Abstract
We aimed to investigate the circRNA–miRNA regulatory network in atrial fibrillation (AF) by using Cytoscape and HMDD v3.0. Finally, 120 differentially expressed circRNAs in peripheral blood monocytes of 4 AF patients were preliminarily screened by circRNA microarray. circRNA_4648, circRNA_4631, and circRNA_2875 were the first four circRNAs with the most binding nodes in the circRNA–miRNA network. The top three most frequent miRNAs for up-regulated circRNAs were hsa-miR-328 that interacted with 5 up-regulated circRNAs, hsa-miR-4685-5p with 4 up-regulated circRNAs, hsa-miR-3150a-3p, hsa-miR-4649-5p, hsa-miR-4783-3p, and hsa-miR-8073 with 3 up-regulated circRNAs,, while the top three most frequent miRNAs for down-regulated circRNAs were hsa-miR-328 that interacted with 14 down-regulated circRNAs, hsa-miR-4685-5p with 11 down-regulated circRNAs and hsa-miR-661 with 9 down-regulated circRNAs. According to HMDD v3.0, five up-regulated and eleven down-regulated circRNAs were found to interact with AF related miRNAs. These results indicated the possible regulatory network between circRNAs and miRNAs in the pathogenesis of AF.
Similar content being viewed by others
Introduction
Atrial fibrillation (AF), one of the most common arrhythmias in clinical practice, with a prevalence about 1–2% in the general population, is characterized with high relative risk of heart failure and embolic stroke. AF is also considered as a potential factor for high mortality and morbidity, especially in elderly individuals1,2. Recent growing reports indicate that structural remodeling and electrical remodeling are important pathophysiological contributors to onset and maintenance of AF 3,4. However, exact mechanism of how AF occurs is still unknown.
To our knowledge, non-coding RNAs (ncRNAs), include a class of RNAs, such as long non-coding RNAs (lncRNAs), micro-RNAs (miRNAs) and circular RNAs (circRNAs), play crucial roles in regulating gene expression under pathological and physiological conditions5,6,7. circRNAs, a novel type of endogenous ncRNAs , have be reported as a key ncRNAs in gene regulation and the pathophysiology of cardiovascular diseases8,9. It has been well-known that dysregulated miRNAs can contribute to the prevalence of AF by deregulating transcription factor, regulating atrial excitability and increasing atrial arrhythmogenicity10,11. Accumulating studies indicate that circRNAs may interact with miRNAs by a sequence-driven sponging effect and the circRNA–miRNA-network has emerging roles in physiological and pathological processes of cardiovascular diseases12,13. However, to our knowledge, there are few studies pointing to the expression of circRNAs in AF, and circRNA–miRNA network in AF remains unclear.
In the present study, we analyzed and predicted the differentially expressed circRNAs in human monocytes from patients with AF and healthy controls using microarray, the potential regulatory network between circRNAs and miRNAs were explored by using Cytoscape and HMDD v3.0. We hypothesized that there were differentially expressed circRNAs in human monocytes and highly possible interaction between circRNAs and miRNAs, which would provide an important landmark for investigating the mechanism of AF.
Materials and methods
Study population and specimen collection
10 patients with AF (AF group) and 10 matched healthy subjects (Control group) who excluded AF were enrolled (Table 1). 10 ml of peripheral blood was collected, monocytes were purified from peripheral blood and frozen for analysis. The diagnosis of AF was consistent with the criteria listed in the 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS14. The Ethics Committee of Taizhou People’s Hospital approved the study, which was conducted according to the principles of the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice guidelines. All the enrolled subjects provided written informed consent before entering this experiment.
The differentially expressed circRNAs of AF detected by microarray analysis
The total RNA in monocytes was extracted using Trizol reagent (Ambion, USA) and purified by QIAGEN RNeasy Mini Kit (QIGEN, Germany). Sample labeling and microarray hybridization were conducted by Outdo Bio‐tech (Shanghai, P.R. China) with the same method as previously described 15. Simply, the circRNAs were measured with the Agilent One-Color Microarray Based Gene Expression Analysis Low. The arrays were scanned by Axon microarray 4000B microarray scanner and extracted using Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and data processing were conducted by the Gene Spring GXv11.5.1 software package (Agilent, USA). The fold-change between AF patients and healthy controls was calculated. The statistical significance was calculated by t test and further filtered with fold change. circRNAs with foldchange > 2 and p < 0.05 were regarded as significant differential expression.
qRT-PCR validation of differentially expressed circRNAs
In order to confirm the results of microarray analysis, four upregulated circRNAs (circRNA_0031, circRNA_1837,circRNA_5901 and circRNA_7571) and four downregulated circRNAs (circRNA_2773, circRNA_5801, circRNA_7386 and circRNA_7577) were selected randomly for validation by qRT-PCR in all study population. Simply, 1 μl of cDNAs was added to 12.5 μl of SYBR‐Green Gene Expression Master Mix (Applied Biosystems, Inc.), 10.5 μl of DEPC‐treated water, and 0.5 μl of reverse and forward primers. The gene expression level of target circRNAs was normalized to the housekeeping gene GAPDH (Sangon Biotech, Shanghai, China) and calculated using the (2−ΔΔCt) method. The primer sequences for RT-PCR were shown in Table 2.
Construction of circRNA–miRNA regulatory networks
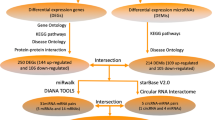
Acting as competing miRNA sponge, the sponging activity of differentially expressed circRNAs over corresponding miRNAs was calculated by the prediction of miRNA target binding sites using the miRanda software. Enrichment results of total differentially expressed circRNAs were sorted by p value, and the potential connections between circRNAs and miRNAs were further explored by using Cytoscape 3.4.0 (http://cytoscape.org/). Finally, the regulatory networks of circRNA–microRNA in AF patients were constructed.
Analyze the AF related circRNAs according to HMDD v3.0
In order to further explore the AF related circRNAs, we used the website of HMDD v3.0. HMDD v3.0, a database for experimentally supported human microRNA–disease associations, integrated many past publications about miRNA–disease associations, and offered evidence-stratified miRNA–disease data based on six categories of 20 evidence codes16. We used the keywords ‘atrial fibrillation’ to obtain AF related miRNAs from HMDD v3.0. If the differentially expressed circRNAs identified by microarray interacted with these reported AF related miRNAs, they were considered to be AF associated circRNAs.
Results
The differentially expressed circRNAs between AF patients and healthy controls
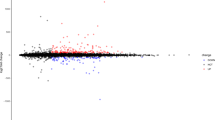
A total of 120 circRNAs was calculated as differentially expressed between AF patients and healthy controls (fold change > 2, and p < 0.05) (Fig. 1). In which, 65 circRNAs were upregulated (Table 3) and 55 circRNAs were downregulated (Table 4).
Differentially expressed circRNAs between AF group and control group. (A) Volcano plots are displayed for visualizing the differential expression of circRNAs. The red and green points in the plot represent the differentially expressed circRNAs with statistical significance. (B) Hierarchical cluster analysis of all the deregulated circRNAs.
qRT-PCR validation of differentially expressed circRNAs
Four upregulated circRNAs (circRNA_0031, circRNA_1837, circRNA_5901 and circRNA_7571) and four downregulated circRNAs (circRNA_2773, circRNA_5801, circRNA_7386 and circRNA_7577) were selected randomly by Random Number Generator Pro V1.79 software for for qRT-PCR validation to confirm the microarray results. As a results, all of 4 upregulated circRNAs (p < 0.05 or p < 0.01 for circRNA_0031, circRNA_1837, circRNA_5901 and circRNA_7571, respectively) and 3 out of 4 downregulated circRNAs (p < 0.05 or p < 0.01 for circRNA_5801, circRNA_7386 and circRNA_7577, respectively) showed a significantly different expression (Fig. 2), which was consistent with microarray results.
Construction of circRNA–miRNA networks
We calculated the terms of miRNAs that targeted these dysregulated circRNAs by using Cytoscape 3.4.0 (http://cytoscape.org/) and conducted the circRNA–miRNA networks (shown in Fig. 3). Results showed that circRNA_7571, circRNA_4648, circRNA_4631, and circRNA_2875 were the first four circRNAs with the most binding nodes in the co-expression network, interacted with 34 miRNAs, 26 miRNAs, 24 miRNAs and 24 miRNAs, respectively (Fig. 4). The top three most frequent miRNAs for up-regulated circRNAs were hsa-miR-328 that interacted with 5 up-regulated circRNAs, hsa-miR-4685-5p with 4 up-regulated circRNAs, hsa-miR-3150a-3p, hsa-miR-4649-5p, hsa-miR-4783-3p, and hsa-miR-8073 that interacted with 3 up-regulated circRNAs, while the top three most frequent miRNAs for down-regulated circRNAs were hsa-miR-328 that interacted with 14 down-regulated circRNAs, hsa-miR-4685-5p that interacted with 11 down-regulated circRNAs and hsa-miR-661 that interacted with 9 down-regulated circRNAs. We predicted that these miRNAs may be more relevant with the differentially expressed circRNAs in AF.
circRNA–miRNA coexpression network explored by using Cytoscape. The size of each node represents functional connectivity of each circRNA. The network consists of 37 circRNAs and 90 miRNAs. The red node represents circRNA and the green node represents miRNA. circRNA_7571, circRNA_4648, circRNA_4631 and circRNA_2875 were the four largest nodes in the network. hsa-miR-328 was the highest positive correlated miRNA in the networks.
Analyze the AF related circRNAs according to HMDD v3.0
We confirmed 100 AF related miRNAs from HMDD v3.0 by using the keywords ‘atrial fibrillation’. If the differentially expressed circRNAs identified by microarray interacted with these reported AF related miRNAs, they were considered to be AF associated circRNAs. Finally, five up-regulated (hsa_circRNA_7571, hsa_circRNA_3448, hsa_circRNA_1402, hsa_circRNA_4284 and hsa_circRNA_1415) and eleven down-regulated circRNAs (hsa_circRNA_2527, hsa_circRNA_4648, hsa_circRNA_4624, hsa_circRNA_1496, hsa_circRNA_3138, hsa_circRNA_3138, hsa_circRNA_6086, hsa_circRNA_2875, hsa_circRNA_3807, hsa_circRNA_4402, hsa_circRNA_4631 and hsa_circRNA_2773) were found to interact with AF related miRNAs. Figures 5 and 6 showed the expression pattern of these dysregulated circRNAs, respectively.
The expression pattern of the five up-regulated circRNAs that interact with AF related miRNAs. (A) The expression pattern of hsa_circRNA_7571 that interact with has-miR-133a. (B) The expression pattern of hsa_circRNA_3448 that interact with has-miR-328. (C) The expression pattern of hsa_circRNA_1402 that interact with has-miR-486. (D) The expression pattern of hsa_circRNA_4284 that interact with has-miR-328. (E) The expression pattern of hsa_circRNA_1415 that interact with has-miR-328.
The expression pattern of the eleven down-regulated circRNAs that interact with atrial fibrillation related miRNAs. (A) The expression pattern of hsa_circRNA_2527 that interact with has-miR-328. (B) The expression pattern of hsa_circRNA_4648 that interact with has-miR-328. (C) The expression pattern of hsa_circRNA_4624 that interact with has-miR-328. (D) The expression pattern of hsa_circRNA_1496 that interact with has-miR-328. (E) The expression pattern of hsa_circRNA_3138 that interact with has-miR-574. (E) The expression pattern of hsa_circRNA_3138 that interact with has-miR-574. (F) The expression pattern of hsa_circRNA_6086 that interact with has-miR-574. (G) The expression pattern of hsa_circRNA_2875 that interact with has-miR-92a. (H) The expression pattern of hsa_circRNA_3807 that interact with has-miR-26b. (I) The expression pattern of hsa_circRNA_4402 that interact with has-miR-328. (J) The expression pattern of hsa_circRNA_4631 that interact with has-miR-199a. (K) The expression pattern of hsa_circRNA_2773 that interact with has-miR-574.
Within the five up-regulated circRNAs, three of them (circRNA_7571, circRNA_3448, circRNA_1415) interacted with hsa-miR-328, one of them (circRNA_1402, circRNA_4284, respectively) interacted with hsa-miR-486 and hsa-miR-133a, respectively. Within the eleven down-regulated circRNAs, five of them (circRNA_4648, circRNA_4624, circRNA_4402, circRNA_2527 and circRNA_1496, respectively) interacted with hsa-miR-328, three of them (circRNA_6086, circRNA_3138 and circRNA_2773, respectively) interacted with hsa-miR-574, while another three (circRNA_2875, circRNA_3807 and circRNA_4631, respectively) interacted with hsa-miR-92a, hsa-miR-26b and hsa-miR-199a, respectively.
Ethical approval
No treatment was tested in patients by the authors for this article. Informed consent was obtained from all individual participants included in the study.
Discussion
In the present study, we provide two experimental findings on circRNAs involved in AF. On the one hand, there was significantly different expression profiles of circRNAs between AF patients and normal controls. On the other hand, five up-regulated (hsa_circRNA_7571, hsa_circRNA_3448, hsa_circRNA_1402, hsa_circRNA_4284 and hsa_circRNA_1415) and eleven down-regulated circRNAs (hsa_circRNA_2527, hsa_circRNA_4648, hsa_circRNA_4624, hsa_circRNA_1496, hsa_circRNA_3138, hsa_circRNA_3138, hsa_circRNA_6086, hsa_circRNA_2875, hsa_circRNA_3807, hsa_circRNA_4402, hsa_circRNA_4631 and hsa_circRNA_2773) were found to interact with AF related miRNAs and considered as the AF associated circRNAs by the construction of circRNA–miRNA network and the analysis using HMDD v3.0.
Atrial electric remodeling associated with profound reduction of L-type Ca2+ current and shortening of the action potential duration was the characteristic of both clinical and experimental AF. It was reported that miR-328, diminished L-type calcium current, shorted the atrial action potential duration, and increased AF vulnerability, would contribute to the atrial electric remodeling in AF and can be used as a diagnosis biomarker of AF17,18. Our findings indicated that hsa-miR-328 interacted with both up-regulated and downregulated circRNAs, which was consistent with the reports and indicated that circRNA_7571, circRNA_3448, circRNA_1415, circRNA_4648, circRNA_4624, circRNA_4402, circRNA_2527 and circRNA_1496 colud be regarded as the diagnosis biomarkers of circRNAs for AF.
miR-486 was related to the accumulation of superoxide anion, induction of DNA damage, reduction of cell proliferation and senescent phenotype in human fibroblasts19. Slagsvold et al. reported that hsa-miR-486 was upregulated in AF within left atria20. Another report from Wang et al. showed that hsa-miR-486 was found to be up-regulated in left atrial appendage in patients with AF21. Thus, hsa-miR-486 was considered as a AF related miRNA. At the same time, circRNA_1402, interacted with hsa-miR-486 in our findings could be considered as one of the AF related circRNAs.
A large number of studies have reported the relationships between the miRNAs (hsa-miR-133a, hsa-miR-574, hsa-miR-92a, hsa-miR-26b and hsa-miR-199a) and AF. For example, miR-133 has a cardioprotective role dependent on AKT serine/threonine kinase (AKT) signaling in control situation, inducing apoptosis in AF patients due to its down-regulation22. hsa-miR-26b increases IK1 current and membrane resting potential, the downregulation of hsa-miR-26b may reduce AF vulnerability23. hsa-miR-574 may promote electrical remodeling via Cav1.2 and contribute to cardiac arrhythmia pathogenesis of AF24.
hsa-miR-92a can attenuate cardiomyocyte apoptosis in AF patients induced by hypoxia/reoxygenation via the up-regulation of SMAD7 and down-regulation of nuclear NF-κB p6525. MiR-26b directly targeted KCNJ2. Both in vivo and in vitro inhibition of miR-26b increased IK1 and AF vulnerability, whereas overexpression of dampened AF vulnerability26. miR-199a down-regulation induces Sirtuin 1, a cardio-protective protein, as a compensatory mechanism to inhibit the process of oxidative stress which contributes to the pathogenesis of AF27. These miRNAs were considered as the potential biomarkers and therapeutic targets related to AF. Therefore, the differentially expressed circRNAs of circRNA_4284, circRNA_6086, circRNA_3138, circRNA_2773, circRNA_2875, circRNA_3807 and circRNA_4631 in the current study were more likely to be AF associated circRNAs.
Study limitations
First, the small sample size does not provide sufficient power for such an analysis. Second, we just preliminarily investigated the circRNA–miRNA regulatory network in AF, the target gene or pathway analysis and functional assays of circRNA–miRNA regulatory network in the AF process should be further explored.
Conclusions
Our study showed that there were differentially expressed circRNAs in AF patients, five up-regulated and eleven down-regulated circRNAs were considered as the AF related circRNAs. The differentially expressed circRNAs had a possible regulatory network with miRNAs, which indicated the possible regulatory network between circRNAs and miRNAs in the pathogenesis of AF.
Change history
23 August 2021
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41598-021-96813-7
References
Dai, H. et al. Global, regional, and national prevalence, incidence, mortality, and risk factors for atrial fibrillation, 1990–2017: results from the Global Burden of Disease Study 2017. Eur. Heart J. Qual. Care Clin. Outcomes 31, 061 (2020).
Butters, A. et al. Epidemiology and clinical characteristics of atrial fibrillation in patients with inherited heart diseases. J. Cardiovasc. Electrophysiol. 31, 465–473 (2020).
Nattel, S. Molecular and cellular mechanisms of atrial fibrosis in atrial fibrillation. JACC Clin. Electrophysiol. 3, 425–435 (2017).
Schotten, U., Verheule, S., Kirchhof, P. & Goette, A. Pathophysiological mechanisms of atrial fibrillation: atranslational appraisal. Physiol. Rev. 91, 265–325 (2011).
Mattick, J. S. & Makunin, I. V. Non-coding RNA. Hum. Mol. Genet. 1, R17-29 (2006).
Danie, C., Lagergren, J. & Öhman, M. RNA editing of non-coding RNA and its role in gene regulation. Biochimie 117, 22–27 (2015).
Dykes, I. M. & Emanueli, C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinform. 15, 177–186 (2017).
Fan, X. et al. Circular RNAs in cardiovascular disease: an overview. Biomed. Res. Int. 2017, 5135781 (2017).
Li, M. et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 285, 220–232 (2018).
Mario, T. et al. A micro-transcription factor blueprint for early atrial arrhythmogenic remodeling. Biomed. Res. Int. 2015, 263151 (2015).
van den Berg, N. W. E. et al. MicroRNAs in atrial fibrillation: from expression signatures to functional implications. Cardiovasc. Drugs Ther. 31, 345–365 (2017).
Fumagalli, M. R., Lionetti, M. C., Zapperi, S., La, C. & Porta, A. M. Cross-talk between circRNAs and mRNAs modulates MiRNA-mediated circuits and affects melanoma plasticity. Cancer Microenviron. 12, 95–104 (2019).
Su, Q. & Lv, X. Revealing new landscape of cardiovascular disease through circular RNA–miRNA–mRNA axis. Genomics S0888–7543, 30565–30568 (2019).
Čihák, R., Haman, L. & Táborský, M. 2016 SC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS Summary of the document prepared by the Czech Society of Cardiology. Cor et Vasa 58, e636–e683 (2016).
Ruan, Z. B., Sun, X. H., Sheng, H. H. & Zhu, L. Long non-coding RNA expression profile in atrial fibrillation. Int. J. Clin. Exp. Pathol. 8, 8402–8410 (2015).
Huang, Z. et al. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 47, D1013–D1017 (2018).
Soeki, T. et al. Relationship between local production of microRNA-328 and atrial substrate remodeling in atrial fibrillation. J. Cardiol. 68, 472–477 (2016).
Michela, M. et al. Upregulation of miR-133b and miR-328 in patients with atrial dilatation: implications for stretch-induced atrial fibrillation. Front. Physiol. 10, 1133 (2019).
Faraonio, R. et al. A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ. 19, 713–721 (2012).
Slagsvold, K. H. et al. Mitochondrial respiration and microRNA expression in right and left atrium of patients with atrial fibrillation. Physiol. Genomics 46, 505–511 (2014).
Wang, J. G. et al. Differential expressions of miRNAs in patients with nonvalvular atrial fibrillation. Zhonghua Yi Xue Za Zhi 92, 1816–1819 (2012).
Tsoporis, J. N. et al. Increased right atrial appendage apoptosis is associated with differential regulation of candidate MicroRNAs 1 and 133A in patients who developed atrial fibrillation after cardiac surgery. J. Mol. Cell Cardiol. 121, 25–32 (2018).
Luo, X. et al. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J. Clin. Investig. 123(5), 1939–1951 (2013).
Allan, B. et al. Molecular mechanisms, diagnostic aspects and therapeutic opportunities of micro ribonucleic acids in atrial fibrillation. Int. J. Mol. Sci. 21, 2742 (2020).
Zhang, B. et al. MicroRNA-92a inhibition attenuates Hypoxia/Reoxygenation-induced myocardiocyte apoptosis by targeting Smad7. PLoS ONE 9, e100298 (2014).
Qi, X.-Y. et al. Fibroblast inward-rectifier potassium current upregulation in profibrillatory atrial remodeling. Circ. Res. 116, 836–845 (2015).
Estefanía, L.-V., Diego, F., Amelia, A. & Houria, D. Genetics and epigenetics of atrial fibrillation. Int. J. Mol. Sci. 21(16), 5717 (2020).
Funding
The study was supported by Jiangsu Provincial Medical Innovation Team (Grant No. CXTDB2017015), Jiangsu Commission of Health, China (Grant No. H201665), the Six Talent Foundation of Jiangsu Province, China (Grant No. WSN-20) and Taizhou science and technology support plan (Grant No. TS201901). The authors are thankful to Hai-Hui Sheng for technical assistance.
Author information
Authors and Affiliations
Contributions
Z.R. and L.Z. conceived the idea and designed the project. Z.R., Q.Y. and G.C. helped in experimentation and data acquisition. F.W. contributed to clinical evaluation and sample provision. Z.R., and G.C. contributed to data analysis and the interpretation of the results. Z.R. took the lead in writing the manuscript along F.W., Q.Y., L.Z. supervised the research. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41598-021-96813-7
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruan, Zb., Wang, F., Yu, Qp. et al. RETRACTED ARTICLE: Integrative analysis of the circRNA–miRNA regulatory network in atrial fibrillation. Sci Rep 10, 20451 (2020). https://doi.org/10.1038/s41598-020-77485-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77485-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.