Abstract
Achieving early molecular response (EMR) has been shown to be associated with better event free survival in patients with chronic phase chronic myeloid leukemia (CP-CML) on Imatinib therapy. We prospectively evaluated the factors influencing the 2-year failure free survival (FFS) and EMR to imatinib therapy in these patients including day29 plasma Imatinib levels, genetic variants and the gene expression of target genes in imatinib transport and biotransformation. Patients with low and intermediate Sokal score had better 2-year FFS compared to those with high Sokal Score (p = 0.02). Patients carrying ABCB1-C1236T variants had high day29 plasma imatinib levels (P = 0.005), increased EMR at 3 months (P = 0.044) and a better 2 year FFS (P = 0.003) when compared to those with wild type genotype. This translates to patients with lower ABCB1 mRNA expression having a significantly higher intracellular imatinib levels (P = 0.029). Higher day29 plasma imatinib levels was found to be strongly associated with patients achieving EMR at 3 months (P = 0.022), MMR at 12 months (P = 0.041) which essentially resulted in better 2-year FFS (p = 0.05). Also, patients who achieved EMR at 3 months, 6 months and MMR at 12 months had better FFS when compared to those who did not. This study suggests the incorporation of these variables in to the imatinib dosing algorithm as predictive biomarkers of response to Imatinib therapy.
Similar content being viewed by others
Introduction
Imatinib mesylate, a tyrosine kinase inhibitor (TKI), has revolutionized the treatment of chronic myeloid leukemia (CML) changing it from a life-threatening disease to a condition that can be controlled in the vast majority of cases by oral medication. For those patients who develop resistance or intolerance to imatinib, choices of second or third generation TKIs are available. However, responses are the best when such changes in therapy are done prior to disease progression to an accelerated phase or blast crisis. This decision process on continuing therapy or changing TKI is compounded by the high cost of these second generation drugs in a predominantly self-paying system as exists in our country. Identifying the sub-optimal responders and early switch of therapy is essential since the achievement of major molecular response (MMR) or deep molecular response (DMR) within 3–6 months post TKI therapy has been shown to significantly improve progression free survival1,2,3,4.
Several factors have been reported to contribute to sub-optimal response or resistance to imatinib5,6. Among the BCR-ABL1 dependent mechanisms, development of mutations in the kinase domain of BCR-ABL17,8,9 plays a major role. Several imatinib independent mechanisms of resistance have also been reported including overexpression of efflux transporters10,11,12, decreased expression of influx transporters13,14,15, decreased plasma levels16,17, binding to plasma proteins18, and genetic polymorphisms in the enzymes and transporters involved in imatinib transport or biotransformation19,20. With the exception of one study reporting an intronic deletion polymorphism in the BIM (BCL2Like11) gene to be associated with imatinib resistance21,22,23, there are no other prospective studies in patients with CML of Asian origin where all the parameters have been systematically evaluated. This is important due to the ethnic differences in genetic polymorphisms, which in turn could influence the differences in systemic exposure to anticancer drugs24,25.
Although there are several studies15,17,21,22,26 exploring the factors influencing MMR and DMR in the international scale, there is no comprehensive analysis of the factors influencing EMR and MMR which in turn influence failure free survival (FFS). Our aim was to prospectively document response to imatinib as first line therapy using serial molecular monitoring to document EMR and MMR to evaluate the factors influencing response to imatinib therapy in newly diagnosed patients with CML-CP.
Patients and methods
Patients
All newly diagnosed, imatinib naïve adult CP- CML patients visiting Department of Haematology, Christian Medical College, Vellore were enrolled in the study after obtaining written informed consent. Ethical approval for this study has been approved by Institutional review board (IRB Min no: IRB (EC)-4–11-06–2008 dated June 19, 2008) at Christian medical college, Vellore -632,004, Tamilnadu, India. All methods in the study were performed in accordance with relevant guidelines and regulations.
Morphology/RT-PCR/Cytogenetics/FISH analysis
All patients at diagnosis were analyzed for the presence of BCR-ABL1 fusion gene by Fluorescence in-situ hybridization (FISH). Interphase FISH analysis was performed using fixed cell suspensions obtained by direct or unstimulated overnight cultures of peripheral blood or bone marrow as reported previously27. Although karyotyping is not routinely performed for CML patients in our centre, it was done in those patients who gave consent for bone marrow aspiration. Peripheral blood was collected at diagnosis and RNA was extracted using Trizol method. cDNA was synthesized from 2 μg RNA using random hexamers and reverse transcriptase enzyme (High capacity c-DNA synthesis kit, Thermo Scientific) followed by RT-PCR to identify the BCR-ABL1 fusion transcript [e13a2/e14a2 or e1a2).
Plasma and intracellular levels of Imatinib
Blood samples were collected on day29 post imatinib therapy in EDTA anticoagulated tubes and plasma was separated and stored immediately. Trough plasma imatinib and desmethyl imatinib (imatinib metabolite) concentration was assessed using HPLC (High Performance Liquid Chromatography) Ultra-violet detection method as reported previously28 with minor modifications. Dasatinib was used as an internal standard.
Peripheral blood mononuclear cells (PBMNCs) (15*106/mL) from CML patients at diagnosis was incubated at 37 °C for 24 h and treated with 5 μM imatinib for 5 h. The cells were washed with phosphate buffered saline (PBS) and the cell pellets were stored at − 80 °C until analysis. Just before processing, the samples were thawed, the cells were lysed by sonication for 10 min on ice. Intracellular levels of Imatinib were analyzed by the same method as that of plasma imatinib incorporating minor changes from a previously published protocol28.
BCR-ABL1 molecular monitoring
Peripheral blood was collected at diagnosis, 3, 6 9, 12 and 18 months’ post imatinib therapy to check the BCR-ABL1 transcript level. Additional samples were analyzed depending on clinical needs. The BCR-ABL1 transcript levels were quantified using real time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) method as reported previously29.
BCR-ABL1 kinase domain mutation detection
Mutations in the BCR-ABL1 kinase domain were evaluated in patients with suboptimal response as reported previously30. The sequences were aligned using SeqScape (Applied Biosystems) and the type of the mutation was predicted using Mutation Taster (https://www.mutationtaster.org/).
RNA expression of influx and efflux transporters
Total RNA was extracted using Trizol method and 1–2 μg RNA was used for c-DNA synthesis (High capacity cDNA Reverse Transcription Kit, Applied Biosystems). RNA expression of various Imatinib efflux and influx transporters were analyzed using TaqMan based assays [assay IDs: ABCB1 (HS01067802-m1), ABCG2 (HS01053787-m1), SLC22A1 (HS00427554-m1), ABCA3 (HS00184543-m1), ABCA5 (HS00363322-m1), ABCA6 (HS00365329-m1), ABCB5 (HS02889060-m1), ABCB6 (HS00180568_m1), ABCB7 (HS00188776-m1), ABCB8 (HS00894817-m1), ABCB10 (HS00429240-m1), ABCB11 (HS00184824-m1), ABCC1 (HS00219905-m1), ABCC3 (HS009784173-m1), ABCC4 (HS00988717-m1), ABCC11 (HS01090768-m1)] and the expression was normalized to the housekeeping gene GAPDH (4352934E).
RNA expression of influx and efflux transporters in CD34+ fraction
CD34+ cells were enriched from the CML patient sample and healthy donors using Easysep magnetic enrichment kit (https://www.stemcell.com/easysep-human-cd34-positive-selection-kit-ii.html). RNA extraction, c-DNA synthesis and expression of influx (hOCT1) and efflux (ABCB1 & ABCG2) transporter analysis was also done as explained previously.
Polymorphisms in imatinib transporters and drug metabolizing enzymes
Genomic DNA was extracted from peripheral blood using standard phenol–chloroform method and 50-100 ng DNA was used for each PCR. All coding exons with flanking introns were screened for SLC22A1 and known single nucleotide polymorphisms (SNP) in Imatinib efflux transporters ABCB1 (exon 26), ABCG2 (promoter, exon 2 and exon 5), OCTN1/SLC22A4 (exon 9) were screened by PCR followed by sequencing and variants were analyzed by SeqScape software. Three SNPs in Cyp3A4/3A5 were analyzed by restriction fragment length polymorphism (RFLP). The primer sequences and the source of the methods are listed in Supplementary Table 1.
BIM deletion polymorphism
Genomic DNA was used to assess the BIM deletion polymorphism by PCR as previously reported21. The PCR conditions used were: 95 °C for 10 min and 40 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 10 min. Emerald Master mix (TaKaRa BIO Inc., Shiga, Japan) was used for this PCR. The amplified products when subjected to 3% agarose gel electrophoresis showed the band size of 216 bp for wild-type and 173 bp if there was deletion.
GSTT1, GSTM1 deletions, GSTA1*B and GSTP1*B polymorphisms
GSTT1 and GSTM1 deletion, GSTA1*B and GSTP1*B ([Ile105val] rs1965) polymorphisms were screened by methods as reported previously31. Primers used for PCR reaction and the conditions are listed in supplementary Table 1.
Assessment of response and definition of outcome
Response criteria including complete hematological response (CHR) and molecular response were defined according to the European Leukaemia Net (ELN) guidelines32. Hematologic response was defined as normalized peripheral blood cell counts (WBC < 10 X109/L and platelet count < 450 X109/L) without evidence of peripheral blasts, promyelocytes, or myelocytes, and without evidence of extramedullary disease including disappearance of palpable splenomegaly lasting for at least 4 weeks. Molecular response was classified based on BCR-ABL1 to control gene transcript ratios, expressed on the International Scale (IS) as reported previously from our lab29. EMR was defined as BCR-ABL1 to control gene ratio of ≤ 10% in the IS at 3 and ≤ 1% at 6 months; MMR was defined as BCR-ABL1 to control gene transcript ratio of ≤ 0.1% in the IS, 12 months after imatinib therapy. In patients without an MMR at 12 months, mutation analysis of the BCR–ABL1 fusion transcript was performed by direct sequencing.
Patients were classified as intolerant to imatinib therapy if the patient meets one or more of the criteria as reported previously33. Patients who progressed to accelerated phase or blast crisis were classified as non-responders at all-time points. FFS was assessed by defining an event as: loss of hematological or cytogenetic response, progression to advanced phase or blast crisis, stem-cell transplant, death from any cause, change in imatinib dose or switch to a second generation TKI due to suboptimal response, as reported previously34. Overall survival (OS) was calculated from the initiation of imatinib therapy until the date of death from any cause or the date of last follow-up.
Statistical analysis
All statistical analyses were performed using SPSS (IBM SPSS statistics version 21.0, Armonk, NY) and GraphPad Prism 5 (GraphPad Software, La Jolla, CA; www.graphpad.com) software; p value < 0.05 was used for significance testing. Genotype distribution was tested for Hardy–Weinberg equilibrium (HWE). Comparison of RNA expression levels of candidate genes was done using one-way ANOVA and student’s t-tests. For analyzing PFS, patients who stopped imatinib or switched treatment during follow-up were censored at the time of stopping or switching.
Results
Patient demographics
The demographics of the CP-CML patients enrolled in this study are listed in Table 1. There were 108 males and 52 females with a median age of 36 (range 18–65) years. One hundred and ten patients had low or intermediate Sokal score while 50 patients had high Sokal Score. Each patient started with an oral dose of 400 mg of imatinib daily. The median follow-up of the patients post therapy was 83 months (range: 12 to 120 months). Of the 146 patients in whom karyotyping was done, 128 had only t(9;22) and 18 had the t(9;22) with additional chromosomal abnormalities.
Response to imatinib therapy
One hundred and fifty of the 160 patients (94%) achieved complete hematological response at 3 months. The proportion of patients who achieved EMR at 3, 6 months and MMR at 12 months’ post imatinib treatment is listed in Table 2. Neither the BCR-ABL1 transcript type nor Sokal score showed significant association with achieving EMR/MMR. The rate of FFS at 24 months’ post imatinib therapy in these patients was 78 ± 3.3%. The events included switch of TKI (n = 18), escalation (n = 33) of imatinib dose due to in-ability to afford or access 2nd/3rd generation TKI, palliative therapy due to severe toxicity (n = 7), imatinib dose reduced due to intolerance (n = 5), stem cell transplantation (n = 1) and progressive disease (n = 3) requiring chemotherapy or death. The decision to switch TKI or change the dose of TKI was entirely upto the discretion of the treating doctor as per the department policy and ELN guidelines.
Spectrum of BCR-ABL1 kinase domain mutation in imatinib non-responders
In patients who failed to achieve milestone responses to imatinib therapy, presence of mutation in the BCR-ABL1 kinase domain was tested. Of the 80 patients tested, 22 patients had mutations in the BCR-ABL1 kinase domain. The spectrum of BCR-ABL1 kinase domain mutation identified in these patients is listed in Supplementary Table 2. T315I and G250E were the common mutations identified.
Genotype/Allele frequencies of genetic variants
The allele frequencies of the 29 genetic variants in 12 genes [drug transporter genes SLC22A1, ABCB1 & ABCG2 and drug metabolizing enzyme genes GST, CYP3A4/A5] screened in this patient cohort is listed (Supplementary Table 3). The genotypes of all the genetic variants were in Hardy–Weinberg equilibrium. The SLC22A1 variants were also screened in normal healthy volunteers (n = 100), as there was no Indian data at the time we started this study (Supplementary Table 4). The allele frequencies of SLC22A1 variants were comparable between patients and normal controls. The SLC22A1 exon5 variant (Arg287Gly) was in complete linkage disequilibrium with an exon 6 variant (Thr340Met). An 8 bp ins polymorphism in intron7 was in complete linkage disequilibrium with an exon7 coding variant (Met408Val; rs628031) which results in a splice variant. When we tried to amplify the full length of SLC22A1cDNA in patients with the intron 7 ins polymorphism in the mutant state, there was a truncated transcript but no full length SLC22A1 transcript while the heterozygotes showed both truncated and full length transcripts; the samples with wild type genotype showed only the full length transcript upon RT-PCR.
RNA expression of imatinib influx transporter hOCT1 and ABC transporters
Expression of Imatinib influx (hOCT1/SLC22A1) and efflux transporters (ABCB1, ABCG2, SLC22A1, ABCA3, ABCA5, ABCA6, ABCB5, ABCB6, ABCB7, ABCB8, ABCB10, ABCB11, ABCC1, ABCC3, ABCC4 and ABCC11) showed wide interpatient variability (Supplementary Table 5).
Plasma and intracellular imatinib and desmethyl imatinib levels
In this cohort, 87.5% of patients (n = 140) were on Glivec and 12.5% patients (n = 20) were on Veenat, a generic imatinib formulation from India. The trough plasma imatinib and desmethyl imatinib levels on day29 was available only in 67 patients. This is due to reasons such as patients not visiting the clinic by the end of one month, taking imatinib at night so trough level sampling was not possible or have withheld imatinib due to intolerance. There was no significant difference in trough plasma imatinib levels between patients receiving Glivec vs. generic imatinib. The median plasma imatinib and desmethyl imatinib concentrations on day29, were 1050 ng/mL (106–5035 ng/mL) and 191 ng/mL (31–2161 ng/mL) respectively. The median intracellular imatinib level after ex-vivo incubation of primary CML cells (n = 64) with imatinib was 1225 ng/mL (range 181-12848 ng/mL).
Plasma imatinib levels influence EMR and MMR to imatinib therapy
The median plasma imatinib level on day29 was significantly higher in those who achieved EMR at 3 months compared to those who did not (1280 ng/ml vs 887 ng/ml; p = 0.022 Fig. 1a). The median plasma imatinib levels on day29 was significantly higher in those who achieved MMR at 12 months compared to those who did not (1207 ng/ml vs 1022 ng/ml; p = 0.0417 respectively; Fig. 1b).
Plasma imatinib levels predict early molecular response at 3 & 6 months and MMR at 12 months post imatinib therapy. (a) Plasma imatinib levels (median, range) on day29 with early molecular response at 3 months and (b) in patients with and without MMR to imatinib at 12 months. Statistical significance was calculated using Mann–Whitney U test.
Plasma Imatinib levels are influenced by genetic variants in ABCB1
When the role of genetic variants in the target imatinib metabolism/ transport genes on trough plasma imatinib levels was evaluated, patients with variant MDR1/ABCB1-C1236T had significantly high day29 plasma imatinib concentration (p = 0.005) compared to those with wild type genotype (Fig. 2a). None of the other variants were significantly associated with plasma imatinib levels.
ABCB1 polymorphism influences plasma imatinib levels and RNA Expression of ABC transporters influence intracellular Imatinib levels (b) Plasma imatinib levels on day29 post imatinib therapy in patients with ABCB1/MDR1 C1236T genotypes (wt (CC) n = 10; Het + Mut (CT + TT) n = 57). (b) Intracellular imatinib levels in patients with above or below median expression of ABCB1 (median expression-21.9), (c) ABCA3 (median expression-505 below median n = 23 vs above median n = 33) and (d) ABCC4 (median expression-28.4; below median n = 30 vs above median n = 26) RNA. The RNA expression of each gene was normalised to GAPDH and relative expression to CML001 using 2-ddCT method. Statistical significance was calculated using Mann–Whitney U test.
RNA expression of ABC transporter genes significantly influence intracellular but not plasma imatinib levels
The expression of influx and efflux transporters was compared with plasma and intracellular imatinib levels. Patients who had ABCB1 expression above median showed significantly lower intracellular imatinib levels (p = 0.0293) (Fig. 2b). Patients who had ABCA3 and ABCC4 expression above median had lower intra cellular imatinib levels (ABCA3 p = 0.0747; ABCC4 p = 0.0536) (Fig. 2c, d).
RNA expression of imatinib transporter genes in the CD34+ fraction and response to therapy
The expression of Imatinib efflux (ABCB1, ABCG2) and influx (SLC22A1) transporters in CD34+ as well as bulk CML cells was compared. There was a significantly increased expression of efflux transporters ABCG2 (p = 0.001) and ABCB1 (p = 0.007) and decreased expression of SLC22A1 (p = < 0.001) in CD34+ fraction compared to the total cellular RNA (Fig. 3a). Decreased expression of hOCT-1 mRNA was observed in CML CD34+ cells compared to CD34+ cells derived from normal healthy donors (Fig. 3b). There was no significant association between RNA expression of these transporters in the CD34+ cells with EMR and MMR.
Expression of imatinib influx and efflux transporters in bulk Vs CD34+ primary CML cells. (a) Expression of hOCT1, ABCB1/MDR1 and ABCG2 in CML bulk cells compared with expression of transporter expression in CML CD34 + (Leukemic stem cells) LSCs cells. Statistical significance was calculated using paired-T test. (b) Expression of hOCT1, ABCB1/MDR1 and ABCG2 in CD34+ cells from normal healthy donors vs. primary CML CD34+ cells where higher the dCt lower the expression and vice versa. Statistical significance was calculated using Mann–Whitney U test.
Genetic polymorphisms in imatinib transporter and drug metabolizing genes influence EMR and MMR to imatinib therapy
We further analyzed the influence of genetic polymorphisms in target genes on the incidence of EMR at 3 & 6 months and MMR at 12 months. The incidence of EMR at 3 months was significantly higher in patients with MDR1-C1236T variant genotype ((p = 0.044) Table 3) compared to those with wild type genotype. The incidence of EMR at 6 months was higher in patients with MDR1-C3435T variant genotype (p = 0.058) (Table 3) compared to those with wild type genotype. Also, the incidence of EMR at 6 months was higher in patients with GSTM1 wild type genotype compared to those with null genotype (p = 0.071).
Factors influencing 2-year FFS post imatinib therapy
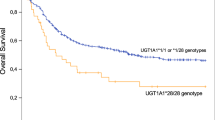
We further analyzed the influence of all the basic demographic (including age, sex, Sokal score, BCR-ABL1 transcript type) as well as variables including imatinib plasma levels, RNA expression and genetic polymorphisms of imatinib influx/efflux transporters on 2-year FFS post imatinib therapy. Patients with low (< 0.8) and intermediate (0.8–1.2) Sokal score showed better 2-year FFS compared to those with high Sokal score (> 1.2) (40/110 vs. 28/50 patients with low/intermediate vs. high Sokal score respectively had failure to imatinib therapy; p = 0.02). None of the other basic demographic factors was significantly associated with FFS or EMR. Patients who had MDR1 variant genotype showed significantly better FFS compared to the wild type genotype (MDR1- 1236; p = 0.005, MDR1- 2677; p = 0.004 and MDR1- 3435; p = 0.004) (Fig. 4a–c). Also, patients with higher median Imatinib levels (> 1757 ng/mL) on day29 had significantly better FFS (p = 0.057) (Fig. 4d). Patients with ABCA6 expression below median showed significantly better FFS compared to those with above median levels (p = 0.007) (Fig. 4e). There was a trend to significantly lower rate of FFS in patients with ABCC4 expression above median compared to below median (p = 0.066) (Fig. 4f.). Patients who achieved EMR at 3 and 6 months had significantly better rate of FFS compared to those who did not achieve EMR (Supplementary Fig. 1a-b). Patients who achieved MMR at 12 months also showed significantly better rate of FFS compared to those who did not achieve MMR (p = 0.002) (Supplementary Fig. 1c). High Sokal score (Hazard ratio: 1.53; p value: 0.05), MDR1 C2677T genotype (Hazard ratio: 0.401; p value: 0.014) and not achieving EMR at 3 months post imatinib therapy (Hazard ratio: 3.875; p value: 0.0001) were significant risk factors for 2-year FFS in multivariate analysis (Tables 4, 5). Plasma imatinib levels were available only in 51 patients for whom molecular response was also available and hence could not be included as a variable in multivariate analysis.
MDR1 polymorphisms, plasma imatinib levels, and RNA expression of efflux transporters influence FFS after imatinib therapy in patients with CP-CML. (a)–(c) Influence of MDR1 genotypes C1236T, G2677T & C3435T on FFS after imatinib therapy. (d) d. Influence of plasma imatinib levels on day29 on FFS. (e)–(f) Influence of ABCA6 and ABCC4 expression on FFS after imatinib therapy. RNA expression was normalised to GAPDH as the housekeeping gene and expressed relative to the expression in CML001 using 2-ddCT method.
Discussion
Targeted therapy with imatinib is still the first line treatment of choice in newly diagnosed patients with CML in chronic phase (CML-CP). However, a proportion of patients do not achieve milestone molecular responses with imatinib and are classified as sub-optimal responders. Identifying these patients followed by early switch of therapy will help improve FFS. Although several studies have evaluated factors influencing attainment of MMR after imatinib therapy, to the best of our knowledge, there is no prospective study evaluating various factors influencing EMR and FFS after imatinib therapy, especially in a uniform cohort of CP-CML patients. We conducted a prospective single centre observational study in CP-CML patients with serial molecular monitoring and comprehensive pharmacogenetic analysis to evaluate factors influencing EMR to imatinib and FFS. Response to imatinib as seen by achieving milestone molecular response post imatinib therapy was similar to previous reports35,36. Mutations in the BCR-ABL1 kinase domain explained only 28% of the patients with sub-optimal response, which is also similar to previous reports37,38,39. None of the basic demographic parameters including age, sex, Sokal score and BCR-ABL1 transcript type showed significant association with achieving EMR/MMR post imatinib therapy in the present study. Similar to previous reports from India, the median age of diagnosis in of CML in our cohort is lower compared to Western patients (36 yrs vs. 47yrs)40, majority of the patients younger than 40 years. This is probably the reason for age not showing significant association with response to imatinib therapy. While the distribution of low, intermediate and high Sokal scores was similar to Western data in CP-CML patients as reviewed by Ganesan and Kumar40, patients with high Sokal score showed significantly poor 2-year FFS in this study, similar to previous reports41,42.
Several studies have reported that genetic variants in the influx and efflux transporters of imatinib namely hOCT1, ABCG2 and MDR1 to be associated with imatinib levels16,20,43,44. In the present study, similar to previous reports, patients with variant MDR1 genotype showed significant association with plasma imatinib levels. The variants of MDR1 C1236T, C3435T and G2326T were shown to result in decreased MDR1 expression45,46,47 which in turn results in higher plasma imatinib levels and hence better EMR/MMR/FFS. Increased RNA expression of ABC transporters ABCB1, ABCA3 and ABCC4 were significantly associated with lower intracellular imatinib levels in our study. It has been reported previously that increased expression of ABCB148; ABCA349 and ABCC450 to be associated with imatinib resistance. Unlike previous reports, none of the genetic variants in hOCT1 were significantly associated with plasma imatinib levels or molecular response in the present study.
Higher trough plasma Imatinib levels have been shown to be associated with significantly better molecular response (Table 4), including the present study. The median plasma imatinib levels in patients with good response vs. suboptimal response in our study is similar to previous reports16,17,26,51,52. However, from the available data from India, although various measures of response including clinical, cytogenetic or MMR were considered, the imatinib levels in good responders seems to be higher than the previous reports from outside India43,44,53,54, and the present study. The two Indian studies that showed association between molecular response and plasma Imatinib levels have assessed either hematological response43,53 or considered BCR-ABL1 ratio below 1% as molecular response. The median trough plasma Imatinib levels on day29 can be a used as biomarker for assessment of response to imatinib therapy to rule out issues related to poor compliance as well as identifying poor or ultra-rapid metabolizers who could benefit from adjustment of imatinib doses.
RNA expression of influx and efflux transporters of imatinib have been shown to influence response to imatinib therapy. Several studies have reported increased expression or functional activity of hOCT1 to be associated with better response to imatinib13,14,55. However, the RNA expression of these transporters did not show any association with EMR or MMR in the present study. Interestingly, when the expression of these transporters in CML CD34+ vs. bulk cells was compared, there was significantly increased ABCB1 and ABCG2 expression and decreased hOCT1 in the CD34 + cells compared to CML bulk cells. This is in line with the fact that imatinib does not eliminate CML stem cells56,57 probably due to this dysregulated expression of the transporters in the CML CD34+ cells.
Polymorphisms in imatinib influx/ efflux transporters have been reported to influence response to imatinib therapy and progression free survival15,19,58,59,60. Similar to these reports, MDR1 and ABCG2 variants were associated with better EMR/MMR in the present study. Higher plasma imatinib levels16,60, attainment of EMR (at 3 or 6 months)61,62,63 & MMR at 12 months64,65 have been reported to result in better FFS in patients with CML on imatinib therapy. In the present study, MDR1 variants, day29 plasma imatinib level of > 1757 ng/mL, lower ABCA6, ABCC4 RNA expression as well as achieving EMR at 3, 6 months and MMR at 12 months were associated with significantly better FFS. While increased ABCC4 RNA expression has been reported previously in suboptimal response to imatinib15,17,21,22,26, the role on increased ABCA6 on imatinib response is not clear.
Our study suggests that factors such as steady state plasma Imatinib levels, MDR1 polymorphisms and ABC transporter expression influence EMR/MMR to imatinib therapy, which in turn influence FFS in patients with CP-CML. The possibility to tailor dose of imatinib considering these factors in order to improve molecular response to imatinib and better FFS, remains to be tested.
References
Steegmann, J. L. et al. European LeukemiaNet recommendations for the management and avoidance of adverse events of treatment in chronic myeloid leukaemia. Leukemia https://doi.org/10.1038/leu.2016.104 (2016).
Kim, D. D. H., Hamad, N., Lee, H. G., Kamel-Reid, S. & Lipton, J. H. BCR/ABL level at 6 months identifies good risk CML subgroup after failing early molecular response at 3 months following imatinib therapy for CML in chronic phase. Am. J. Hematol. 89, 626–632 (2014).
Branford, S. et al. Early molecular response and female sex strongly predict stable undetectable BCR-ABL1, the criteria for imatinib discontinuation in patients with CML. Blood 121, 3818–3824 (2013).
Hanfstein, B. et al. Early molecular and cytogenetic response is predictive for long-term progression-free and overall survival in chronic myeloid leukemia (CML). Leukemia 26, 2096–2102 (2012).
Hochhaus, A., Erben, P., Ernst, T. & Mueller, M. C. Resistance to targeted therapy in chronic myelogenous leukemia. Semin. Hematol. 44, S15-24 (2007).
Volpe, G., Panuzzo, C., Ulisciani, S. & Cilloni, D. Imatinib resistance in CML. Cancer Lett. 274, 1–9 (2009).
Khorashad, J. S. et al. BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships. Blood 121, 489–498 (2013).
Hughes, T. et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood 108, 28–37 (2006).
O’Hare, T., Eide, C. A. & Deininger, M. W. N. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood 110, 2242–2249 (2007).
Mahon, F.-X. et al. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood 101, 2368–2373 (2003).
Jordanides, N. E., Jorgensen, H. G., Holyoake, T. L. & Mountford, J. C. Functional ABCG2 is overexpressed on primary CML CD34+ cells and is inhibited by imatinib mesylate. Blood 108, 1370–1373 (2006).
Illmer, T. et al. P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia 18, 401–408 (2004).
White, D. L. et al. Functional activity of the OCT-1 protein is predictive of long-term outcome in patients with chronic-phase chronic myeloid leukemia treated with imatinib. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 28, 2761–2767 (2010).
White, D. L. et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood 110, 4064–4072 (2007).
Grinfeld, J. et al. A common novel splice variant of SLC22A1 (OCT1) is associated with impaired responses to imatinib in patients with chronic myeloid leukaemia. Br. J. Haematol. 163, 631–639 (2013).
Larson, R. A. et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood 111, 4022–4028 (2008).
Picard, S. et al. Trough imatinib plasma levels are associated with both cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood 109, 3496–3499 (2007).
Gambacorti-Passerini, C. et al. Alpha1 acid glycoprotein binds to imatinib (STI571) and substantially alters its pharmacokinetics in chronic myeloid leukemia patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 9, 625–632 (2003).
Angelini, S. et al. Association between imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica 98, 193–200 (2013).
Giannoudis, A. et al. The hOCT1 SNPs M420del and M408V alter imatinib uptake and M420del modifies clinical outcome in imatinib-treated chronic myeloid leukemia. Blood 121, 628–637 (2013).
Ng, K. P. et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat. Med. 18, 521–528 (2012).
Ko, T. K. et al. The BIM deletion polymorphism: A paradigm of a permissive interaction between germline and acquired TKI resistance factors in chronic myeloid leukemia. Oncotarget 7, 2721–2733 (2016).
Ying, H.-Q. et al. The effect of BIM deletion polymorphism on intrinsic resistance and clinical outcome of cancer patient with kinase inhibitor therapy. Sci. Rep. 5, 11348 (2015).
Lee, W., Lockhart, A. C., Kim, R. B. & Rothenberg, M. L. Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. Oncologist 10, 104–111 (2005).
Bhatia, S. et al. Systemic Exposure to Thiopurines and Risk of Relapse in Children With Acute Lymphoblastic Leukemia: A Children’s Oncology Group Study. JAMA Oncol. 1, 287–295 (2015).
Ishikawa, Y. et al. Trough plasma concentration of imatinib reflects BCR-ABL kinase inhibitory activity and clinical response in chronic-phase chronic myeloid leukemia: a report from the BINGO study. Cancer Sci. 101, 2186–2192 (2010).
Jain, P. P. et al. Fluorescence in situ hybridization patterns of BCR/ABL1 fusion in chronic myelogenous leukemia at diagnosis. Indian J. Pathol. Microbiol. 55, 347 (2012).
Velpandian, T. et al. Development and validation of a simple liquid chromatographic method with ultraviolet detection for the determination of imatinib in biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 804, 431–434 (2004).
Balasubramanian, P. et al. International Reporting Scale of BCR-ABL1 Fusion Transcript in Chronic Myeloid Leukemia: First Report from India. Acta Haematol. 127, 135–142 (2012).
Markose, P. et al. Spectrum of BCR-ABL kinase domain mutations in patients with chronic myeloid leukemia from India with suspected resistance to imatinib-mutations are rare and have different distributions. Leuk. Lymphoma 50, 2092–2095 (2009).
Balasubramanian, P. et al. Population pharmacokinetics of cyclophosphamide in patients with thalassemia major undergoing HSCT. Bone Marrow Transplant. 47, 1178–1185 (2012).
Baccarani, M., Castagnetti, F., Gugliotta, G. & Rosti, G. A review of the European LeukemiaNet recommendations for the management of CML. Ann. Hematol. 94(Suppl 2), S141-147 (2015).
Jabbour, E., Saglio, G., Hughes, T. P. & Kantarjian, H. Suboptimal responses in chronic myeloid leukemia: implications and management strategies. Cancer 118, 1181–1191 (2012).
Gene expression signature that predicts early molecular response failure in chronic-phase CML patients on frontline imatinib | Blood Advances | American Society of Hematology. https://ashpublications.org/bloodadvances/article/3/10/1610/246652/Gene-expression-signature-that-predicts-early.
Kantarjian, H. M. et al. Nilotinib versus imatinib for the treatment of patients with newly diagnosed chronic phase, Philadelphia chromosome-positive, chronic myeloid leukaemia: 24-month minimum follow-up of the phase 3 randomised ENESTnd trial. Lancet Oncol. 12, 841–851 (2011).
Larson, R. A. et al. Nilotinib vs imatinib in patients with newly diagnosed Philadelphia chromosome-positive chronic myeloid leukemia in chronic phase: ENESTnd 3-year follow-up. Leukemia 26, 2197–2203 (2012).
Etienne, G. et al. Incidence and outcome of BCR-ABL mutated chronic myeloid leukemia patients who failed to tyrosine kinase inhibitors. Cancer Med. 8, 5173–5182 (2019).
Soverini, S. et al. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leuk. Res. 38, 10–20 (2014).
Khorashad, J. S. et al. Finding of kinase domain mutations in patients with chronic phase chronic myeloid leukemia responding to imatinib may identify those at high risk of disease progression. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 26, 4806–4813 (2008).
Ganesan, P. & Kumar, L. Chronic Myeloid Leukemia in India. J. Glob. Oncol. 3, 64–71 (2016).
Mahon, F.-X. et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 11, 1029–1035 (2010).
Etienne, G. et al. Long-Term Follow-Up of the French Stop Imatinib (STIM1) Study in Patients With Chronic Myeloid Leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 35, 298–305 (2017).
Malhotra, H. et al. Correlation of plasma trough levels of imatinib with molecular response in patients with chronic myeloid leukemia. Leuk. Lymphoma 55, 2614–2619 (2014).
Arora, B. et al. Therapeutic drug monitoring for imatinib: Current status and Indian experience. Indian J. Med. Paediatr. Oncol. Off. J. Indian Soc. Med. Paediatr. Oncol. 34, 224–228 (2013).
Hoffmeyer, S. et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. U. S. A. 97, 3473–3478 (2000).
Fung, K. L. & Gottesman, M. M. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta 1794, 860–871 (2009).
Wang, D., Johnson, A. D., Papp, A. C., Kroetz, D. L. & Sadée, W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet. Genomics 15, 693–704 (2005).
Eadie, L. N. et al. The clinical significance of ABCB1 overexpression in predicting outcome of CML patients undergoing first-line imatinib treatment. Leukemia 31, 75–82 (2017).
Chapuy, B. et al. ABC transporter A3 facilitates lysosomal sequestration of imatinib and modulates susceptibility of chronic myeloid leukemia cell lines to this drug. Haematologica 94, 1528–1536 (2009).
Giannoudis, A. et al. The clinical significance of ABCC3 as an imatinib transporter in chronic myeloid leukaemia. Leukemia 28, 1360–1363 (2014).
Takahashi, N. et al. Correlation between imatinib pharmacokinetics and clinical response in Japanese patients with chronic-phase chronic myeloid leukemia. Clin. Pharmacol. Ther. 88, 809–813 (2010).
Forrest, D. L. et al. Cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia are correlated with Sokal risk scores and duration of therapy but not trough imatinib plasma levels. Leuk. Res. 33, 271–275 (2009).
Singh, N., Kumar, L., Meena, R. & Velpandian, T. Drug monitoring of imatinib levels in patients undergoing therapy for chronic myeloid leukaemia: comparing plasma levels of responders and non-responders. Eur. J. Clin. Pharmacol. 65, 545–549 (2009).
Harivenkatesh, N. et al. Influence of MDR1 and CYP3A5 genetic polymorphisms on trough levels and therapeutic response of imatinib in newly diagnosed patients with chronic myeloid leukemia. Pharmacol. Res. 120, 138–145 (2017).
White, D. L. et al. OCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood 108, 697–704 (2006).
Corbin, A. S. et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Invest. 121, 396–409 (2011).
Perl, A. & Carroll, M. BCR-ABL kinase is dead; long live the CML stem cell. J. Clin. Invest. 121, 22–25 (2011).
Dulucq, S. et al. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood 112, 2024–2027 (2008).
Kim, D. H. D. et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 15, 4750–4758 (2009).
Jiang, Z.-P. et al. Trough concentration and ABCG2 polymorphism are better to predict imatinib response in chronic myeloid leukemia: a meta-analysis. Pharmacogenomics 18, 35–56 (2017).
Hughes, T. P. et al. Early molecular response predicts outcomes in patients with chronic myeloid leukemia in chronic phase treated with frontline nilotinib or imatinib. Blood 123, 1353–1360 (2014).
Akard, L. P. & Bixby, D. Considering baseline factors and early response rates to optimize therapy for chronic myeloid leukemia in chronic phase. Leuk. Lymphoma 57, 1002–1014 (2016).
Stella, S. et al. Clinical Implications of Discordant Early Molecular Responses in CML Patients Treated with Imatinib. Int. J. Mol. Sci. 20, (2019).
Cortes, J. et al. Molecular responses in patients with chronic myelogenous leukemia in chronic phase treated with imatinib mesylate. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 11, 3425–3432 (2005).
Hughes, T. P. et al. Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS). Blood 116, 3758–3765 (2010).
Acknowledgements
This study is supported by Department of Biotechnology India-Programme support Grant: BT/01/COE/08/03; Centre of Excellence grant from Department of Biotechnology India: BT/COE/34/SP13432/2015 and Indian Council of Medical Research Centre for Advanced Research Grant 70/14/14-CAR to Dr. Poonkuzhali Balasubramanian. RVS, VM and PB are supported by Wellcome DBT India Alliance (IA/S/17/1/503118, IA/CPHS/18/1/503930 and IA/S/15/1/501842) respectively. UK is supported by an early career fellowship program of Wellcome DBT India Alliance (IA/CPHE/17/1/503351), Government of India. SK is supported by University Grants Commission, ESB by DST Inspire fellowship and SV by Indian Council of Medical Research, Government of India. We sincerely acknowledge the encouragement and support provided by Dr. Mammen Chandy, Professor and Former Head, Department of Haematology, CMC Vellore, currently Director, Tata Medical Centre, Kolkata in the initial stages of this study. The help provided by Ms. Preetha Markose & Dr. Ajay Abraham in the initial stages of this study, Mr. Christopher Benjamin, Mr. Selvakumar, Ms. Kalaiselvi in CML patient recruitment for the study and by Dr. Eunice S. Edison in managing the DNA sequencing core facility are gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
B.M.R., E.S.B., S.G., S.A., S.K., S.V., E.M. and P.B. performed experiments, analysed the results. P.B. designed the research. B.M.R. and P.B. wrote the manuscript. K.M.L. contributed to statistical analyses. N.B.J., V.M.S., S.R.V., A.A., U.K., A.J.D., F.N.A., A.K., B.G., A.S., V.M. – enrolled patients, managed them and provided clinical data. P.B. and V.M. reviewed the manuscripts and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rajamani, B.M., Benjamin, E.S.B., Abraham, A. et al. Plasma imatinib levels and ABCB1 polymorphism influences early molecular response and failure-free survival in newly diagnosed chronic phase CML patients. Sci Rep 10, 20640 (2020). https://doi.org/10.1038/s41598-020-77140-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77140-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.