Abstract
The oral cavity may comprise a significant reservoir for Staphylococcus aureus but the data on molecular epidemiology and clonal distribution of oral strains are really scarce. This study aimed to evaluate the clonal relatedness in S. aureus isolated from oral cavity and their relationship with carriage of virulence genes, and antimicrobial resistance profiles. A total of 139 oral S. aureus isolates were obtained from 2327 analysed oral samples of dental patients. Antimicrobial susceptibility testing was performed. Isolates were characterized using protein A gene (spa) typing, spa-CC clonal complexes, toxin genes and SCCmec typing for MRSA. High resistance rates for penicillin, tetracycline and gentamicin were detected, respectively 58.3%, 42.4%, and 35.2%. Twelve (8.6%) S. aureus isolates were identified as MRSA. All of MRSA isolates were mecA-positive and mecC-negative. SCCmec IV was the most common type (66.7%), which was typical for community-acquired MRSA (CA-MRSA). Overall, the enterotoxin gene cluster (egc) was the most frequent detected virulence factor (44.9%), both in MSSA and MRSA isolates. Presence of genes encoding for the enterotoxins (sea, seb, sec, seh, sek), exfoliative toxin A (eta), and toxic shock syndrome toxin-1 (tst) was also observed. Strains carrying lukS-PV/lukF-PV genes belonged to SCCmecV- spa type t437. The most prevalent spa types were t091, t015, t084, t002, t571, and t026 among all 57 identified. Spa types, including 3 new ones, grouped in 6 different spa-CC clonal complexes, with four major dominated; CC45, CC30, CC5, and CC15. This study demonstrated that both methicillin-susceptible and methicillin-resistant major European clones of S. aureus could be isolated from the oral cavity of dental patients, with the emergence of PVL-positive CA-MRSA strains. The oral cavity should be considered as a possible source of toxigenic egc-positive S. aureus strains, in terms of potential risk of cross-infection and dissemination to other body sites.
Similar content being viewed by others

Introduction
Staphylococcus aureus is responsible for a wide variety of human infections ranging from mild symptoms in superficial skin infections to life-threatening systemic disease, such as infective endocarditis and sepsis1.
Staphylococcus aureus may also be associated with some oral conditions and infections in dentistry. These include angular cheilitis, mucositis, some endodontic infections, osteomyelitis of the jaw, and parotitis2,3,4. Recent reports suggest that S. aureus is possibly also involved in the pathogenesis of periodontal lesions5,6.
However, the role of S. aureus in oral infections still raises some controversies since the presence of S. aureus in the oral cavity has the commensal asymptomatic character in the healthy carriers7,8. Although anterior nares are considered a primary ecological niche for S. aureus, it is estimated that 15–50% of persons colonized by these microorganisms are non-nasal carriers9. The oral cavity is frequently colonized by S. aureus, either as a primary location or as a consequence of migration from the anterior nares10,11. Recent evidence suggests that the colonization may be not only transient but also persistent12,13. According to Kearney, the oral cavity represents a significant and underappreciated reservoir for S. aureus10.
The oral carriage may give rise to staphylococcal infection, whether endogenous or cross-infection. Oral carriers, especially immunocompromised persons, such as hemodialyzed patients, subjects with haematological malignancies, rheumatoid arthritis, diabetes mellitus, etc., have increased risk of severe endogenous infections14,15,16,17. According to Terpenning et al., the presence of S. aureus in saliva was a significant risk factor for aspiration pneumonia18. Also, a strong relationship was found between the isolation of S. aureus and the occurrence of other severe infections, such as bacteraemia and infective endocarditis19,20,21. As the majority of these infections are endogenous, the risk among S. aureus colonized patients is 11.5 times higher than in non-colonized persons22.
Furthermore, the oral cavity may also constitute a reservoir for transmission events23. Small et al. emphasized that this site can be often overlooked for screening and subsequent decolonization24. A large body of evidence suggests that the transmission of S. aureus may occur between patients and dentist via the clinical environment3,4,25.
Thus understanding the distribution and relatedness of staphylococcal clones colonizing oral cavity is essential for the strategies to control its dissemination and to reduce the incidence of infections26.
Currently, genotyping using protein A gene (spa) typing is the most popular method for the epidemiological analysis of S. aureus isolates, their genetic relatedness and diversity27,28. spa typing is a useful tool for discriminating S. aureus from different sources and nosocomial infection control29. The method is based on spa gene polymorphism in the X-region, with variable number of 24-bp repeat sequences. The results of spa typing correlate with other genotyping methods, especially with worldwide used clonal grouping based on multilocus sequence typing (MLST)30,31.
The studies of molecular epidemiology and clonal distribution of S. aureus isolated from the oral cavity are really scarce. This study aimed to evaluate the clonal relatedness in S. aureus isolated from oral cavity and their relationship with carriage of virulence genes, as well as antimicrobial resistance profiles.
Results
Prevalence of S. aureus isolates
A total 139 S. aureus were isolated from 2327 oral samples of 750 (18.5%) dental patients with symptoms of infection. S. aureus were detected among 128 patients, 71 females and 57 males aged between 17 and 82 years (mean 56 years). The number of collected samples from each patients varied, from one sample per patients (118 patients) to two (9 patients) or three samples (1 patient). Sixty seven isolates derived from dorsum of the tongue swabs, 38 isolates from buccal mucosa swabs, 19 isolates from denture swabs and 15 isolates from the corners of the mouth swabs.
Antimicrobial susceptibility
Overall, 139 isolates demonstrated resistance to penicillin (58.3%), tetracycline (42.4%), gentamicin (35.2%), clindamycin (19.4%), erythromycin (19.4%), amoxicillin/clavulanic acid (16.8%), cefoxitin (8.6%), oxacillin (8.6%), chloramphenicol (4.3%), trimethoprim/sulfamethoxazole (4.3%), and ciprofloxacin (1.4%). None of the isolates were resistant to vancomycin. A high proportion (68%) of resistance to gentamicin obtained in disk diffusion method was verified by E-tests (35.2%). D-test demonstrated that 15.1% of isolates represented the inducible phenotype of clindamycin resistance (MLSBi) (Table 1).
Twelve (8.6%) S. aureus isolates were identified as MRSA. All of MRSA isolates were mecA-positive, while none harboured mecC. Twenty-five percent of isolates showed MLSBi resistance and this proportion was higher than in MSSA isolates. Resistance to tetracycline, clindamycin, erythromycin, gentamicin, and trimethoprim/sulfamethoxazole was found in 41.7%, 25%, 25%, 16.6% and 8.3% of the MRSA isolates, respectively. All isolates were sensitive to ciprofloxacin, chloramphenicol, and vancomycin (Table 1). Staphylococcal cassette chromosome mec (SCCmec) types IV (66.7%) and V (33.3%) were detected, suggesting a community origin (CA-MRSA). No isolate represented I, II and III SCCmec types.
Multidrug-resistance (MDR) was observed among 38 (27.3%) of all isolated S. aureus, in MRSA turned out to be higher (41.7%) than in MSSA (26%). MDR isolates were resistant to 3, 4 and 5 groups of antibiotics; 57.9%, 18.4%, and 23.7% respectively.
Distribution of toxin genes
Analysis of the distribution of virulence genes among the 139 oral S. aureus isolates evidenced that 66.9% of them contained genes encoding toxins, with a high percentage in MRSA isolates (83.3%). Overall, the gene cluster egc (seg, sei, sem, sen, seo, seu) were the most common detected superantigen (44.9%). Genes encoding for the enterotoxins sea (9.4%), seb (3.6%), sec (12.9%), seh (2.2%), sek (3.6%), exfoliative toxin A eta (2.2%), and toxic shock syndrome toxin-1 tst (7.2%) were also identified. None of the isolates tested positively for sed and see enterotoxin genes. Detection of the Panton-Valentine leukocidin genes (lukS-PV/lukF-PV) (p = 0.007), the enterotoxin genes seb and sek were significantly more prevalent (16.6%) among MRSA than among MSSA isolates. While enterotoxin genes sea, seh, sej, sel and exfoliative toxin B gene (etb) were solely present in MSSA isolates, 10.2%, 2.4%, 9.4%, 4.7%, 0.8%, respectively (Table 2).
spa typing
The spa typing analysis revealed distinct 57 spa types within 139 tested S. aureus isolates, the most common were: t091 (10.8%), t015 (7.9%), t002 (5.0%), t012 (5.0%), t084 (4.3%), t005 (3.6%), t230 (2.9%), t571 (2.2%) ,t026 (2.2%). t065 (2.2%), t148 (2.2%), t160 (2.2%), and t242 (2.2%). The other spa types were less frequent, such as t045, t056, t085, t127, t131, t209, t437, t688, t693, t711, t1268, and t3297. The determined spa types included three new ones: t18952, t18953, and t18954; all of them were registered in the international database, Ridom SpaServer (https://spaserver.ridom.de/) (Table 3).
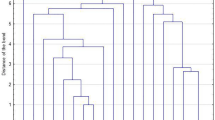
The identified spa types were clustered into six spa-clonal complexes (spa-CCs) by BURP repeat analysis; spa-CC 571/1451, spa-CC 021, spa-CC 084, spa-CC 888, spa-CC 015, and spa-CC 267. Forty-six (33.1%) isolates represented 19 spa types and belonged to singletons, ten (7.2%) isolates were excluded from BURP cluster analysis due to presence of less than four spa repeats. The newly described spa types were distributed across various clusters and singletons (Fig. 1, Table 3).
Population structure of 139 S. aureus oral isolates after BURP analysis with a cost of 4. Clusters of linked spa types correspond to spa-CCs. The spa types that were defined as founders of particular clusters are indicated in blue. % of strains based on 139 strains collection; % of spa-types based on 57 spa-types (including excluded ones).
Twelve MRSA isolates were assigned to three clonal complexes (spa-CC 084, spa-CC 888 and spa-CC 021) and five singletons, comprising of 9 spa types (t012, t091, t156, t189, t437, t888, t5644, t13670, t18953). Two isolates were excluded due to the low number of repeats. Particular attention should be given to two isolates of the spa type t437 having lukS-PV/lukF-PV genes as well as seb and sek (Table 3).
Overall, most isolates (10%) were assigned to the spa-CC 015 clonal complex and carried the most enterotoxin genes, egc (78.6%), sec (71.4%), sel, (42.8%), and sej (28.6%). egc-positive S. aureus isolates were not limited to a single clonal group. All isolates belonged to the spa-CC 021 also had superantigen genes, such as egc (72.7%), sea (72.7%) and tst (27.3%). All of them showed resistance to penicillin and most to tetracycline (81.8%). Another toxigenic clonal complex, spa-CC571/1451 carried egc genes and showed inducible phenotype of erythromycin and clindamycin resistance. spa-CC 084 contained 7.2% of isolates, 90% of which did not have any toxin genes and were penicillin-resistant (Tables 3, 4, Fig. 1).
Most of isolates belonging to major MLST clonal complexes (CC45, 30, 5, and 22) were egc-positive. The differences in prevalence of egc-positive isolates were statistically significant (p < 0.001), as shown in Table 5.
Discussion
The epidemiology of S. aureus infections constantly changes, with novel clones emerging in various geographical regions19,27,32. This warrants continuous surveillance of staphylococcal isolates from different sources, especially from the oral cavity. Recent evidence suggests that the latter constitutes a significant yet underrated reservoir of S. aureus10.
Staphylococcus aureus isolation rates differ considerably depending on the population. In healthy adults, oral carriage rates vary from 12 to 36%, with more frequent isolation (17–48%) among students4,33. The carriage rate documented in our study (18.5%) was similar as reported by other authors34,35,36, and remained within a typical range for European and American adult dental patients. It was, however, lower than in dentistry students examined by Ohara-Nemoto (46.6%)7, and in patients with periodontitis included in another study6. Acrylic denture wearers also seem to be predisposed to the oral carriage37, with S. aureus isolation rates of 27–48%2,38.
Nearly 9% of isolates included in this study were resistant to methicillin. The prevalence of MRSA in the oral cavity of adult dental patients is known to be relatively low, 5–12%2,5,36,39. Smith found MRSA strains among 6% of studied patients, more often in those over 70 years39. Long-term MRSA carriage was also reported in 6 out of 10 complete denture wearers participating in a Finnish study, and 10.3% of Asian denture wearers38,40.
MRSA are typically isolated in a hospital setting; such hospital-acquired MRSA (HA-MRSA) are believed to spread primarily via human-to-human transmission41. However, a growing number of infections caused by non-nosocomial MRSA, referred to as community-acquired MRSA (CA-MRSA) was also observed in the last decade42,43. CA-MRSA genetically differ from HA-MRSA, are less resistant to non-β-lactam antibiotics and carry a smaller version of staphylococcal cassette chromosome mec (SCCmec)44,45. All MRSA isolates in this study harbored SCCmec type IV or V and were more susceptible to non-β-lactam antimicrobials, which suggested their community origin (CA-MRSA). None of the MRSA were resistant to ciprofloxacin, chloramphenicol or vancomycin. The proportions of strains resistant to gentamycin, clindamycin, and erythromycin were similar as in other types of staphylococcal infections27,46. The analyzed MRSA showed multidrug resistance (MDR) more often than MSSA, which is consistent with the characteristics of MRSA strains47,48.
spa typing demonstrated a broad genetic diversity of our staphylococcal isolates. We did not find any published report about the clonal variety of oral S. aureus. Nevertheless, the heterogeneity observed in our series corresponds well with recent European and American data about MRSA involved in other infections49,50. Rather than showing clustering, a typical feature of MRSA, most of our methicillin-susceptible isolates displayed extensive genetic diversity. Our MSSA corresponded to major clones widespread in Poland and other European countries19,49,51,52,53,54. In contrast, different predominant clonal complexes were reported in Africa and Asia, indicating geographical variation53,55,56.
The most toxigenic clonal complexes in our series were CC45, CC30 and CC22, which included the largest proportion of staphylococci testing positively for toxin genes in egc locus. The enterotoxin gene cluster (egc) encodes up to six enterotoxins (SEG, SEI, SEM, SEN, SEO, and SEU) being superantigens. The superantigens induce T lymphocytes and antigen-presenting cells causing massive cytokine production, with lethal effects dependent on direct toxic and cytokine effects on the cardiovascular system57. Recent studies showed that egc-clustered enterotoxins are the most prevalent virulence factors in S. aureus isolated nowadays27,46,58. However, longitudinal studies are needed to better elucidate the role of locus egc in oral S. aureus strains.
The predominant clonal complex in our material was CC45, with the most common spa type being spa-CC t015. CC45 are widely distributed among both nasal colonization and bloodstream infections strains in Europe19,59, and recent results suggest that the nasal isolates carry the potential to cause an invasive disease59. However, no reports on severe infections caused by oral CC45 strains have been published to date. Bonnet observed predominance of spaCC t015 among S. aureus associated with infective endocarditis60, and Deasy pointed to this spa type as an emerging etiological factor of bloodstream infections19. Other authors reported on infective endocarditis after dental extraction and treatment 61,62. These findings imply that peroral spread of endogenous S. aureus should be considered in at least some instances. Our findings suggest that spa-CC t015 seems to be particularly prone to the acquisition of virulence factors, including superantigens genes, sec, sel, sej and egc. These findings are consistent with the data from a report on bloodstream infections63.
The second most common clonal complex in our series was CC30. Many previous studies analyzed CC30 strains and their link with endocarditis64; and some authors demonstrated their role in the development of an invasive disease19,20. Our spa-CC 021 belonging to CC30 clone characterized high proportion of egc-, sea-and tst-positive strains, especially CC30-t012, which is consistent with the results of previous studies28,52,65. Also, according to an American report, CC30 strains from patients with infective endocarditis were significantly more likely to contain these enterotoxin genes and had the potential to cause hematogenous complications64.The high resistance rates to penicillin and tetracycline was observed among our CC30 strains. To this date, resistance to tetracycline was not reported in S. aureus from the oral cavity but in the strains from other human or animal sources66,67,68,69,70. However, tetracycline is also used in the treatment of some oral infections in humans, especially in patients with periodontitis71. Thus, our observation on the potential emergence of tetracycline-resistant oral strains warrants further investigation.
To the best of our knowledge, present study was the first to demonstrate the prevalence of spa type t437 SCCmec-V-pvl-positive S. aureus strains in dental patients. Panton-Valentine leukocidin (PVL) genes are considered a stable genetic marker for CA-MRSA strains carrying SCCmec type IV or V72,73. More than half (66.6%) of strains identified in this study were assigned to SCCmec IV, whereas the strains represented spa t437 harbored SCCmec V. Similar strains were isolated in Germany and Taiwan, and according to a recent Polish report, t437 SCCmec-IV-pvl-positive strains predominated in specimens from diabetic patients74,75,76. The PVL -positive strains were associated with purulent skin infections, necrotizing pneumonia, pyomyositis and other S. aureus infections74,77,78,79. PV leukocidin is considered a potent inducer of inflammation and cytotoxicity but its role in oral infections is still little unknown80,81.
Our study has several limitations. First, the proportion of MRSA strains was small in comparison with number of MSSA strains. It should be taken account in the interpretation of the results. Second, we did not analyze clinical data of the swabbed patients, such as type of oral infection and its manifestations. However, even considering these drawbacks, the results of our study add to current knowledge about oral S. aureus strains.
In conclusion, this study demonstrated that both methicillin-susceptible and methicillin-resistant S. aureus strains major European clones could be isolated from the oral cavity of dental patients, with the emergence of PVL-positive CA-MRSA strains. The oral cavity should be considered as a possible source of toxigenic egc-positive S. aureus strains, in terms of potential risk of cross-infection and dissemination to other body sites.
Materials and methods
Isolation and identification of S. aureus
The study included oral S. aureus isolated from all 2327 oral microbiological samples analysed consecutively at the Laboratory of Department of Oral Microbiology of the Medical University of Gdansk during routine clinical laboratory procedures, over a period of three years. The samples were obtained with sterile cotton swabs from the oral mucosa, the dorsal surface of the tongue, denture surface and angular cheilitis lesions. The analysed S. aureus were not specifically isolated for this research, they were part of the diagnostic laboratory procedure and no humans were involved in the experiments.
All samples were plated onto Columbia blood agar (GrasoBiotech, Starogard Gd., Poland) and mannitol salt agar (bioMérieux, Marcy l'Etoile, France) and were incubated 18–24 h at 37 °C. Suspected staphylococcal colonies were identified by standard methods, on the basis of colony characteristics, pigment production, Gram-staining, haemolysis and Pastorex StaphPlus latex agglutination kit (Bio-Rad, Marnes la Coquette, France). Further, all isolates eventually identified as S. aureus based on PCR amplification of species-specific thermostable nuclease gene (nuc)82.
After final identification, the isolates were stored at − 80 °C in Trypticase Soy Broth (Becton Dickinson, Franklin Lakes, NJ, USA) supplemented with 20% glycerol.
Antimicrobial susceptibility testing
The antimicrobial susceptibility was determined on Mueller–Hinton agar plates (Becton Dickinson, Franklin Lakes, NJ, USA) by the disk diffusion method and interpreted according to the EUCAST83. The following antimicrobial agents were tested: oxacillin, cefoxitin, gentamicin, erythromycin, clindamycin, tetracycline, chloramphenicol, ciprofloxacin, amoxicillin/clavulanic acid, trimethoprim/sulfamethoxazole (Bio-Rad, Marnes la Coquette, France) and penicillin G (Oxoid, Basingstoke, England). The inducible resistance to macrolide-lincosamide-streptogramin B (MLSB) was detected by disk diffusion method with use the clindamycin (2 μg) and erythromycin (15 μg) disks positioned 15–26 mm apart83. Resistance to gentamicin was verified by using E-tests (bioMérieux, Marcy l'Etoile, France), an isolate with MIC value > 1 µg/ml was considered as a resistant83. Vancomycin susceptibility was determined with E-test strips (bioMerieux, Marcy-l’Etoile, France), in line with the manufacturer’s instruction. Multidrug resistance (MDR) was defined as a resistance to three or more classes of antimicrobials. Resistance to methicillin was first identified using cefoxitin (30 µg) and oxacillin (1 µg) disks, and then confirmed by the detection of PBP2a protein (Latex Agglutination Test Kit, Oxoid, Basingstoke, England), and verified by the detection of the mecA gene according to Khairalla et al.25 and mecC according to Stegger et al.84. S. aureus ATCC25923 (methicillin-susceptible) and S. aureus ATCC43300 (MDR) were used as the reference strains.
Molecular characterization
Isolation of staphylococcal DNA
Genomic Micro AX Staphylococcus Gravity kit (A&A Biotechnology, Gdynia, Poland) was used to isolate genomic DNA from bacteria by gravity according to the manufacturer's instructions.
Detection of toxin genes
Genes of the enterotoxins (sea, seb, sec, sed, see), toxic shock syndrome toxin-1 (tst), and exfoliative toxins (eta, etb) were detected as described by Becker et al.85, for the other enterotoxins genes (seg, seh, sei, sej, sek, sel, sem, sen, seo, seu) according to Bania et al.86. Detection of Panton-Valentine leukocidin genes (lukS-PV/lukF-PV) was performed as described by Lina et al.87.
SCCmec typing
Typing of the five (I–V) major staphylococcal chromosomal cassette mec (SCCmec) in MRSA strains was determined by PCR as described by Oliveira et al.88 and by Milheiriço et al.89. The SCCmec type was determined on the basis of the band pattern profiles obtained.
spa typing
The spa typing, based on amplification of the variable X region of protein A gene, was performed as described previously90. The spa types were assigned using the Ridom StaphType software version 2.2.1 (https://www.ridom.de/ Ridom GmbH, Wurzburg, Germany) and the Ridom SpaServer database (https://spaserver.ridom.de/). The predicted MLST were assigned based on Ridom SpaServer. The based upon repeat pattern (BURP) algorithm was used to calculate spa clonal complexes (spa-CCs) with following parameters: (I) exclude spa types shorter than 5 repeats; (II) cost less or equal to 4; (III) cluster composed of 2 or more related spa types was regarded as CC; (IV) a spa type that was not grouped into a CC was considered as singleton.
Statistical analysis
All calculations were performed with Statistica 10 package (StatSoft, Tulsa, OK, USA). The significance of between-group differences in the percentages of positive isolates was verified with Pearson chi-squared test or Fisher exact test. The threshold of statistical significance was set at p ≤ 0.05, with Bonferroni correction applied whenever multiple comparisons had to be carried out.
References
Asgeirsson, H., Thalme, A. & Weiland, O. Staphylococcus aureus bacteraemia and endocarditis—epidemiology and outcome: A review. Infect. Dis. 50, 175–192 (2018).
Lewis, N. et al. Colonisation of dentures by Staphylococcus aureus and MRSA in out-patient and in-patient populations. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1823–1826 (2015).
Kim, G. Y. & Lee, C. H. Antimicrobial susceptibility and pathogenic genes of Staphylococcus aureus isolated from the oral cavity of patients with periodontitis. J. Periodontal. Implant. Sci. 45, 223–228 (2015).
Smith, A. J., Jackson, M. S. & Bagg, J. The ecology of Staphylococcus species in the oral cavity. J. Med. Microbiol. 50, 940–946 (2001).
Passariello, C., Lucchese, A., Virga, A., Pera, F. & Gigola, P. Isolation of Staphylococcus aureus and progression of periodontal lesions in aggressive periodontitis. Eur. J. Inflamm. 10, 501–513 (2012).
Zhuang, L. F. et al. Periodontal and peri-implant microbiota in patients with healthy and inflamed periodontal and peri-implant tissues. Clin. Oral. Implant Res. 27, 13–21 (2016).
Ohara-Nemoto, Y., Haraga, H., Kimura, S. & Nemoto, T. K. Occurrence of staphylococci in the oral cavities of healthy adults and nasal–oral trafficking of the bacteria. J. Med. Microbiol. 57, 95–99 (2008).
Passariello, C., Puttini, M., Iebba, V., Pera, P. & Gigola, P. Influence of oral conditions on colonization by highly toxigenic Staphylococcus aureus strains. Oral Dis. 18, 402–409 (2012).
Wertheim, H. F. et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762 (2005).
Kearney, A. et al. The oral cavity revealed as a significant reservoir of Staphylococcus aureus in an acute hospital by extensive patient, healthcare worker and environmental sampling. J. Hosp. Infect. https://doi.org/10.1016/j.jhin.2020.03.004 (2020).
Patil, A. K. et al. Prevalence of community-associated methicillin-resistant Staphylococcus aureus in oral and nasal cavities of 4 to 13-year-old Rural School Children: A cross-sectional study. Contemp. Clin. Dent. 10, 99–104 (2019).
Percival, R. S., Challacombe, S. J. & Marsh, P. D. Age-related microbiological changes in the salivary and plaque microflora of healthy adults. J. Med. Microbiol. 35, 5–11 (1991).
Bueris, V., Pimenta, F. C., Ito, I. Y. & Marin, J. M. Oral incidence of Staphylococcus aureus and antimicrobials agents resistance. Braz. J. Oral. Sci. 4, 676–679 (2005).
Venditti, M. et al. Staphylococcus aureus bacteremia in patients with hematologic malignancies: A retrospective case-control study. Haematologica 88, 923–930 (2003).
Scheuch, M. et al. Staphylococcus aureus colonization in hemodialysis patients: A prospective 25 months observational study. BMC Nephrol. 20, 153. https://doi.org/10.1186/s12882-019-1332-z (2019).
Bassetti, S. et al. Staphylococcus aureus in patients with rheumatoid arthritis under conventional and anti-tumor necrosis factor-alpha treatment. J. Rheumatol. 32, 2125–2129 (2005).
Smit, J. et al. Diabetes and risk of community-acquired Staphylococcus aureus bacteremia: A population-based case-control study. Eur. J. Endocrinol. 174, 631–639 (2016).
Terpenning, M. S. et al. Aspiration pneumonia: Dental and oral risk factors in an older veteran population. J. Am. Geriatr. Soc. 49, 557–563 (2001).
Deasy, E. C. et al. A molecular epidemiological investigation of methicillin-susceptible Staphylococcus aureus causing bloodstream infections in Ireland, 2006–2017. Eur. J. Clin. Microbiol. Infect. Dis. 38, 927–936 (2019).
Pérez-Montarelo, D. et al. Molecular epidemiology of Staphylococcus aureus bacteremia: Association of molecular factors with the source of infection. Front. Microbiol. 9, 2210. https://doi.org/10.3389/fmicb.2018.02210 (2018).
Saeed, K. et al. An update on Staphylococcus aureus infective endocarditis from the International Society of Antimicrobial Chemotherapy (ISAC). Int. J. Antimicrob. Agents. 53, 9–15 (2019).
Zacharioudakis, I. M., Zervou, F. N., Ziakas, P. D. & Mylonakis, E. Meta-analysis of methicillin-resistant Staphylococcus aureus colonization and risk of infection in dialysis patients. J. Am. Soc. Nephrol. 25, 2131–2141 (2014).
Lam, O. L., McGrath, C., Bandara, H. M., Li, L. S. & Samaranayake, L. P. Oral health promotion interventions on oral reservoirs of Staphlococcus aureus: A systematic review. Oral. Dis. 18, 244–254 (2012).
Small, H. et al. The oral cavity—An overlooked site for MRSA screening and subsequent decolonisation therapy?. J. Infect. 55, 378–379 (2007).
Khairalla, A., Wash, R. & Ashour, H. M. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental healthcare personnel, patients, and environment. Sci. Rep. 7, 7390. https://doi.org/10.1038/s41598-017-07713-8 (2017).
Gompelman, M. et al. Long-term Staphylococcus aureus decolonization in patients on home parenteral nutrition: Study protocol for a randomized multicenter trial. Trials. 19, 346. https://doi.org/10.1186/s13063-018-2732-2 (2018).
Garbacz, K. et al. Emergence and spread of worldwide Staphylococcus aureus clones among cystic fibrosis patients. Infect. Drug Resist. 11, 247–255 (2018).
Ilczyszyn, W. M. et al. Clonal structure and characterization of Staphylococcus aureus strains from invasive infections in pediatric patients from South Poland: Assotiation between age, spatypes, clonal complexes, and genetic markers. PLoS ONE 11, e0154485. https://doi.org/10.1371/journal.pone.0154485 (2016).
Asadollahi, P. et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: A review. Front. Microbiol. 9, 163. https://doi.org/10.3389/fmicb.2018.00163 (2018).
Wang, X. Spa typing of Staphylococcus aureus isolates. Methods Mol. Biol. 2069, 89–94 (2020).
O’Hara, F. P. et al. spa Typing and multilocus sequence typing show comparable performance in a macroepidemiologic study of Staphylococcus aureus in the United States. Microb. Drug Resist. 22, 88–96 (2016).
Lisowska-Łysiak, K. et al. New insight into genotypic and phenotypic relatedness of Staphylococcus aureus strains from human infections or animal reservoirs. Pol. J. Microbiol. 68, 93–104 (2019).
Blomqvist, S., Leonhardt, A., Arirachakaran, P., Carlen, A. & Dahlѐn, G. Phenotype, genotype, and antibiotic susceptibility of Swedish and Thai oral isolates of Staphylococcus aureus. J. Oral. Microbiol. 7, 26250. https://doi.org/10.3402/jom.v7.26250 (2015).
McCormack, M. G. et al. Staphylococcus aureus and the oral cavity: An overlooked source of carriage and infection?. Am. J. Infect. Control. 43, 35–37 (2015).
Koukos, G. et al. Prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus (MRSA) in the oral cavity. Arch. Oral. Biol. 60, 1410–1415 (2015).
Cuesta, A. I., Jewtuchowicz, V. M., Brusca, M. I., Mujica, M. T. & Rosa, A. C. Antibiotic susceptibility of Staphylococcus aureus isolates in oral mucosa and pockets of patients with gingivitis-periodontitis. Acta Odontol. Latinoam. 24, 35–40 (2011).
Garbacz, K., Jarzembowski, T., Kwapisz, E., Daca, A. & Witkowski, J. Do the oral Staphylococcus aureus strains from denture wearers have a greater pathogenicity potential?. J. Oral. Microbiol. 11, 1536193. https://doi.org/10.1080/20002297.2018.1536193 (2018).
Tawara, Y., Honma, K. & Naito, Y. Methicillin-resistant Staphylococcus aureus and Candida albicans on denture surfaces. Bull. Tokyo Dent. Coll. 37, 119–128 (1996).
Smith, A. J. et al. Staphylococcus aureus in the oral cavity: A three-year retrospective analysis of clinical laboratory data. Br. Dent. J. 195, 701–703 (2003).
Rossi, T. et al. Eradication of the long-term carriage of methicillin-resistant Staphylococcusaureus in patients wearing dentures: A follow-up of 10 patients. J. Hosp. Infect. 34, 311–320 (1996).
Harris, S. R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010).
Nelson, L. et al. Community case of methicillin-resistant Staphylococcus aureus infection. Emerg. Infect. Dis. 12, 172–174 (2006).
Otter, J. A. & French, G. L. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Europe. Lancet Infect. Dis. 10, 227–239 (2010).
Chambers, H. & DeLeo, F. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641 (2009).
Ma, X. X. et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46, 1147–1152 (2002).
Katkowska, M., Garbacz, K., Kopala, W., Schubert, J. & Bania, J. Genetic diversity and antimicrobial resistance of Staphylococcus aureus from recurrent tonsillitis in children. AMPIS. https://doi.org/10.1111/apm.13007 (2019) (Epub ahead of print).
Li, X. et al. Molecular characteristics and virulence gene profiles of Staphylococcus aureus isolates in Hainan, China. BMC Infect. Dis. 19, 873. https://doi.org/10.1186/s12879-019-4547-5 (2019).
Chen, X. et al. Molecular and virulence characteristics of methicillin-resistant Staphylococcus aureus in burn patient. Front. Lab. Med. 1, 43–47 (2017).
Grundmann, H. et al. Geographic distribution of Staphylococcus aureuscausing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 7, e1000215. https://doi.org/10.1371/journal.pmed.1000215 (2010).
Miko, B. A. et al. Molecular characterization of methicillin-susceptible Staphylococcus aureus clinical isolates in the United States, 2004 to 2010. J. Clin. Microbiol. 51, 874–879 (2013).
Wiśniewska, K., Piórkowska, A., Kasprzyk, J., Bronk, M. & Świeć, K. Clonal distribution of bone sialoprotein-binding gene among Staphylococcus aureus isolates associated with bloodstream infections. Folia Microbiol. 59, 465–471 (2014).
Blomfeldt, A., Aamot, H. V., Eskesen, A. N., Müller, F. & Monecke, S. Molecular characterization of methicillin-sensitive Staphylococcus aureus isolates from bacteremic patients in a Norwegian University Hospital. J. Clin. Microbiol. 51, 345–347 (2013).
Ruffing, U. et al. Community-associated Staphylococcus aureus from Sub-Saharan Africa and Germany: A cross-sectional geographic correlation study. Sci. Rep. 7, 154. https://doi.org/10.1038/s41598-017-00214-8 (2017).
Tavares, A., Faria, N. A., Lencastre, H. & Miragaia, M. Population structure of methicillin-susceptible Staphylococcus aureus (MSSA) in Portugal over a 19-year period (1992–2011). Eur. J. Clin. Microbiol. Infect. Dis. 33, 423–432 (2014).
Vandendriessche, S. et al. Characterisation of Staphylococcus aureus isolates from bloodstream infections, Democratic Republic of the Congo. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1163–1171 (2017).
Dekker, D. et al. Antibiotic resistance and clonal diversity of invasive Staphylococcus aureus in the rural Ashanti Region, Ghana. BMC Infect. Dis. 16, 720. https://doi.org/10.1186/s12879-016-2048-3 (2016).
Stach, C. S. et al. Novel tissue level effects of the Staphylococcus aureus enterotoxin gene cluster are essential for infective endocarditis. PLoS ONE 11, e0154762. https://doi.org/10.1371/journal.pone.0154762 (2016).
Fischer, A. J. et al. High prevalence of Staphylococcus aureus enterotoxin gene cluster superantigens in cystic fibrosis clinical isolates. Genes. 10, 1036. https://doi.org/10.3390/genes10121036 (2019).
Roe, Ch. et al. Genomic analyses of Staphylococcus aureus clonal complex 45 isolates does not distinguish nasal carriage from bacteraemia. Microbial. Genom. https://doi.org/10.1099/mgen.0.000403 (2020).
Bonnet, I. et al. High prevalence of spatype t571 among methicillin-susceptible Staphylococcus aureusfrom bacteremic patients in a French University Hospital. PLoS ONE 13, e0204977. https://doi.org/10.1371/journal.pone.0204977 (2018).
Bayliss, R., Clarke, C., Oakley, C., Somerville, W. & Whitfield, A. G. W. The teeth and infective endocarditis. Br. Heart J. 50, 506–512 (1983).
Etienne, J., Fleurette, J., Ninet, J. F., Favet, P. & Gruer, L. D. Staphylococcal endocarditis after dental extraction. Lancet 2, 511–512 (1986).
Roetzer, A. et al. Genotypic and phenotypic analysis of clinical isolates of Staphylococcus aureus revealed production patterns and haemolytic potentials unlinked to gene profiles and source. BMC Microbiol. 16, 13. https://doi.org/10.1186/s12866-016-0630-x (2016).
Nienaber, J. J. et al. Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins [published correction appears in J Infect Dis. 2013 Feb 1;207(3):546]. J. Infect. Dis. 204, 704–713 (2011).
Sharma, H. et al. Clinical and molecular epidemiology of Staphylococcal Toxic Shock Syndrome in the United Kingdom. Emerg. Infect. Dis. 24, 258–266 (2018).
Valasco, V. et al. Characterization of Staphylococcus aureus from humans and a comparison with isolates of animal origin, in North Dakota, United States. PLoS ONE 10, e0140497. https://doi.org/10.1371/journal.pone.0140497 (2015).
Ye, X. et al. Livestock-associated methicillin and multidrug resistant S. aureus in humans is associated with occupational pig contact, not pet contact. Sci. Rep. 6, 19184. https://doi.org/10.1038/srep19184 (2016).
Benrabia, I., Hamdi, T. M., Shehata, A. A., Neubauer, H. & Wareth, G. Methicillin-resistant Staphylococcus aureus (MRSA) in poultry species in Algeria: Long-term study on prevalence and antimicrobial resistance. Vet. Sci. 7, 54. https://doi.org/10.3390/vetsci7020054 (2020).
Li, H. et al. Antimicrobial resistance and virulence gene profiles of methicillin-resistant and susceptible Staphylococcus aureus from food products in Denmark. Front. Microbiol. 10, 2681. https://doi.org/10.3389/fmicb.2019.02681 (2019).
Jamali, H., Radmehr, B. & Ismail, S. Short communication: Prevalence and antibiotic resistance of Staphylococcus aureus isolated from bovine clinical mastitis. J. Dairy Sci. 97, 2226–2230 (2014).
Akrivopoulou, C., Green, I. M., Donos, N., Nair, S. P. & Ready, D. Aggregatibacter actinomycetemcomitans serotype prevalence and antibiotic resistance in a UK population with periodontitis. J. Glob. Antimicrob. Resist. 10, 54–58 (2017).
Vandenesch, F. et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: Worldwide emergence. Emerg. Infect. Dis. 9, 978–984 (2003).
D’Souza, N., Rodrigues, C. & Mehta, A. Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. J. Clin. Microbiol. 48, 1806–1811 (2010).
Klein, S., Menz, M.-D., Zanger, P., Heeg, K. & Nurjadi, D. Increase in the prevalence of Panton-Valentine leukocidin and clonal shift in community-onset methicillin-resistant Staphylococcus aureus causing skin and soft-tissue infections in the Rhine-Neckar Region, Germany, 2012–2016. Int. J. Antimicrob. Agents. 53, 261–267 (2019).
Chen, F. J. et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J. Clin. Microbiol. 50, 1679–1683 (2012).
Stańkowska, M., Garbacz, K., Piechowicz, L. & Bronk, M. Dissemination of t437-SCCmecIV and coagulase-negative t037-SCCmecIII types among borderline oxacillin-resistant Staphylococcus aureus isolated from skin infections and diabetic foot ulcers. Infect. Drug Resist. 12, 3197–3203 (2019).
Gillet, Y. et al. Factors predicting mortality in necrotizing community-acquired pneumonia caused by Staphylococcus aureus containing Panton-Valentine leukocidin. Clin. Infect. Dis. 45, 315–321 (2007).
Young, B. C. et al. Panton-Valentine leucocidin is the key determinant of Staphylococcus aureus pyomyositis in a bacterial GWAS. eLife. 8, e42486. https://doi.org/10.7554/eLife.42486 (2019).
Boyle-Vavra, S. & Daum, R. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab. Investig. 87, 3–9 (2007).
Elizur, A. et al. Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus lung infections in patients with cystic fibrosis. Chest 131, 1718–1725 (2007).
Zhang, C., Guo, Y. & Chu, X. In vitro generation of Panton-Valentine leukocidin (PVL) in clinical methicillin-resistant Staphylococcus aureus (MRSA) and its correlation with PVL variant, clonal complex, infection type. Sci. Rep. 8, 7696. https://doi.org/10.1038/s41598-018-26034-y (2018).
Baron, F. et al. Development of a PCR test to differentiate between Staphylococcus aureus and Staphylococcus intermedius. J. Food Prot. 67, 2302–2305 (2004).
The European Committee on Antimicrobial Susceptibility Testing (EUCAST) https://www.eucast.org/ (2018).
Stegger, M. et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or new mecA homologue mecALGA251. Clin. Microbiol. Infect. 18, 395–400 (2012).
Becker, K., Roth, R. & Peters, G. Rapid and specific detection of toxigenic Staphylococcus aureus: Use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36, 2548–2553 (1998).
Bania, J. et al. Distribution of newly described enterotoxin-like genes in Staphylococcus aureus from food. Int. J. Food Microbiol. 108, 36–41 (2006).
Lina, G. et al. Involvement of Panton-Valentine Leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132 (1999).
Oliveira, D. C. & de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46, 2155–2161 (2002).
Milheirico, C., Oliveira, D. C. & de Lancastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 3374–3377 (2007).
Aires-de-Sousa, M. et al. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44, 619–621 (2006).
Acknowledgements
The study was financially supported by the Medical University of Gdansk, statutory Grant, no. ST02-0099/07/402. We wish to thank mgr inż. Małgorzata Jarosiewicz for her technical assistance.
Author information
Authors and Affiliations
Contributions
E.K. and K.G. conceived the study and designed the experiments. E.K. collected the strains. E.K. and K.G. performed the experiments. E.K., K.G., M.K.S., J.S., J.B., and J.M. analyzed the data. M.K.S. performed spa analyses and prepared figure. E.K. and K.G. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kwapisz, E., Garbacz, K., Kosecka-Strojek, M. et al. Presence of egc-positive major clones ST 45, 30 and 22 among methicillin-resistant and methicillin-susceptible oral Staphylococcus aureus strains. Sci Rep 10, 18889 (2020). https://doi.org/10.1038/s41598-020-76009-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76009-1
This article is cited by
-
Whole genome sequencing of methicillin-resistant Staphylococcus aureus clinical isolates from Terengganu, Malaysia, indicates the predominance of the EMRSA-15 (ST22-SCCmec IV) clone
Scientific Reports (2024)
-
Antimicrobial resistance and virulence of subgingival staphylococci isolated from periodontal health and diseases
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.