina propria mucosa at the level of the villi and crypts (Fig. 2C), and these innate cells were the most abundant. In addition, single macrophages were observed in moderate numbers in the submucosa overlying the Peyer’s patches and in the follicles in all age groups. A significant increase in macrophage number was detected at 28 and 96 DOA compared to birth (P < 0.05) and macrophages remained the most abundant cell type throughout the 96 day sampling period. In contrast, eosinophils displayed s significant decrease at 7 and 14 DOA relative to birth (P < 0.05, Fig. 2C).
Numbers of adaptive cells were lowest at birth but T lymphocyte populations steadily increased from birth with significant increases at 21, 28 and 96 DOA in T cells (P < 0.01). Adaptive T-cells showed a similar mucosal distribution and also infiltrated the mucosal epithelial lining cells (referred to as intra-epithelial lymphocytes). From 14 DOA, the number of intra-epithelial lymphocytes significantly increased up to and including 96 DOA (P < 0.01, Fig. 2C). Moderate numbers of T-cells were seen in the submucosa overlying the Peyer’s patches and few single T-cells were seen within the follicles at D28 and D96. In the lamina propria mucosa only single B-cells were detected in all age groups and these cells were therefore not included in the cell count. Dilated lymphatics in the villi were occasionally filled with B-cells at D7, D14 and D28. Further, B-cells were diffusely seen in the lymphoid follicles of the Peyer’s patches. In addition, moderate numbers of B-cells were also observed in the submucosa overlying the Peyer’s patches (data not shown). In the dome area single T-cells were observed at day 0 which increased with age (Supplementary Fig. 1). Moderate numbers of B-cells populated the dome area and infiltrated the FAE. These numbers increased slightly with age as did the number of B-cells infiltrating the FAE. Similarly, macrophages within the lymphoid tissue of the dome increased in number with age.
Representative images of the ileal villi of calves are shown in Fig. 3 across age groups from birth—0 (a to c), 7 (d to f), 14 (g to i), 21 (j to l), 28 (m to o) and 96 (p to q) DOA. The number of eosinophils infiltrating the lamina propria mucosa of the villi is decreased by approximately 50% on 7 and 14 DOA compared to at birth. The number of eosinophils increases on 28 DOA with highest levels seen at 96 DOA. T-cells, observed in the lamina propria mucosa and in the mucosal epithelium, clearly increase with age. The numbers of macrophages infiltrating the lamina propria mucosa of the villi also increase in number until 28 DOA of age.
Calf ileum plate; Representative images of the ileal villi of calves from 6 different age groups; birth—0 (a to c), 7 (d to f), 14 (g to i), 21 (j to l), 28 (m to o) and 96 (p to q) days of age (DOA). First column H&E staining. Second column, Anti-CD3 immunohistochemical staining (brown), counterstained with Mayer’s haematoxylin. Third column, Anti-Iba1 immunohistochemical staining (brown), counterstained with Mayer’s haematoxylin. Scale bar = 50 µm.
Correlations between immune cell abundance and relative abundance of bacteria in the ileum of calves during early life
Correlations between the three dominant immune cell types (eosinophils, macrophages and T lymphocytes) and dominant bacteria revealed some informative correlations at both phylum and genus level. Average T lymphocyte number in the ileal villi was positively correlated with the relative abundance of phyla Candidatus Saccharibacteria but negatively correlated with Verrucomicrobia. In contrast, average eosinophil number per villi was negatively correlated with the relative abundance of the phyla Bacteroidetes (Table 1).
The abundance of intraepithelial T cells, which play an important role in pathogen resistance and tissue tolerance, was negatively correlated with the relative abundance of the phyla Proteobacteria, Verrucomicrobia, Tenericutes, Synergistes and Fusobacteria in the ileal digesta (Supplementary Fig. 2). A low positive correlation was detected between intraepithelial T cell abundance and the relative abundance of unclassified bacteria, but no other statistically significant correlations were observed.
At genus level, average T cell number in the villi was positively correlated with the relative abundance of Olsenella, Saccharofermentans, Turicibacter and unclassified genera of Candidatus Saccharibacteria and Lachnispiraceae and a negative correlation with Lactococcus, Streptococcus, Succiniclasticum, Campylobacter, Gallibacterium and Akkermansia (Supplementary Fig. 3).
T cell abundances in the intraepithelial layers were also negatively correlated with many of the same bacterial genera (Lactococcus, Streptococcus, Succiniclasticum, Campylobacter, Gallibacterium and Akkermansia). Some moderate positive correlations were observed between T cell populations in the intra-epithelial layers of the villi and the relative abundance of Turicibacter and unclassified genera of Lachnospiraceae and Candidatus Saccharibacteria (Supplementary Fig. 4). In contrast, eosinophil abundance in the villi had no strong positive correlation with any bacteria genera and showed only moderate negative correlations with Blautia and Subdoligranulum.
Likewise macrophage abundance in the villi showed no strong positive correlations with the abundance of specific bacterial genera in the ileum digesta, and only moderate negative correlations were observed with Subdoligranulum (Supplementary Fig. 3). No statistically significant correlations were detected between the remaining bacterial phyla or genera and the abundance of any of the immune cells counted in the villi of calves between birth and 96 DOA.
Host gene expression in ileum over time
Expression levels of genes encoding cytokines associated with pro- and anti-inflammatory pathways of the host immune system were measured by qPCR. A consistent trend of increased expression of the chemokine IL8 was observed from birth and the increase reached statistical significance at 96 DOA (Fig. 4). A similar increase in expression from birth was also detected for the members of the IL1 family of proinflammatory cytokines. Expression of IL1A was significantly increased at 7 and 14 DOA, relative to birth; and IL1B expression was significantly increased at the same time points (P < 0.05). In contrast to this pattern of activation, both anti-inflammatory genes analysed showed a distinct reduction in levels present from birth with a significant reduction in IL10 by 7 DOA. Interestingly, a biphasic response pattern was detected for the gene encoding cytokine IL33 with two significant increases from birth to 7 and 28 DOA, respectively (P < 0.05). Expression of the inflammasome sensor NLRP3 gene was also significantly increased at 7 DOA relative to birth (P < 0.05). Pearson’s correlation analysis showed no strong relationships between the abundance of different immune cells and the expression of genes encoding cytokines in the ileal villi (Supplementary Fig. 5A). However, a strong positive correlation was detected between intraepithelial T cell abundance and the expression of IL1β and a negative correlation between IEL T cell abundance and IL10 (Supplementary Fig. 5B).
Relative expression of immune genes (A) IL8; (B) IL1A; (C) IL1B; (D) IL10; (E) TGFB; (F) IL33 and (G) NLRP3 in the ileum of calves at birth (D0), day 7 (D07), day 14 (D14), day 28 (D28) and day 96 (D96). N = 3 calves per group. Statistical significance is denoted as *, **, *** and **** for P values < 0.05, < 0.01, < 0.001 and < 0.0001, respectively. For clarity of interpretation, only significant changes from birth (0 DOA) are shown.
Discussion
Research on the ruminant gut microbiome has increased in recent years, with a wealth of information now available in relation to microbial abundance and environment-related shifts in bacterial populations5,16,23,24,25. However, establishment of the site-specific microbiome in conjunction with structural post-natal adaptations, as well as the implications for neonatal immunity have to-date generally been studied as separate aspects of ruminant biology. The majority of studies investigating the developmental ontogeny of the digestive tract in the neonatal calf have tended to focus only on a single time point and usually on the rumen, which is understandable given that it accounts for approximately 70% of the entire stomach of the adult animal and represents a unique digestive environment. Furthermore, the reliance on analysis from faecal samples as a proxy to indicate which microbes may be present in the lower GI tract26,27,28 does not reveal regional specification and it limits our ability to understand region-specific relationships.
Initial colonisation
The initial microbial colonisation of the GIT in the neonatal calf is unique, as at this stage the rumen is non-functional and in this regard, the vast majority of what is known has been extrapolated from mouse and other monogastric animal models. Although the basic tissue structure of the GI tract is formed during gestation, rapid morphological and functional development occurs immediately after birth29,30. From a purportedly sterile environment in utero, the peri-natal calf must immediately adapt to an extra-uterine environment of immense microbial complexity. In a range of animals, Firmicutes, Proteobacteria and Bacteroidetes are known to inhabit the vaginal canal of the dam pre-parturition and account for the majority of bacteria present in the neonate intestine immediately after birth31,32, consistent with the findings for the ileal digesta in this study. In this study, Firmicutes was present in the highest relative abundance in new-born calves and this agrees with the bacterial profile in the ileum of young calves reported in other studies23 and also in goats33. Furthermore, the majority of other microorganisms detected in the ileum immediately after birth in the current study belonged to the phyla Proteobacteria, Verrucomicrobia, Bacteroidetes, Actinobacteria, Chlamydiae and Synergistes. A similar composition was reported in the ileum of mature cows8. This vertical transmission suggests that there is a strong maternal influence on microbial composition in the gut in early life. Yeoman et al. compared the microbiota in the lumen of calves to the microbiota in maternal colostrum, udder skin, and vaginal scrapings and found 46% of microorganisms detected in the calves were also detected in one of the maternal sources. In this study, 8 families (Streptococcaceae, Verrucomicrobiaceae, Pasteurellaceae, Enterobacteriaceae, Campylobacteriaceae, Lachnospiraceae, Veillonellaceae and Synergistaceae) together comprised approximately 91% of all bacterial sequences detected in the ileum of young calves, with Streptococcaceae by far the most dominant9. At the genus level, members of the genus Streptococcus were found to be dominant. It has been suggested that initial colonisers such as Streptococcus and Enterococcus utilise available oxygen in the gut creating an anaerobic environment which facilitates ileal colonisation by strict anaerobic bacteria5. At genus level, the majority of all sequences are assigned to 6 genera all of which are anaerobes, facultative anaerobes or microaerophilic microorganisms and associated with the gut environment. The most abundant, Streptococcus (71.21% ± 4.47%) are known to reside in the gut of young calves and be dominant in the 24 h after birth34.
In humans, Bacteroides and Bifidobacterium are two of the main gut bacterial genera present and contribute to development of immunotolerance to commensal organisms35, which demonstrates the importance of host-microbial interactions during early life to the developing gut and immune system. However, our data indicated that Bacteroides and Bifidobacterium represented less than 1% of the total bacterial community, indicating distinct species-specific differences in colonisation. In calves, Firmicutes remained the most abundant phylum at all time points within ileal digesta and contributed only modestly to the overall dissimilarity in bacterial community structure between calves from different age cohorts. Although low in abundance, Bacteroidetes were the main contributors to dissimilarity between the bacterial community structure in new-born calves and week-old calves, increasing in relative abundance between these time points. Bacteroidetes produce glycoside hydrolases for glycan degradation and therefore this increase after birth may be linked to increased digestive requirements in the host36. This indicates that host-microbe interactions and the mechanisms prompting GI tract development and stimulating immunotolerance may be species-specific and therefore it cannot be assumed that findings from human or rodent studies apply to interactions taking place in the ruminant gut.
Immunological development
Immune cells within the Peyer’s patches, as well as the associated epithelium are well endowed with pattern recognition receptors including Toll-like receptors which lead to cytokine expression and the production of host defence peptides37, and it is these functional molecules that determine the course of the immune response. However, regulation of innate immune responses in the neonate has been suggested as one mechanism by which the host avoids unnecessary inflammatory responses during the early stages of colonisation commensal microbiota. In addition to innate mechanisms, the development of anti-inflammatory Th2 type responses, anergy and T cell deletion have all been described as contributing to immunological tolerance38. This immune tolerance is important to allow colonisation with beneficial non-pathogenic bacteria39,40, establishment of homeostasis and also to protect the mucosal tissue from detrimental inflammatory responses.
NLRP3 is a member of multi-protein innate immune complex known as the inflammasome which senses a variety of endogenous and environmental stimuli, and thereby regulates homeostasis or damage control41. The enhanced expression of NLRP3 suggests that it plays a conserved regulatory role in ileal function. Innate cells like macrophages are also key regulators of intestinal homeostasis42 and they were detected here in large numbers in the ileum of calves immediately after birth. The increased number of macrophages shown here infiltrating the lamina propria mucosa of the villi are a likely source of the cytokines which are significantly increased. IL1A and IL1B encode key inflammatory proteins which were increased in the ileum in this study, and their expression is known to be mediated via the NLRP3 inflammasome. The potent chemotactic IL8 gene is increased and reaches statistical significance at 96 DOA. TGFβ is a strong inducer of tolerogenic responses and the expression of the anti-inflammatory cytokine IL10 was significantly reduced by 7 DOA showing a role at birth. This suggests that IL10 could play a role in modulating inflammation in the neonatal ileum. A significant influx of eosinophils to the ileum was recorded over time, with maximum numbers identified at birth. In the villi, the section of the ileum with closest contact to the external environment and microbial communities, eosinophils accounted for roughly one third of total host cells present. Although the presence of eosinophils in the neonatal ileal villi has not been previously reported in any species, regulatory subsets of these cells have been recently reported. Eosinophils have now been shown to be important for the regulation of the gut microbiota as well as the development of regulatory T cells43,44. A new appreciation of the breadth of eosinophil function is now starting to emerge, including their function in the intestine45 as key regulators of homeostasis46. IL-33 is a key mediator of eosinophil recruitment as well as regulatory T cell expansion47, thereby promoting Th2-biased immune responses. Therefore the significantly enhanced expression of this cytokine could play a role in this regard in cattle.
This influx of innate cells and the obvious presence of immune cells in the ileum supports previous reports that the ileum is one of the key sites of immune-microbe interaction in the mammalian gut13,48,49,50. From an adaptive cell viewpoint, T cells were present in low relative abundances in the ileum villi of new-born calves compared to innate cells but T cell numbers increased markedly at 7, 14 and 21 DOA. In our study the eosinophil and T cell population show an inverse relationship and this may be regulated by the eosinophil population51.
Host–microbe interactions
Members of the Bacteroidetes genus have been shown to have a role in inducing regulatory T cells which promote epithelial repair, promote tolerance to microbes and initiate suppression of immune responses to self and bacterial antigens52. In agreement with these findings, in the current study it was recorded that relative abundance of Bacteroidetes in the ileum was positively correlated with the abundance of T cells. T cells in the intraepithelial spaces of the intestine provide a first line of defence against invading bodies at the barrier and are linked to pathogen resistance and tissue tolerance through cytotoxic T cell-related and anti-inflammatory molecules53,54. Dysregulation of intraepithelial lymphocytes is correlated with loss of mucosal barrier integrity, susceptibility to infections and chronic inflammatory processes. In this study, intraepithelial T cell populations in the ileum markedly increased at 7, and again at 28 DOA. Fluctuations could represent attempts to balance cytotoxic responses triggered by the influx of microorganisms with tissue tolerant responses necessary to avoid prolonged inflammation of the tissue and subsequent tissue damage.
Conclusion
Distinctive species-specific features of ileal tissue, and specifically in Peyer’s patches have been described in terms of cellular composition between mouse and humans13. Here age-related structural, cellular and molecular changes associated with initial colonization and adaptation of the neonatal bovine ileum have been documented for the first time. Neonatal ruminants are highly susceptible to a variety of viral and bacterial enteric infections within the first few weeks of life55 and the role of the gut microbiome in priming the morphological and immune development of the host GIT is crucial56. Here we document the presence of large numbers of innate immune cells (eosinophils ad macrophages) in the ileum as an important portal of host-microbe interaction and education of the neonatal immune system. Maladapted early neonatal immune system development could have negative consequences for lifetime immunity57,58 and therefore these integrated findings provide an important baseline for future intervention studies. Appropriate activation of the NLRP3 inflammasome and the expansion of eosinophils in ileal tissue are likely to be important in terms of regulating maladaptive inflammation immediately after birth and warrants further investigation.
Methods
Experimental design
All procedures described were approved by the Teagasc Animal Ethics Committee and conducted under experimental license from the Irish Health Products Regulatory Authority in accordance with the Cruelty to Animals Act 1876 and the European Communities (Amendments of the Cruelty to Animals Act 1876) Regulations, 1994. This research was carried out as part of a larger study designed to investigate the establishment and development of the rumen microbiome in early life. Full details of this animal model has been recently published59. Briefly, commercially purchased 18-month-old oestrous synchronised Aberdeen Angus heifers (n = 21) were artificially inseminated with frozen thawed semen from one Aberdeen Angus sire to provide 70 calves. Calves were assigned at random to one of six age groups. For this study calves were sampled following euthansia at 0 (n = 11), 7 (n = 8), 14 (n = 9), 21 (n = 9), 28 (n = 9) and 96 days of age (n = 9) DOA). Calves were born over four replicated periods; December 2014 (n = 13), January 2015 (n = 27), May 2015 (n = 14), July 2015 (n = 11), and treatments were assigned as evenly as possible across all replicates. Pregnant heifers were held together in a pen until immediately prior to calving and induced to calve by administration of 2 ml prostaglandin F2α analogue (Estrumate, Merck), to facilitate a staggered calving schedule. Calves not assigned to a D0 treatment were allowed to suckle their dam for 48 h post birth at which point they were removed and penned individually. Calves were then artificially reared on milk replacer (5L/d), offered as two equal feeds and offered concentrates from 7 DOA. Calves to be euthanised at 96 DOA were weaned at 8 weeks, when they were consuming 1 kg concentrates per day for 3 consecutive days. Subsequently they were offered 2 kg concentrate per day, with ad libitum access to water and hay.
Following euthanasia, the gastrointestinal tract was removed, and samples of ileal content were collected using sterilized and RNase-free instruments, and immediately snap-frozen in liquid nitrogen. Cross sections of the ileum were washed in PBS and snap frozen in liquid nitrogen. These samples were subsequently stored at − 80 °C. A cross section from the ileum was fixed in 10% neutral buffered formalin. The tissues and content samples were collected within 20 min of euthanisation.
DNA extraction and microbial community analysis
The room was cleaned before sampling commenced and the area was cleaned again in between each calf. Each calf was sampled in isolation using sterile scalpel blades. All surfaces and sampling equipment were treated with boiling water, ethanol and RNase inhibitor prior to the processing of samples from each calf. Sterile vessels were used to store all samples. Day 0 calves were taken for sampling immediately after birth without further contact with the cow or the external environment.
DNA was extracted from ileal content samples collected from 37 animals at 6 sampling time points; 0 (n = 8), 7 (n = 5), 14 (n = 5), 21 (n = 6), 28 (n = 10) and 96 (n = 3) DOA, using the PCSA method with phenol chloroform added before the bead beating step 60. The DNA concentration and purity were assessed using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA) and all samples had a 260/280 value of > 1.8 and a 260/230 value > 2. Purified DNA was stored at − 20 °C.
Bacterial community structures were determined using amplicon sequencing on an Illumina Miseq platform (Illumina, San Diego, CA), using the method described by Kozich et al. (2013). Briefly, 1 μL of purified DNA was added to a well on a 96-well PCR plate already containing 17 μL of Accuprime Pfx Supermix (Invitrogen, Thermo Fisher Scientific, Dublin, Ireland) and 2 μL of a primer set targeting the V4 region of the 16S rRNA gene61. The PCR conditions consisted of a hot start at 95 °C for 2 min, followed by denaturation at 95 °C for 20 s, annealing at 55 °C for 15 s, and extension at 72 °C for 5 min (30 cycles) with a final extension at 72 °C for 10 min. The PCR products were visualized on a 1.2% (w/v) agarose gel (Roche Diagnostic, Basel, Switzerland); PCR products were then purified, and concentrations normalized using the SequalPrep Normalization Plate Kit (Invitrogen) according to manufacturer’s instructions and an amplicon pool was created using 5 μL of PCR product from each sample. The concentration of the pool was then determined using a Qubit fluorometer and dsDNA kit (Thermo Fisher Scientific) to ensure that at least 100 ng of DNA was present. The amplicon pool was then sent to the Centre for Genomic Research, University of Liverpool (Liverpool, UK), for sequencing on an Illumina Miseq platform. Negative controls were included in library preparation for amplicon sequencing. After PCR the negative controls, like all other samples, were evaluated by gel electrophoresis and were shown to contain no DNA indicating that reagents and materials used up until this point were not introducing contaminants.
MiSeq sequencing data were initially processed using the mothur program v.1.32.162. Illumina adapter sequences were trimmed by cutadapt ver.2.1.1 using option -O 363. Sickle ver.1.264 was used to further trim the data with a quality score of ≥ 20. Reads < 10 bp after trimming were removed. Each read was then trimmed to a maximum of 275 bp and ambiguous bases were removed. Sequences which contained homo-polymer runs > 8 bases were discarded. After trimming, identical sequences were grouped into unique sequences. Chimeric sequences were identified using the UCHIME algorithm65 within mothur and were then removed. Sequences were assigned to OTU using the cluster command and the average neighbour algorithm. All subsequent OTU-based analyses were performed using a cutoff of 0.03. Taxonomy was assigned to all remaining aligned sequences by comparing processed data to the silva128 databases for bacteria and archaea independently (arb -silva .de/ silva -license-information66).
Histology
Following formalin fixation ileal tissues were dehydrated with increasing concentrations of ethyl-alcohol (50–100%) using an automated processer (Tissue-TEK VIP, Sakura, Finetek) and embedded in paraffin wax. Sections of 4 µm were cut using a microtome (Leitz RM2255), mounted on glass slides and stained with Hematoxylin and Eosin (H&E) using a Leica auto stainer (Leica ST5020) for histology examination using a light microscope (Leica DM3000).
Immunohistochemistry (IHC)
For identification of the immune cells, formalin fixed paraffin embedded tissue sections of 4 µm mounted on to negatively charged glass slides were investigated by immunohistochemistry (IHC) using the manual technique with an ImmPRESS HRP anti-goat IgG (Iba1), anti-rabbit IgG (CD3) or anti-mouse-IgG (Pax5) (peroxidase) polymer detection kit (Vector Laboratories, Peterborough, UK). Primary antibodies specific for the macrophage marker Iba1 (AIFI/Iba1 goat anti-human polyclonal (internal) (unconjugated) antibody (Lifespan Biosciences, Nottingham, UK), no antigen retrieval, dilution 1 in 500), the T-cell marker CD3 (CD3 polyclonal rabbit anti-human antibody (Agilent Technologies Ltd, Cork, Ireland); antigen retrieval by citrate buffer at pH 6 heat treatment for 20 min (dilution 1 in 300) and the B-cell marker Pax5 (BD BioSciences, Oxford, England, monoclonal mouse antibody, antigen retrieval by citrate buffer at pH 6 heat treatment for 20 min (dilution 1 in 40) were used.
All sections, with or without antigen retrieval, were rinsed with H2O, and incubated with 3% (v/v) hydrogen peroxide, an endogenous enzyme block, for 30 min. Sections were rinsed with PBS before incubation in horse serum (from kit) for 30 min followed by incubation with the primary antibody. Sections were again rinsed with PBS, incubated with ImmPRESS goat, rabbit or mouse serum for 30 min and then rinsed again before application of the substrate chromogen, 3,3′-Diaminobenzidine (DAB). The DAB was washed off after approximately 40 s and the sections were counterstained with haematoxylin, rinsed and dehydrated before cover slipped.
Villi measurements and cell counts
Measurements of villi length and width were taken on H&E stained ileal sections at 40× using a 1 cm graticule, with 10 well-orientated villi measured per section for length and width and 10 fields of vision assessed for frequency. Villus height was measured from the tip of the villus to the crypt-villus junction. Villus width was measured at the midpoint of the villus length, from the outside epithelial edges. Eosinophils, identified by their lobed nuclei and bright red large intracytoplasmic granules, and macrophages, and lymphocytes, identified by IHC, were examined at 400× magnification in the lamina propria mucosa. The cells of each cell type were counted in five representative full length villi in each case. Equally sized villi were chosen in all cases to allow comparison between the different age groups. In addition, T cells were counted in the mucosal epithelial lining. The composition of cells in the dome area was assessed separately.
RNA extraction and cDNA synthesis
The frozen tissue samples of ileum were homogenised in TRIzol (Life Technologies, Paisley, UK) using a Pro Scientific homogeniser model PRO200 (PRO Scientific Inc., Oxford, CT, USA) with 3 s pulses repeated 10 times. RNA was then extracted and cleaned using the RNeasy Plus mini kit (Qiagen Ltd., Manchester, UK).
RNA quality and quantity were determined by measuring the absorbance at 260 nm using a NanoDrop spectrophotometer ND-100 (Thermo Scientific, Waltham, MA, United States), all samples had an absorbance above 1.8. RNA quality was further assessed on an Agilent Bioanalyser 2100 (Agilent Technologies Ireland Ltd., Dublin, Ireland) using the RNA 6000 Nano lab chip kit, all samples had RIN values between 8 and 10. cDNA was synthesised using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. 2 µg of total RNA was reverse transcribed into cDNA using MultiScribe reverse transcriptase. The quantity of converted cDNA was determined by absorbance at 260 nm using a NanoDrop spectrophotometer and stored at – 20 °C.
Quantitative PCR
Genes involved in the immune response, specifically genes for cytokines involved in recruiting and promoting immune cells were targeted and are listed in Supplementary Table 5. All primers for quantitative PCR (qPCR) were designed using Primer3 and analysed through BLAST to check specificity and homology to the bovine sequence. Primers for reference and target genes were commercially synthesised (Sigma-Aldrich Ireland, Dublin, Ireland). Five genes were assessed in the ileal tissue, all of which promoted cytokine expression in T cells or macrophages. The gene expression levels were measured by qPCR and the expression stabilities were evaluated by the M value calculated in GeNORM67.
Real-time PCR was carried out using an ABI 7500 Fast Real-time PCR System with SYBR green master mix (Applied Biosystems, Warrington, UK). Reactions were carried out in PCR tubes and prepared in a total volume of 20 µl with 2 µl cDNA, 10 µl SYBR green master mix, 7 µl nuclease free H2O and 1 µl of forward and reverse primer mix. Thermal cycling conditions remained constant for each assay, with an initial denaturation step at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing and extension at 60 °C for 40 s. SYBR green fluorescence was detected at the end of each cycle to track the quantity of PCR product. The efficiency of the qPCR reaction was calculated for each gene by creating a standard curve from two-fold serial dilutions of cDNA. For each gene, efficiency correction of the raw Ct values, normalisation to the reference genes, calculations of quantities relative to the max Ct value and log transformation of Ct values were all carried out using GenEx 5.2.1.3 (MultiD Analyses AB, Gothenburg, Sweden).
Statistical analysis
Analysis of variance (ANOVA) with a Tukey HSD post-hoc test were carried out within the R software environment using the ‘lsmeans’ package to analyse differences between means for villi length, width, frequency and immune cell populations in the bovine ileum from birth to 96 days of life. Immune cell (eosinophil, macrophage and T cell) counts in the lamina propria of ileum harvested from calves were averaged and compared between age groups. Significant differences in gene expression levels in the ileum of calves were detected using one way ANOVA performed in Graphpad Prism (v7.02). For bacterial populations and immune cell changes across time, a 2 way ANOVA was performed using the same software.
Bacterial phylum, family and genus richness, evenness and diversity (Shannon–Wiener index and Simpson index) were calculated for each age cohort. ANOVA and post hoc, Tukey HSD tests were carried out within the R software environment using the ‘lsmeans’ package to analyze differences between means. Multivariate analysis was carried out using PRIMER-E v.7 software with the Distance-based permutational multivariate analysis of variance (PERMANOVA) add on68. Similarity matrices were constructed for samples using Bray–Curtis similarities on standardized, fourth root transformed abundance data. PERMANOVA was then performed to test the null hypothesis that there were no differences in microbial community structure across treatments at a significance level of α = 0.05 based on 9999 possible permutations. Nonmetric multidimensional scaling (nMDS) plots were constructed to visualize the data. Similarity percentages (SIMPER) were calculated using Bray–Curtis similarities to evaluate the level to which each phylum, family and genus contributed to the difference in community structures between groups. Effects and differences were declared as significant when P ≤ 0.05.
Pearson’s correlation coefficient (r) analysis was conducted to determine if any significant correlations could be observed between immune cell population levels in the villi and intra-epithelial layers in the ileum of calves at different age points and any of the most relatively abundant bacterial phyla and genera contributing to differences in bacterial community structure of calves between age points. This method was also used to determine if any statistically significant correlations existed between immune cell population profiles in the ileum villi of calves and gene expression levels of a subset of cytokines at different age points.
Data availability
Sequence files associated with each sample have been submitted to the NCBI Sequence Read Archive (Accession number PRJNA453736).
References
Wampach, L. et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front. Microbiol. 8, 738. https://doi.org/10.3389/fmicb.2017.00738 (2017).
Fanaro, S., Chierici, R., Guerrini, P. & Vigi, V. Intestinal microflora in early infancy: Composition and development. Acta Paediatr. Suppl. 91, 48–55 (2003).
Jost, T., Lacroix, C., Braegger, C. P. & Chassard, C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 7, e44595. https://doi.org/10.1371/journal.pone.0044595 (2012).
Arrieta, M. C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M. & Finlay, B. The intestinal microbiome in early life: Health and disease. Front. Immunol. 5, 427. https://doi.org/10.3389/fimmu.2014.00427 (2014).
Malmuthuge, N., Griebel, P. J. & le Guan, L. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front. Vet. Sci. 2, 36. https://doi.org/10.3389/fvets.2015.00036 (2015).
Kararli, T. T. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm. Drug Dispos. 16, 351–380. https://doi.org/10.1002/bdd.2510160502 (1995).
Santaolalla, R. & Abreu, M. T. Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 28, 124–129. https://doi.org/10.1097/MOG.0b013e3283506559 (2012).
Mao, S., Zhang, M., Liu, J. & Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 5, 16116. https://doi.org/10.1038/srep16116 (2015).
Yeoman, C. J. et al. Biogeographical differences in the influence of maternal microbial sources on the early successional development of the bovine neonatal gastrointestinal tract. Sci. Rep. 8, 3197. https://doi.org/10.1038/s41598-018-21440-8 (2018).
Blattler, U. et al. Feeding colostrum, its composition and feeding duration variably modify proliferation and morphology of the intestine and digestive enzyme activities of neonatal calves. J. Nutr. 131, 1256–1263. https://doi.org/10.1093/jn/131.4.1256 (2001).
Chow, J., Lee, S. M., Shen, Y., Khosravi, A. & Mazmanian, S. K. Host-bacterial symbiosis in health and disease. Adv. Immunol. 107, 243–274. https://doi.org/10.1016/B978-0-12-381300-8.00008-3 (2010).
Backhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 852. https://doi.org/10.1016/j.chom.2015.05.012 (2015).
Jung, C., Hugot, J. P. & Barreau, F. Peyer’s patches: The immune sensors of the intestine. Int. J. Inflamm. 2010, 823710. https://doi.org/10.4061/2010/823710 (2010).
Hormann, N. et al. Gut microbial colonization orchestrates TLR2 expression, signaling and epithelial proliferation in the small intestinal mucosa. PLoS ONE 9, e113080. https://doi.org/10.1371/journal.pone.0113080 (2014).
Lotz, M. et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203, 973–984. https://doi.org/10.1084/jem.20050625 (2006).
Malmuthuge, N., Li, M., Fries, P., Griebel, P. J. & Guan, L. L. Regional and age dependent changes in gene expression of Toll-like receptors and key antimicrobial defence molecules throughout the gastrointestinal tract of dairy calves. Vet. Immunol. Immunopathol. 146, 18–26. https://doi.org/10.1016/j.vetimm.2012.01.010 (2012).
Surlis, C. et al. Birth delivery method affects expression of immune genes in lung and jejunum tissue of neonatal beef calves. BMC Vet. Res. 13, 391. https://doi.org/10.1186/s12917-017-1310-2 (2017).
Liebler-Tenorio, E. M., Riedel-Caspari, G. & Pohlenz, J. F. Uptake of colostral leukocytes in the intestinal tract of newborn calves. Vet. Immunol. Immunopathol. 85, 33–40. https://doi.org/10.1016/s0165-2427(01)00404-4 (2002).
Khare, S. et al. Systems biology analysis of gene expression during in vivo Mycobacterium avium paratuberculosis enteric colonization reveals role for immune tolerance. PLoS ONE 7, e42127. https://doi.org/10.1371/journal.pone.0042127 (2012).
Aho, A. D., McNulty, A. M. & Coussens, P. M. Enhanced expression of interleukin-1alpha and tumor necrosis factor receptor-associated protein 1 in ileal tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 71, 6479–6486 (2003).
Allen, A. J. et al. Development of a bovine ileal cannulation model to study the immune response and mechanisms of pathogenesis of paratuberculosis. Clin. Vac. Immunol. 16, 453–463. https://doi.org/10.1128/CVI.00347-08 (2009).
Charavaryamath, C. et al. Host responses to persistent Mycobacterium avium subspecies paratuberculosis infection in surgically isolated bovine ileal segments. Clin. Vac. Immunol. 20, 156–165. https://doi.org/10.1128/CVI.00496-12 (2013).
Malmuthuge, N., Griebel, P. J. & le Guan, L. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl. Environ. Microbiol. 80, 2021–2028. https://doi.org/10.1128/AEM.03864-13 (2014).
Malmuthuge, N. & Guan, L. L. Understanding the gut microbiome of dairy calves: Opportunities to improve early-life gut health. J. Dairy Sci. 100, 5996–6005. https://doi.org/10.3168/jds.2016-12239 (2017).
Malmuthuge, N. et al. Distinct commensal bacteria associated with ingesta and mucosal epithelium in the gastrointestinal tracts of calves and chickens. FEMS Microbiol. Ecol. 79, 337–347. https://doi.org/10.1111/j.1574-6941.2011.01220.x (2012).
Rada, V., Vlkova, E., Nevoral, J. & Trojanova, I. Comparison of bacterial flora and enzymatic activity in faeces of infants and calves. FEMS Microbiol. Lett. 258, 25–28. https://doi.org/10.1111/j.1574-6968.2006.00207.x (2006).
Uyeno, Y., Sekiguchi, Y. & Kamagata, Y. rRNA-based analysis to monitor succession of faecal bacterial communities in Holstein calves. Lett. Appl. Microbiol. 51, 570–577. https://doi.org/10.1111/j.1472-765X.2010.02937.x (2010).
Oikonomou, G. et al. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS ONE 8, e63157. https://doi.org/10.1371/journal.pone.0063157 (2013).
Tourneur, E. & Chassin, C. Neonatal immune adaptation of the gut and its role during infections. Clin. Dev. Immunol. 2013, 270301. https://doi.org/10.1155/2013/270301 (2013).
Meale, S. J., Chaucheyras-Durand, F., Berends, H., Guan, L. L. & Steele, M. A. From pre- to postweaning: Transformation of the young calf’s gastrointestinal tract. J. Dairy Sci. 100, 5984–5995. https://doi.org/10.3168/jds.2016-12474 (2017).
Laguardia-Nascimento, M. et al. Vaginal microbiome characterization of nellore cattle using metagenomic analysis. PLoS ONE 10, e0143294. https://doi.org/10.1371/journal.pone.0143294 (2015).
Greenhalgh, K., Meyer, K. M., Aagaard, K. M. & Wilmes, P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ. Microbiol. 18, 2103–2116. https://doi.org/10.1111/1462-2920.13318 (2016).
Mao, S. Y., Huo, W. J. & Zhu, W. Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18, 525–541. https://doi.org/10.1111/1462-2920.12724 (2016).
Alipour, M. J. et al. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 8, 10437. https://doi.org/10.1038/s41598-018-28733-y (2018).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. https://doi.org/10.1038/nature07008 (2008).
Patrascu, O. et al. A fibrolytic potential in the human ileum mucosal microbiota revealed by functional metagenomic. Sci. Rep. 7, 40248. https://doi.org/10.1038/srep40248 (2017).
Larsson, E. et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61, 1124–1131. https://doi.org/10.1136/gutjnl-2011-301104 (2012).
Mowat, A. M., Millington, O. R. & Chirdo, F. G. Anatomical and cellular basis of immunity and tolerance in the intestine. J. Pediatr. Gastroenterol. Nutr. 39(Suppl 3), S723-724. https://doi.org/10.1097/00005176-200406003-00003 (2004).
Teran, R. et al. Immune system development during early childhood in tropical Latin America: Evidence for the age-dependent down regulation of the innate immune response. Clin. Immunol. 138, 299–310. https://doi.org/10.1016/j.clim.2010.12.011 (2011).
Mowat, A. M. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 3, 331–341. https://doi.org/10.1038/nri1057 (2003).
Hirota, S. A. et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Dis. 17, 1359–1372. https://doi.org/10.1002/ibd.21478 (2011).
Bain, C. C. & Mowat, A. M. Macrophages in intestinal homeostasis and inflammation. Immunol. Rev. 260, 102–117. https://doi.org/10.1111/imr.12192 (2014).
Chu, V. T. et al. Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity 40, 582–593. https://doi.org/10.1016/j.immuni.2014.02.014 (2014).
Jung, Y. et al. IL-1beta in eosinophil-mediated small intestinal homeostasis and IgA production. Mucosal Immunol. 8, 930–942. https://doi.org/10.1038/mi.2014.123 (2015).
Jung, Y. & Rothenberg, M. E. Roles and regulation of gastrointestinal eosinophils in immunity and disease. J. Immunol. 193, 999–1005. https://doi.org/10.4049/jimmunol.1400413 (2014).
Marichal, T., Mesnil, C. & Bureau, F. Homeostatic eosinophils: Characteristics and functions. Front. Med. 4, 101. https://doi.org/10.3389/fmed.2017.00101 (2017).
Zaiss, D. M., Minutti, C. M. & Knipper, J. A. Immune- and non-immune-mediated roles of regulatory T-cells during wound healing. Immunology 157, 190–197. https://doi.org/10.1111/imm.13057 (2019).
Cebra, J. J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046S-1051S. https://doi.org/10.1093/ajcn/69.5.1046s (1999).
Fries, P. et al. Mucosal dendritic cell subpopulations in the small intestine of newborn calves. Dev. Comp. Immunol. 35, 1040–1051. https://doi.org/10.1016/j.dci.2011.04.003 (2011).
Liang, G. et al. Transcriptome analysis reveals regional and temporal differences in mucosal immune system development in the small intestine of neonatal calves. BMC Genom. 17, 602. https://doi.org/10.1186/s12864-016-2957-y (2016).
Lingblom, C., Andersson, J., Andersson, K. & Wenneras, C. Regulatory eosinophils suppress T cells partly through galectin-10. J. Immunol. 198, 4672–4681. https://doi.org/10.4049/jimmunol.1601005 (2017).
Reinoso Webb, C., Koboziev, I., Furr, K. L. & Grisham, M. B. Protective and pro-inflammatory roles of intestinal bacteria. Pathophysiology 23, 67–80. https://doi.org/10.1016/j.pathophys.2016.02.002 (2016).
Sheridan, B. S. & Lefrancois, L. Intraepithelial lymphocytes: To serve and protect. Curr. Gastroenterol. Rep. 12, 513–521. https://doi.org/10.1007/s11894-010-0148-6 (2010).
Denning, T. L. et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 187, 733–747. https://doi.org/10.4049/jimmunol.1002701 (2011).
Uetake, K. Newborn calf welfare: A review focusing on mortality rates. Anim. Sci. J. 84, 101–105. https://doi.org/10.1111/asj.12019 (2013).
Guarner, F. Enteric flora in health and disease. Digestion 73(Suppl 1), 5–12. https://doi.org/10.1159/000089775 (2006).
Agace, W. W. & McCoy, K. D. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46, 532–548. https://doi.org/10.1016/j.immuni.2017.04.004 (2017).
Meade, K. G. & O’Farrelly, C. beta-Defensins: Farming the microbiome for homeostasis and health. Front. Immunol. 9, 3072. https://doi.org/10.3389/fimmu.2018.03072 (2018).
O’Hara, E. et al. Investigating temporal microbial dynamics in the rumen of beef calves raised on two farms during early life. FEMS Microbiol. Ecol. https://doi.org/10.1093/femsec/fiz203 (2020).
Lueders, T., Manefield, M. & Friedrich, M. W. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. https://doi.org/10.1046/j.1462-2920.2003.00536.x (2004).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1), 4516–4522. https://doi.org/10.1073/pnas.1000080107 (2011).
Schloss, P. D. et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. https://doi.org/10.1128/AEM.01541-09 (2009).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Joshi, N. A. & J.N, F. (2011).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. https://doi.org/10.1093/bioinformatics/btr381 (2011).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590-596. https://doi.org/10.1093/nar/gks1219 (2013).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Anderson, L. P., Paterson, J. A., Ansotegui, R. P., Cecava, M. & Schmutz, W. The effects of degradable and undegradable intake protein on the performance of lactating first-calf heifers. J. Anim. Sci. 79, 2224–2232. https://doi.org/10.2527/2001.7982224x (2001).
Acknowledgements
The authors would like to acknowledge funding from the JPI FACCE program (RumenStability project) and the Irish Department of Agriculture Fisheries and Marine’s Agricultural Greenhouse Gas Research Initiative for Ireland (Project no. 10/RD/GHGIRELAND/716). We would also like to thank the staff in Teagasc for assistance with animal care.
Author information
Authors and Affiliations
Contributions
Designed the overall study and collected samples: D.K., S.W., E.O.H. and T.L. Conceived and designed this analysis: K.G.M., E.D. and T.L. Veterinary pathology analysis and expertise: H.J. and J.B. Contributed to analysis of samples and interpretation of results: H.J., T.L., E.D. and K.G.M. Wrote and edited the paper: All authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lyons, T., Jahns, H., Brady, J. et al. Integrated analyses of the microbiological, immunological and ontological transitions in the calf ileum during early life. Sci Rep 10, 21264 (2020). https://doi.org/10.1038/s41598-020-77907-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77907-0
This article is cited by
-
Potential benefits of a blend of essential oils on metabolism, digestibility, organ development and gene expression of dairy calves
Scientific Reports (2023)
-
The impact of vitamin D3 supplementation on the faecal and oral microbiome of dairy calves indoors or at pasture
Scientific Reports (2023)
-
High fat diet–induced hyperlipidemia and tissue steatosis in rabbits through modulating ileal microbiota
Applied Microbiology and Biotechnology (2022)
-
Low serum vitamin D concentrations in Spring-born dairy calves are associated with elevated peripheral leukocytes
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.