Abstract
The aim of the present study was to analyze copy number variations (CNV) of multiple oncogenes and tumor suppressor genes in genomic DNA from primary tumor tissue, lymph node metastasis and cell-free DNA (cfDNA) from serum of 72 urothelial carcinoma of bladder (UCB) patients treated with radical cystectomy (RC), using multiplex ligation-dependent probe amplification (MLPA). We hypothesized that primary tumor and lymph node metastasis show similar CNV profiles, and CNV are more present in lymph node metastasis compared to primary tumor tissue. Samples from 43 (59.7%) patients could be analyzed. In total, 35 (83%), 26 (68%) and 8 (42%) patients had CNV in primary tumor, serum and lymph node metastasis, respectively. MYC, CCND1, ERBB2 and CCNE1 displayed the most frequent amplifications. In particular, CNV in ERBB2 was associated with aggressive tumor characteristics. CNV in both ERBB2 and TOP2A were risk factors for disease recurrence. The current findings show that CNV are present in various oncogenes and tumor suppressor genes in genomic DNA from primary tumor, lymph node metastasis and cfDNA from serum. CNV were more present in genomic DNA from primary tumor tissue compared to cfDNA from serum and genomic DNA from lymph node metastasis. Patients with CNV in ERBB2 and TOP2A are at increased risk for disease recurrence following RC. Further studies are necessary to validate, whether these genes may represent promising candidates for targeted-therapy.
Similar content being viewed by others

Introduction
Urothelial carcinoma of the bladder (UCB) is the second leading genitourinary cancer and a potentially lethal malignancy, with an incidence of over 80,000 new cases and over 17,000 estimated deaths in 2019 in the United States1. Radical cystectomy (RC) with bilateral pelvic lymphadenectomy represents the golden standard surgical treatment for muscle-invasive and recurrent high-risk non-muscle invasive UCB2. Outcomes have remained stable over the past decades3, and a relevant number of patients experience disease recurrence and progression within 2 years after RC4. Various clinic-pathologic UCB features and biomarkers have been investigated to allow identifying those patients, who are at the highest risk of suffering from poor outcome5. Genetic analyses, including copy number variations (CNV), have the potential to elucidate the cellular mechanisms involved in the pathogenesis of UCB, and by offering targets to emerging targeted-therapy may contribute to improve outcome.
With the emergence of high-throughput genomic profiling methods like next generation sequencing, the genomic landscape of UCB has come into focus of research. Comprehensive molecular characterization by The Cancer Genome Atlas (TCGA) revealed specific RNA and DNA alterations, such as CNV in UCB6. DNA deletions, insertions and duplications lead to CNV ranging in size from several dozens of bases to megabases. They either exhibit no phenotypic effect and implicate adaptive traits, or cause severe diseases. It has been estimated that up to 9.5% of the genome accounts for CNV and surprisingly, approximately 100 genes can be completely deleted without generating apparent phenotypic consequences indicating that these genes may be functionally redundant7. CNV are a hallmark of different cancer types, including UCB, and affect the activity of tumor-associated signaling pathways and anticancer drug sensitivity as well as toxicity8. CNV can be investigated in different sources, such as tissue and liquid biopsies, by different techniques, including array comparative genomic hybridization (array-CGH)9, droplet digital PCR10,11, whole genome sequencing12 and multiplex ligation-dependent probe amplification (MLPA)13.
To date, most studies have analyzed CNV in genomic DNA derived from tissues and only few studies in cell-free DNA (cfDNA) derived from plasma or serum14,15, because of its low quantity and quality. CfDNA is released into the blood circulation by cell death (apoptosis and necrosis) and active secretion (integrated in exosomes)16,17,18 and is highly fragmented19. CNV may additionally contribute to its high fragmentation. Moreover, the majority of cfDNA originates from leukocytes that mask the small fraction of tumor-derived cfDNA in peripheral blood16,17.
Previously, we established an efficient method to detect CNV in serum cell-free DNA (cfDNA) of cancer patients using MLPA20,21,22. MLPA is a semi-quantitative technique for determining the relative CNV, including copy number gain and loss of tumor suppressor genes and oncogenes in a multiplex PCR22. In addition, MLPA with its custom-developed data analysis software represents an easy, rapid and inexpensive method without the need of complex statistics. The present study aimed to evaluate CNV of various oncogenes and tumor suppressor genes in primary tumor tissue, cfDNA and lymph node metastasis; and to compare CNV profiles in serum with those in primary tumor and lymph node metastasis. Tumor progression is accompanied by additional genetic alterations23, and therefore CNV may be more present in genomic DNA of lymph node metastasis compared to primary tumor tissue. In this respect, we hypothesized that although primary tumor and lymph node metastasis show similar CNV profiles, we might detect more presence of CNV in lymph node metastasis. Since DNA is released into the blood circulation among others from primary tumor and metastasis by cell death, serum may reflect the CNV profiles from primary tumor and metastasis. This prompted us to hypothesize that CNV profiles of cfDNA from primary tumor and lymph node metastasis can be detected in serum.
Results
Copy number variations in primary tumor, serum and lymph node metastasis
In the present study, we analyzed 46 chromosomal regions containing 2–3 exons of 16 tumor-associated genes (Table S1) in tumor tissue, serum and lymph node metastasis of 72 UCB patients for CNV using the MLPA assay. In total, 29 (40.3%) patients were excluded due to insufficient amounts of the primary tumor, which impeded our analysis, resulting in 43 (59.7%) patients available for analyses. Table 1 summarizes the clinico-pathologic features of the study cohort. Patients' median age was 71 years, and 32 (74.4%) patients were male. As expected, leukocyte DNA did not show any CNV, and was used for data normalization. Figure 1 shows an exemplary box plot for CNV evaluation in primary tumor, lymph node metastasis and leukocytes (control) of a single patient who harbored CNV in both tissues.
Example of CNV in primary tumor and lymph node metastasis of one patient. The box plot shows data of leukocytes (reference), primary tumor and lymph node metastasis (as calculated by Coffalyser.Net software). The DNA probes are arranged by chromosomal locations. The target-specific probes have a blue and orange background in different hues (left), whereas the reference probes have a grey background (right). Only the dark blue points indicate significant CNV gains, whereas light blue and yellow points are ambiguous and not considered. As expected the leukocyte DNA does not show any CNV. The data were calculated by intra- and inter-sample comparisons. Intra-sample normalization was performed by dividing the fluorescence signal of each target-specific probe by the signal of every single reference probe in this probe. The median of all these ratios of this probe is the normalization constant. Subsequently, inter-sample comparison was performed by dividing the normalization constant of each probe of this sample by the average normalization constant of all reference (leukocyte) samples.
As shown in Table 2, 42 primary tumor tissues were available from 43 patients, while only metastasis tissue and serum were available from the one remaining patient. In total, 35 (83.3%) patients harbored CNV in the primary tumor. In the majority of UCB patients, CNV was detected in MYC and CCND1 in 27 (64.3%) patients and 17 (40.5%) patients, respectively. In the primary tumor, 15 (35.7%), 11 (26.2%), 4 (9.5%) and 4 (9.5%) patients harbored amplifications in all 3 exons of MYC, CCND1, ERBB2 and CCNE1, respectively. In each case, 2 patients (4.8%) harbored CNV in all 3 exons of EGFR and FGFR1, and only one patient (2.4%) had CNV in both exons of TOP2A, suggesting that the whole genes were possibly amplified in the primary tumor. Particularly, 4 (9.3%), 2 (4.7%) and one (2.3%) patient had even the copy number gain in all 3 exons of MYC together with the 3 exons of CCND1, ERBB2 or CCNE1, respectively. In each case, only one (2.3%) patient had CNV in all 3 exons of CCND1 and ERBB2 in the corresponding primary tumor and serum, while 2 (4.7%) patients had CNV in all 3 exons of CCND1 in the corresponding primary tumor and lymph node metastasis. The CNV in all 3 exons of MYC could not be found in the corresponding specimens. A heterogeneous CNV profile in serum cfDNA was revealed in 26 of 38 (68.4%) patients, with a low CNV (usually 0.5 to 2 fold), indicating that tumor cfDNA in serum was masked by normal wild type cfDNA. CNV in all 3 exons of MYC in the serum of one (2.6%) patient was not detected in primary tumor and lymph node metastasis. However, CNV in all 3 exons of CCND1 and ERBB2 in serum of one (2.6%) patient, respectively, were present in the primary tumor. Only 8 (42.1%) of 19 UCB patients harbored CNV in their lymph node metastasis. Lymph node metastasis of 3 (15.8%), 2 (10.5%) and 2 (10.5%) patients exhibited amplifications in all 3 exons of ERBB2, CCND1 and MYC, respectively. One (5.3%) patient had amplifications in all 3 exons of the 3 genes plus the both exons of TOP2A, which is localized together with ERBB2 on chromosome 17 (Table 2).
Associations of copy number variations with clinico-pathologic UCB characteristics
Table 3 presents CNV in various genes and their association with clinico-pathologic UCB characteristics: In the primary tumor, CNV in FGFR1 and ERBB2 were associated with variant histology (p ≤ 0.027); CNV in KLF5, PTP4A, MYC and ERBB2 with pathologic tumor stage (p ≤ 0.038); CNV in ERBB2 exon 13 and exon 30 with advanced pathologic tumor stage (p ≤ 0.048); CNV in ERBB2 with LVI (p ≤ 0.048); CNV in GATA4, MYC, KLF5, ERBB2 with MVI (p ≤ 0.045); CNV in GATA4 and TOP2A with a positive soft tissue surgical margin (STSM, p ≤ 0.039); CNV in ERBB2 and CCNE1 with the presence of incidental prostate cancer (p ≤ 0.029).
In serum, CNV in CCND1 was associated with MVI (p = 0.004); CNV in PTP4A, KRAS, ERBB2, TOP2A with positive STSM (p ≤ 0.035); CNV in CCND1 with the presence of incidental prostate cancer (p = 0.018).
Outcomes according to copy number variations
The median follow-up was 15.5 months (IQR: 4.5; 22.8). Actuarial two-year recurrence-free, cancer-specific and overall survival estimates were 68% ± 8% (standard error), 89% ± 6% and 82% ± 8% respectively.
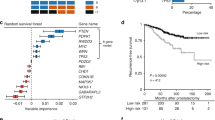
In Kaplan–Meier analyses there was no difference in cancer-specific and overall survival according to the CNV status in primary tumor, serum and lymph node metastasis. However, patients with CNV in exon 13, exon 30 and all 3 exons of ERBB2 in the primary tumor had significantly reduced recurrence-free survival, compared to patients without CNV in ERBB2 (pairwise p-values ≤ 0.008; Fig. 2A–D). In univariate Cox regression analysis, CNV in exon 13 (HR: 4.983, p = 0.018), exon 30 (HR: 5.374, p = 0.015) and all 3 exons (HR: 5.374; p = 0.015) of ERBB2 in the primary tumor were risk factors for disease recurrence.
Kaplan–Meier plots of recurrence-free survival stratified by CNV in ERBB2 (A–D) and TOP2A (E) in 43 UCB patients treated with RC. Top curves (in blue) show UCB patients with no CNV (no genomic aberrations), and bottom curves (in red) show patients with CNV comprising DNA gains in the 13th exon (ERBB2_13. A), 23th exon (ERBB2_23, B), 30th exon (ERBB2_30, C) and all 3 exons patients of ERBB2 (D), as well as in both exons of TOP2A (E).
In addition, one patient with CNV in exon 20 and 14 of TOP2A in the primary tumor had reduced recurrence-free survival, compared to the remaining patients without CNV in TOP2A (pairwise p = 0.002, Fig. 2E). In univariate Cox regression analysis, CNV in both exons of TOP2A in the primary tumor was a risk factor for disease recurrence (HR: 17.134, p = 0.021).
Discussion
For the first time, the present study analyzed CNV in genomic DNA derived from primary tumor and lymph node metastasis, as well as in cfDNA from serum of UCB patients treated with RC, using MLPA. We found that MLPA is an efficient method for the detection of CNV in genomic DNA from primary tumor and metastasis, as well as cfDNA from serum. MLPA is a high throughout analysis, allowing simultaneous evaluation of up to 96 samples, with results being available within 24 h. Therefore, MLPA represents a promising tool for further investigations of the genomic landscape and metastatic cascade of UCB patients. We found most CNV in DNA of primary tumor, and to a lesser extent in lymph node metastasis and in cfDNA from serum. Thus, we have to reject our hypothesis. The lower extent of CNV in cfDNA may be due to dilution of tumor-derived cfDNA by wild type cfDNA in the blood of cancer patients that camouflages the detection of CNV in tumor-derived cfDNA. Thus, the heterogeneous CNV profile may be due to the low prevalence of tumor-derived cfDNA in the serum of UCB patients18,24. The lower extent of CNV in genomic DNA of lymph node metastasis may be due to limited amount of metastatic tumor tissue in lymph nodes, possibly influencing the efficiency of MLPA.
In the primary tumor, most CNV were found in MYC, CCND1, ERBB2 and CCNE1. Importantly, CNV in these genes were associated with aggressive clinico-pathologic UCB features. Only one patient had CNV in all 3 exons of CCND1 and ERBB2 in both, serum and primary tumor. In contrast, CNV in all 3 exons of MYC were exclusively detected in the serum of one patient, but not in the primary tumor. Although it remains speculative, it is possible that in this patient CNV in all 3 exons of MYC may not originate from the primary tumor, but rather from circulating tumor cells, which have a deleterious impact on outcome in UCB25. In UCB, alterations of chromosome 8, especially DNA deletions of the 8p arm and gains of the 8q arm, belong to the most frequent cytogenetic changes26,27. MYC is located on 8q24.21, and displayed copy number gains in all 3 exons in our study. Its amplification and overexpression have been described as main events, by which MYC is deregulated in various cancer entities28,29. Deregulation of MYC activity contributes to cancer progression, metastasis and resistance to therapy. MYC is a transcription factor and activates the expression of multiple genes that encode for proteins involved in cell proliferation, differentiation, adhesion and survival28,30. Using high-throughput tissue microarray, Zaharieva et al.31 showed that MYC gains are associated with genetically unstable UCB which is characterized by high histologic grade and/or invasive growth. Conconi et al.9 applied array-CGH and detected that bladder cancer stem cell (CSC) subpopulations retained CNV in MYC. These researchers postulated MYC to be a therapeutic target for bladder CSC subpopulations. We found that 15 UCB patients harbored CNV in all 3 exons of MYC gene in the tumor tissue. In addition, some patients harbored CNV in all 3 exons of MYC together with the 3 exons of CCND1, ERBB2 or CCNE1, respectively. Our analyses in primary tumor showed that CNV in all 3 exons and CNV in the second exon of MYC were associated with unfavorable UCB features, e.g. pathologic tumor stage and MVI. In addition, CNV in exons of other genes, including CCND1, FGFR1 ERBB2, KLF5, PTP4A, GATA4, TOP2A and CCNE1 were associated with aggressive clinico-pathologic UCB characteristics, e.g. presence of variant histology, LVI and positive STSM. Previously, it has been suggested that KLF5 facilitates angiogenesis and that KLF5 might represent a therapeutic target in UCB32. Correspondingly, GATA4 may potentially represent a therapeutic target, since whole exome sequencing showed that UCB patients with high expression of GATA4 have worse survival compared to patients with low GATA4 expression33. CCNE1 is located on 19q12 and has the same functions as CCND1. It induces S phase entry of the cell cycle and specifically interacts with CDK234. The molecular signature of CCNE1 defined by CNV and expression changes has been reported to be an independent risk factor for disease progression in UCB patients. Moreover, gene network and upstream regulator analyses revealed that disease progression is potentially mediated by the CCND1-CCNE1-SP1 pathway9,35,36. Our analyses showed that in primary tumor CNV in CCNE1 is associated with presence of incidental prostate cancer.
Watters et al.29 showed that the majority of bladder carcinomas are polysomic for chromosome 8 and 11. These characteristics were reflected by the high CNV detected in MYC and CCND1. Amplifications of both genes have been reported to be associated with muscle-invasive UCB29. CCND1 regulates the cell cycle transition from G1 to S phase by the cyclin-dependent kinases CDK4 and CDK6. Its overexpression results in a dysregulated CDK activity followed by a rapid cell growth37. CCND1 is located on 11q13.2, and amplification of this chromosomal band is a common event in cancer38. In our investigations on DNA of the primary tumor, we found prominent copy number gains in all 3 exons of CCND1, in line with the findings by Chekaluk et al., who also used MLPA39, and Weltman et al., who used array-CGH36.
We found CNV in ERBB2 in DNA of primary tumor and lymph node metastasis. ERBB2 is a member of the ErbB receptor family, which is often overexpressed, amplified or mutated in various cancer types. ERBB2 regulates epithelial-mesenchymal transition (EMT), migration and tumor invasion by modulating extracellular matrix (ECM) components40. In metastases, a considerable heterogeneity in centromere 17 and CNV in ERBB2 located on 17q12 have been described, supporting genomic instability of these cells41. Our analyses on primary tumor showed that CNV in different exons of ERBB2 were associated with aggressive UCB features, e.g. presence of variant histology, pathologic tumor stage, LVI and MVI, and incidental prostate cancer. In addition, CNV in the exons of TOP2A, a topoisomerase which is also located on chromosome 1742, was associated with positive STSM. As TOP2A gene is located adjacent to the ERBB2 gene, it is frequently either co-amplified with or independent of ERBB2 in many cancer types, including UCB43. In addition, simultaneous amplification of both genes caused by different mechanisms was described in breast cancer, and contributed to a higher level of amplification of ERBB244. In our cohort, patients with CNV in these genes in the primary tumor were at increased risk for disease recurrence. However, the patient number in the subgroup of patients with CNV in ERBB2 and TOP2A is very low. Thus, it has to be emphasized, that these results have to be interpreted very carefully. Although it remains speculative, patients with CNV in ERBB2 and CNV in TOP2A in the primary tumor and/or lymph node metastasis may benefit from HER2- or TOP2A-targeted therapy. Previously, a negative impact of ERBB2 amplifications on outcomes have been reported also in non-muscle invasive UCB45,46. Similar to our analyses, Kim et al.47 revealed that the levels of TOP2A expression are a risk factor for disease recurrence.
The present study is not devoid of limitations. A relevant number of patients was excluded from analyses, which might have influenced findings. The resulting total patient number was low and follow-up was limited. Thus, a multivariable analysis was not possible to identify independent risk factors for disease recurrence, cancer-specific and overall mortality. Nevertheless, our study remains the first for the analysis of CNV in genomic DNA from primary tumor, lymph node metastasis and tumor-derived cfDNA from serum using MLPA in UCB patients treated with RC. The MLPA assay, which was used in the present study, was not specifically manufactured or validated for analyses of UCB patients. However, we have previously shown that MLPA is an efficient method for the detection of CNV in UCB patients treated with RC20. The MLPA assay did contain sequences to analyze a limited number of genes, but of several exons; some genes that may have an important role in bladder cancer progression were not included in this assay, e.g. the androgen receptor, which may contribute to disease progression by EGFR signaling48. Lastly, since the amount of cfDNA was low, we could not perform single analyses on every chromosomal region by real-time Taqman PCR. Further studies are warranted to complete such evaluations in the future.
In conclusion, MLPA using genomic DNA derived from primary tumor tissue and lymph node metastasis, as well as cfDNA may offer a less time-consuming tool for the quick analysis of CNV in oncogenes and tumor suppressor genes in UCB. CNV are present in various genes of genomic DNA derived from primary tumor, lymph node metastasis and cfDNA from serum in UCB patients treated with RC. Most CNV were detected in all 3 exons of MYC, CCND1, ERBB1 and CCNE1. CNV in specific genes of genomic DNA derived from primary tumor, lymph node metastasis, and cfDNA are associated with unfavorable clinico-pathologic UCB features. In addition, patients with CNV in ERBB2 and TOP2A in the primary tumor are at increased risk for disease recurrence. Further studies in larger patient cohorts are necessary to validate our findings, and to evaluate whether these genes are suitable biomarkers for disease progression and candidates for targeted-therapy.
Material and methods
Patient cohort
The cohort of 72 UCB patients has been described in detail previously20: Briefly, patients were treated with RC and bilateral pelvic lymphadenectomy without neoadjuvant chemotherapy. Recurrent Ta, T1, or carcinoma in situ (CIS), refractory to transurethral resection of the bladder (TURBT) with or without intravesical immunotherapy or chemotherapy, or muscle invasive UCB were indications for RC. Preoperative staging consisted of computed tomography (CT) of the thorax and abdomen/pelvis, and bone scan and cranium imaging when clinically indicated. Exclusion criteria included metastatic disease at preoperative staging, a history of any other malignancy, previous systemic chemotherapy or radiation, and incomplete clinico-pathologic or follow-up data. Patients received adjuvant chemotherapy based on tumor stage, overall health status, renal function and patients' desire. Platinum-based adjuvant chemotherapy generally started within 90 days after RC. All experiments have been carried out in accordance with relevant regulations: all patients have signed written informed consent, and the study was approved by the local ethics committee "Universitätsklinikum Hamburg-Eppendorf; Ethik-Kommission der Ärztekammer Hamburg" (No. PV3962).
Pathological evaluation
As described in detail previously20, the complete surgical RC specimen was inked, and multiple sections were obtained from the bladder and the tumor in addition to the regional lymph nodes and ureters. Tumor stage and nodal status were assessed according to the tumor, lymph node and metastasis (TNM) system. Tumor grade was assessed according to the 1998 World Health Organization (WHO) grading system49. Concomitant CIS was defined as the presence of CIS in conjunction with another tumor other than CIS alone. Lymphovascular invasion (LVI) was defined as the unequivocal presence of tumor cells within an endothelium-lined space without underlying muscular walls50. Micro-vascular invasion (MVI) was defined as the presence of tumor cells within a vessel with a vascular wall and red blood cells in the lumen51. A positive soft tissue surgical margin (STSM) was defined as the presence of tumor at inked areas of soft tissue on the RC specimen52. Presence of variant UCB histology was defined as the presence of UCB combined with any variant histology. Variant UCB histologies were classified corresponding to the WHO Classification of Tumors53. Incidental prostate cancer was defined as presence of prostate cancer in the RC specimens54.
DNA extraction from tumor tissues
Unstained paraffin‐embedded primary tumor tissue and lymph node metastasis blocks were incubated at 50 °C for 30 min and de-paraffinized twice with xylol and 6 times with ethanol. After washing with water, the tumor areas were scrapped off from the slides with lysis buffer, as indicated by the slices stained with haematoxylin and eosin (Merck, Darmstadt, Germany). Genomic DNA was isolated using the Innu Prep DNA Micro Kit (Analytik Jena, Germany) and corresponding to the manufacturer´s recommendations. Briefly, the tumor tissues were incubated with 200 µl lysis buffer TLS and 20 µl proteinase K at 50 °C till the sample was completely lysed. After incubating the lysed samples at 90 °C for 60 min, they were supplemented with 200 µl Binding Solution TBS and added to spin filters. After washing the filters, DNA was eluted in 30 µl Elution Buffer (Analytik Jena).
DNA extraction from serum and leukocytes
Preoperative blood samples were usually collected on the day prior to RC at a median of 39 days [interquartile range (IQR): 27; 61] after the preceding TURB. Serum was prepared from 6 ml whole blood by 2 centrifugation steps of 3000 g and 16,000 g each for 10 min. Leukocytes (reference) were extracted from 6 ml EDTA blood supplemented up to 50 ml with lysis buffer containing 0.3 M sucrose, 10 mM Tris–HCl pH 7.5, 5 mM MgCl2 and 1% Triton X100 (Sigma, Taufkirchen, Germany). Following incubation for 15 min on ice, the isolation and purification of the leukocytes were carried out by 2 centrifugation steps at 2500 g, at 4 °C for 20 min. cfDNA was extracted from 2 ml serum using the PME free-circulating DNA Extraction kit (Analytik Jena), while DNA was extracted from leukocytes using the Qiamp DNA Blood Mini kit (Qiagen, Hilden, Germany). These DNA extractions were carried out according to the manufacturer´s instructions and similar to the procedure as described above. Quantification and quality of the extracted cfDNA were determined spectrophotometrically using the NanoDrop Spectrometer ND-1000 (Thermo Fisher Scientific, Wilmington, DE, USA).
MLPA assay
MLPA experiments were already described in our both previous studies20,22 and are briefly described in the following paragraphs, again: CNV were determined using 5 µl (50 ng) cfDNA and 5 µl of each (100 ng) leukocytes (reference), tumor tissues and metastasis DNA from 46 bladder cancer patients and the SALSA MLPA probemix SALSA MLPA Probemix P458-B1 kit (MRC Holland, Amsterdam, The Netherlands). This kit contains a probe mix of 46 sequences of 16 genes to be analyzed (Table S1), 15 reference genes (Table S2) and 9 quality control fragments (Table S3). According to the manufacturer’s instructions, 5 µl DNA samples were denatured at 98 °C for 5 min, and hybridized with 1.5 probemix and 1.5 µl MLPA buffer at 60 °C for about 18 h. Next day, the hybridization reaction was ligated with Ligase-65 master mix containing 25 µl water, 3 µl ligase buffer A, 3 µl ligase buffer B and 1 µl Ligase-65 enzyme at 54 °C for 15 min and at 98 °C for 5 min. Then, the MLPA reaction was mixed with polymerase master mix containing 7.5 µl water, 2 µl SALSA PCR primer mix and 0.5 µl SALSA polymerase. PCR was carried out at 95 °C for 30 s, at 60° for 30 s and at 72 °C for 60 s in 35 cycles, with a last step of 72 °C for 20 min on MJ Research PTC-200 Peltier Thermal Cycler (Global Medical Instrumentation, Ramsey, Minnesota, USA). During the PCR, all MLPA samples were amplified simultaneously using the same PCR primer pair, of which one PCR primer was fluorescently labelled.
Capillary electrophoresis
For fragment analysis, 1.4 µl PCR was mixed with 0.6 µl 500-ROX size marker which served as an internal standard (ThermoFisher, Darmstadt, Germany) and 18 µl HiDi formamide (ThermoFisher). After heating at 86 °C for 3 min and cooling down at 4 °C, fragment separation was done by capillary electrophoresis on an automated ABI 3130 DNA analyzer (Applied Biosystems, Freiburg, Germany). Fragment length and fluorescence intensity were evaluated by the Coffalyser.Net software (MRC). The Coffalyser.Net software could be downloaded for free after buying the MLPA kit which contained the corresponding chromosomal regions referring to the software.
Data normalization
As previously described20, data normalization was carried out by Coffalyser.Net analysis software (www.mlpa.com). It consists of 2 steps: intra- and intersample normalization. For intrasample normalization, within each sample, each probe peak was compared with the peaks of the reference probes. Reference probes located on various chromosomes detect sequences that are expected to have a normal copy number in all samples. The determined relative probe signals were then used for intersample normalization (Table S1). Final probe ratios were determined by comparing the relative probe peak in the cfDNA sample of interest with those of all leukocyte DNA samples. Leukocyte DNA samples are expected to have a normal copy number for both the reference and target probe. To avoid false positive data due to the quality and quantity of the serum cfDNA, only unambiguous values were used (Fig. 1), and PCR was repeated.
MLPA was carried out using the panel MRC kit following the provided protocol. Data analysis was carried out using the Coffalyser program. Raw data of all leukocytes with adequate quality was taken as reference for calculating the 95% confidential interval reference range for each probe (Fig. 1). For each patient, ratio charts of leukocytes, tumor, metastasis and serum were compared. Probes with increase or increase in more than two standard deviations to the mean reference value were identified as duplication or deletion.
Follow-up regimen
Follow-up has been outlined in detail previously20: For the first year, patients were seen every 3 months, from the second to fifth years every 6 months, and annually thereafter. Diagnostic imaging of the abdomen including the urinary tract (e.g. ultrasonography and/or intravenous urography, CT of the abdomen/pelvis with intravenous contrast) and chest radiography were performed at least annually or when clinically indicated.
Disease recurrence was defined as local failure in the operative site, regional lymph nodes, or distant metastasis. Upper tract urothelial carcinoma was not considered as disease recurrence but metachronous tumor. Cancer-specific mortality was defined as death from UCB. Overall mortality was defined as death from any cause. The cause of death was determined by the treating physician, by chart review corroborated by death certificates, or by death certificates alone55. Perioperative mortality (i.e., death within 30 days of surgery) was censored at time of death for bladder cancer-specific survival analyses.
Statistical analyses
The co-primary endpoints of the present study were disease recurrence, cancer-specific and overall mortality according to CNV profile in 16 tumor suppressor genes and oncogenes. The indicator variable (i.e., CNV) was analyzed as categorical variables. Associations between categorical variables were assessed using the Fisher exact and χ2-test. Differences in continuous variables were analyzed using the Mann–Whitney-U test (two categories) and the Kruskal–Wallis test (three or more categories). Recurrence-free, cancer-specific and overall survival probabilities were estimated using the Kaplan–Meier method and differences between groups were assessed using the Log rank statistic. Univariable Cox regression models assessed time to disease recurrence, cancer-specific and overall mortality. All tests are two-sided and a p-value of < 0.05 was set to be statistically significant. All analyses were performed with SPSS 20 (SPSS Inc., IBM Corp., Armonk, NY).
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34. https://doi.org/10.3322/caac.21551 (2019).
Witjes, J. A. et al. EAU guidelines on muscle-invasive and metastatic bladder cancer: Summary of the 2013 guidelines. Eur. Urol. 65, 778–792. https://doi.org/10.1016/j.eururo.2013.11.046 (2014).
Stein, J. P. et al. Radical cystectomy in the treatment of invasive bladder cancer: Long-term results in 1054 patients. J. Clin. Oncol. 19, 666–675 (2001).
Rink, M. et al. Predictors of cancer-specific mortality after disease recurrence following radical cystectomy. BJU Int. 111, E30-36. https://doi.org/10.1111/j.1464-410X.2012.11433.x (2013).
Riethdorf, S., Soave, A. & Rink, M. The current status and clinical value of circulating tumor cells and circulating cell-free tumor DNA in bladder cancer. Transl. Androl. Urol. 6, 1090–1110. https://doi.org/10.21037/tau.2017.09.16 (2017).
Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540-556.e525. https://doi.org/10.1016/j.cell.2017.09.007 (2017).
Zarrei, M., MacDonald, J. R., Merico, D. & Scherer, S. W. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183. https://doi.org/10.1038/nrg3871 (2015).
Mikhail, F. M. Copy number variations and human genetic disease. Curr. Opin. Pediatr. 26, 646–652. https://doi.org/10.1097/MOP.0000000000000142 (2014).
Conconi, D. et al. Using copy number alterations to identify new therapeutic targets for bladder carcinoma. Int. J. Mol. Sci. 17, 271. https://doi.org/10.3390/ijms17030271 (2016).
Gevensleben, H. et al. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin. Cancer Res. 19, 3276–3284. https://doi.org/10.1158/1078-0432.CCR-12-3768 (2013).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610. https://doi.org/10.1021/ac202028g (2011).
Hehir-Kwa, J. Y., Pfundt, R. & Veltman, J. A. Exome sequencing and whole genome sequencing for the detection of copy number variation. Exp. Rev. Mol. Diagn. 15, 1023–1032. https://doi.org/10.1586/14737159.2015.1053467 (2015).
Kondrashova, O. et al. High-throughput amplicon-based copy number detection of 11 genes in formalin-fixed paraffin-embedded ovarian tumour samples by MLPA-seq. PLoS ONE 10, e0143006. https://doi.org/10.1371/journal.pone.0143006 (2015).
Molparia, B., Nichani, E. & Torkamani, A. Assessment of circulating copy number variant detection for cancer screening. PLoS ONE 12, e0180647. https://doi.org/10.1371/journal.pone.0180647 (2017).
Xia, S. et al. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer 90, 78–84. https://doi.org/10.1016/j.lungcan.2015.07.002 (2015).
Rink, M., Schwarzenbach, H., Riethdorf, S. & Soave, A. The current role and future directions of circulating tumor cells and circulating tumor DNA in urothelial carcinoma of the bladder. World J. Urol. https://doi.org/10.1007/s00345-018-2543-9 (2018).
Rink, M., Schwarzenbach, H., Vetterlein, M. W., Riethdorf, S. & Soave, A. The current role of circulating biomarkers in non-muscle invasive bladder cancer. Transl. Androl. Urol. 8, 61–75. https://doi.org/10.21037/tau.2018.11.05 (2019).
Schwarzenbach, H., Hoon, D. S. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437. https://doi.org/10.1038/nrc3066 (2011).
Muller, I. et al. Identification of loss of heterozygosity on circulating free DNA in peripheral blood of prostate cancer patients: potential and technical improvements. Clin. Chem. 54, 688–696 (2008).
Soave, A. et al. Copy number variations of circulating, cell-free DNA in urothelial carcinoma of the bladder patients treated with radical cystectomy: A prospective study. Oncotarget 8, 56398–56407. https://doi.org/10.18632/oncotarget.17657 (2017).
Schwarzenbach, H., Chun, F. K., Isbarn, H., Huland, H. & Pantel, K. Genomic profiling of cell-free DNA in blood and bone marrow of prostate cancer patients. J. Cancer Res. Clin. Oncol. 137, 811–819. https://doi.org/10.1007/s00432-010-0941-5 (2011).
Schwarzenbach, H. Copy number variation analysis on cell-free serum DNA. Methods Mol. Biol. 85–93, 2019. https://doi.org/10.1007/978-1-4939-8973-7_6 (1909).
McConkey, D. J. et al. Molecular genetics of bladder cancer: Emerging mechanisms of tumor initiation and progression. Urol Oncol 28, 429–440. https://doi.org/10.1016/j.urolonc.2010.04.008 (2010).
Diaz, L. A. Jr. & Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 32, 579–586. https://doi.org/10.1200/JCO.2012.45.2011 (2014).
Soave, A. et al. Detection and oncological effect of circulating tumour cells in patients with variant urothelial carcinoma histology treated with radical cystectomy. BJU Int. 119, 854–861. https://doi.org/10.1111/bju.13782 (2017).
Wagner, U. et al. Chromosome 8p deletions are associated with invasive tumor growth in urinary bladder cancer. Am. J. Pathol. 151, 753–759 (1997).
Prat, E. et al. Comparative genomic hybridization analysis reveals new different subgroups in early-stage bladder tumors. Urology 75, 347–355. https://doi.org/10.1016/j.urology.2009.04.080 (2010).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35. https://doi.org/10.1016/j.cell.2012.03.003 (2012).
Watters, A. D. et al. Genetic aberrations of c-myc and CCND1 in the development of invasive bladder cancer. Br. J. Cancer 87, 654–658. https://doi.org/10.1038/sj.bjc.6600531 (2002).
Sears, R. C. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3, 1133–1137 (2004).
Zaharieva, B. et al. High-throughput tissue microarray analysis of CMYC amplificationin urinary bladder cancer. Int. J. Cancer 117, 952–956. https://doi.org/10.1002/ijc.21253 (2005).
Gao, Y. et al. Beyond proliferation: KLF5 promotes angiogenesis of bladder cancer through directly regulating VEGFA transcription. Oncotarget 6, 43791–43805. https://doi.org/10.18632/oncotarget.6101 (2015).
Wu, S. et al. Novel variants in MLL confer to bladder cancer recurrence identified by whole-exome sequencing. Oncotarget 7, 2629–2645. https://doi.org/10.18632/oncotarget.6380 (2016).
Moroy, T. & Geisen, C. Cyclin E. Int. J. Biochem. Cell Biol. 36, 1424–1439. https://doi.org/10.1016/j.biocel.2003.12.005 (2004).
Song, B. N., Kim, S. K. & Chu, I. S. Bioinformatic identification of prognostic signature defined by copy number alteration and expression of CCNE1 in non-muscle invasive bladder cancer. Exp. Mol. Med. 49, e282. https://doi.org/10.1038/emm.2016.120 (2017).
Veltman, J. A. et al. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Can. Res. 63, 2872–2880 (2003).
Qie, S. & Diehl, J. A. Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. (Berl) 94, 1313–1326. https://doi.org/10.1007/s00109-016-1475-3 (2016).
Huang, X., Gollin, S. M., Raja, S. & Godfrey, T. E. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc. Natl. Acad. Sci. U.S.A. 99, 11369–11374. https://doi.org/10.1073/pnas.172285799 (2002).
Chekaluk, Y. et al. Identification of nine genomic regions of amplification in urothelial carcinoma, correlation with stage, and potential prognostic and therapeutic value. PLoS ONE 8, e60927. https://doi.org/10.1371/journal.pone.0060927 (2013).
Appert-Collin, A., Hubert, P., Cremel, G. & Bennasroune, A. Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 6, 283. https://doi.org/10.3389/fphar.2015.00283 (2015).
Sauter, G., Moch, H., Gasser, T. C., Mihatsch, M. J. & Waldman, F. M. Heterogeneity of chromosome-17 and Erbb-2 gene copy number in primary and metastatic bladder-cancer. Cytometry 21, 40–46. https://doi.org/10.1002/cyto.990210109 (1995).
Mano, M. S., Rosa, D. D., De Azambuja, E., Ismael, G. F. & Durbecq, V. The 17q12-q21 amplicon: Her2 and topoisomerase-IIalpha and their importance to the biology of solid tumours. Cancer Treat. Rev. 33, 64–77. https://doi.org/10.1016/j.ctrv.2006.10.001 (2007).
Simon, R. et al. HER-2 and TOP2A coamplification in urinary bladder cancer. Int. J. Cancer 107, 764–772. https://doi.org/10.1002/ijc.11477 (2003).
Nielsen, K. V. et al. Aberrations of ERBB2 and TOP2A genes in breast cancer. Mol. Oncol. 4, 161–168. https://doi.org/10.1016/j.molonc.2009.11.001 (2010).
Ding, W. et al. Human epidermal growth factor receptor 2: a significant indicator for predicting progression in non-muscle-invasive bladder cancer especially in high-risk groups. World J. Urol. 33, 1951–1957. https://doi.org/10.1007/s00345-015-1557-9 (2015).
Abdelrahman, A. E. et al. Fatty acid synthase, Her2/neu, and E2F1 as prognostic markers of progression in non-muscle invasive bladder cancer. Ann. Diagn. Pathol. 39, 42–52. https://doi.org/10.1016/j.anndiagpath.2019.01.002 (2019).
Kim, E. J. et al. Clinical implications and prognostic values of topoisomerase-II alpha expression in primary non-muscle-invasive bladder cancer. Urology 75(1516), e1519–e1513. https://doi.org/10.1016/j.urology.2009.08.055 (2010).
Tripathi, A. & Gupta, S. Androgen receptor in bladder cancer: A promising therapeutic target. Asian J. Urol. 7, 284–290. https://doi.org/10.1016/j.ajur.2020.05.011 (2020).
Epstein, J. I., Amin, M. B., Reuter, V. R., Mostofi, F. K. & Committee, T. B. C. C. The World Health Organization/International Society of Urological pathology consensus classification of urothelial (transitional cell) neoplasms of the urinary bladder. Am. J. Surg. Pathol. 22, 1435–1448 (1998).
Lotan, Y. et al. Lymphovascular invasion is independently associated with overall survival, cause-specific survival, and local and distant recurrence in patients with negative lymph nodes at radical cystectomy. J. Clin. Oncol. 23, 6533–6539. https://doi.org/10.1200/jco.2005.05.516 (2005).
Hong, S. K., Kwak, C., Jeon, H. G., Lee, E. & Lee, S. E. Do vascular, lymphatic, and perineural invasion have prognostic implications for bladder cancer after radical cystectomy?. Urology 65, 697–702. https://doi.org/10.1016/j.urology.2004.10.048 (2005).
Novara, G. et al. Soft tissue surgical margin status is a powerful predictor of outcomes after radical cystectomy: A multicenter study of more than 4,400 patients. J. Urol. 183, 2165–2170. https://doi.org/10.1016/j.juro.2010.02.021 (2010).
Eble, J., Sauter, G., Epstein, J. I. & Sesterhenn, I. A. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. World Health Organization Classification of Tumours Lyon: IARCC Press (2004).
Bruins, H. M. et al. Incidental prostate cancer in patients with bladder urothelial carcinoma: Comprehensive analysis of 1,476 radical cystoprostatectomy specimens. J. Urol. 190, 1704–1709. https://doi.org/10.1016/j.juro.2013.05.034 (2013).
Rink, M. et al. Death certificates are valid for the determination of cause of death in patients with upper and lower tract urothelial carcinoma. Eur. Urol. 61, 854–855. https://doi.org/10.1016/j.eururo.2011.12.055 (2012).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
A.S. conception of the work, interpretation of data, acquisition, analysis, drafted the work or substantively revised it. L.K. statistical analysis of data, interpretation of data. H.Y. substantively revised the work. M.R. substantively revised the work. P.G. substantively revised the work. M.W.V. substantively revised the work. P.M. substantively revised the work. G.S. acquisition of data, analysis of data. M.F. substantively revised the work. C.P.M. substantively revised the work. T.L. substantively revised the work. R.D. substantively revised the work. S.M. acquisition of data, analysis of data. K.P. substantively revised the work. B.S. did the laboratory analysis. H.S. design of the work, interpretation of data, acquisition, analysis, drafted the work or substantively revised it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Soave, A., Kluwe, L., Yu, H. et al. Copy number variations in primary tumor, serum and lymph node metastasis of bladder cancer patients treated with radical cystectomy. Sci Rep 10, 21562 (2020). https://doi.org/10.1038/s41598-020-75869-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75869-x
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.